The protozoan parasite Trypanosoma brucei is responsible for sleeping sickness and alternates between mammal and tsetse fly hosts. Two proteins of the ALBA family associate to mRNA in cytoplasmic granules during starvation stress, are stage regulated, and contribute to trypanosome development in the tsetse fly.
Abstract
The protozoan parasite Trypanosoma brucei is responsible for sleeping sickness and alternates between mammal and tsetse fly hosts, where it has to adapt to different environments. We investigated the role of two members of the ALBA family, which encodes hypothetical RNA-binding proteins conserved in most eukaryotes. We show that ALBA3/4 proteins colocalize with the DHH1 RNA-binding protein and with a subset of poly(A+) RNA in stress granules upon starvation. Depletion of ALBA3/4 proteins by RNA interference in the cultured procyclic stage produces cell modifications mimicking several morphogenetic aspects of trypanosome differentiation that usually take place in the fly midgut. A combination of immunofluorescence data and videomicroscopy analysis of live trypanosomes expressing endogenously ALBA fused with fluorescent proteins revealed that ALBA3/4 are present throughout the development of the parasite in the tsetse fly, with the striking exception of the transition stages found in the proventriculus region. This involves migration of the nucleus toward the posterior end of the cell, a phenomenon that is perturbed upon forced expression of ALBA3 during the differentiation process, showing for the first time the involvement of an RNA-binding protein in trypanosome development in vivo.
INTRODUCTION
The protozoan parasite Trypanosoma brucei is responsible for the fatal disease sleeping sickness in Central Africa (Brun et al., 2009). The transmission of T. brucei to a mammalian host takes place by the bite of an infected tsetse fly of the Glossina genus. African trypanosomes live exclusively as extracellular parasites and are found in the lymphatic system, the bloodstream, and the cerebrospinal fluid of the mammalian hosts or in the alimentary tract and the salivary glands of the fly. The parasite is characterized by a complex developmental cycle comprising at least 10 distinct morphological forms. Culture conditions are available for two proliferating stages: the long, slender bloodstream form and the procyclic stage from the tsetse midgut. The transition between these two forms is ensured by the production of nonproliferating short, stumpy parasites in the blood, which are preadapted to differentiate into procyclics once transferred to the tsetse fly. This transition can be reproduced in vitro, and molecular mechanisms have begun to be unveiled (Reuner et al., 1997; Vassella et al., 1997; Engstler and Boshart, 2004; Szoor et al., 2006; Dean et al., 2009).
A third proliferating stage is encountered in the salivary gland of the fly, but this cannot be reproduced in the laboratory (Vickerman et al., 1988). The transition to the salivary gland stage represents a bottleneck in the parasite cycle (Oberle et al., 2010) and involves multiple morphological differentiation steps that have been investigated at the morphological level (Vickerman et al., 1988; Van Den Abbeele et al., 1999; Sharma et al., 2008; Rotureau et al., 2011). Only low numbers of differentiating cells can be recovered from the fly, and their colocalization with various bacterial populations (Aksoy et al., 2003) restricted molecular and biochemical analysis. So far, only one specific molecular marker (brucei alanine-rich protein [BARP], a cell surface protein) has been identified in the proliferating parasites in the salivary glands (Urwyler et al., 2007), and molecular events controlling the transition from midgut to salivary gland parasites are completely unknown.
Trypanosomes are characterized by the presence of the kinetoplast, the concatenated DNA of the single mitochondrion (de Souza et al., 2009) that is linked to the basal body of the flagellum (Robinson and Gull, 1991). It can be found posterior to the nucleus, and the cell is called a trypomastigote, or it can be present in an anterior position, defining the epimastigote configuration. During the journey in the tsetse fly, trypanosomes transform from trypomastigote to epimastigote in the proventriculus and back again before producing the infective trypomastigote form in the saliva. They need to adapt rapidly to changing environments (temperature, nutritional resources, physical properties of the surrounding medium) and to deal with the immune systems of each host. Expression of appropriate surface proteins and exhaustive alteration of metabolic pathways appear to be tightly controlled (Besteiro et al., 2005; Urwyler et al., 2005).
In T. brucei, most genes are transcribed by RNA polymerase II, which generates polycistronic transcripts in a run-through manner (Siegel et al., 2009; Wright et al., 2010). However, genes present in the same cistron are not functionally related (Berriman et al., 2005). Mature mRNA is obtained by trans-splicing and polyadenylation (Kolev et al., 2010; Nilsson et al., 2010; Siegel et al., 2010), and its fate is largely controlled at the posttranscriptional level (Daniels et al., 2010; Fernández-Moya and Estévez, 2010; Martinez-Calvillo et al., 2010), a process often dependent on the 3′ untranslated region (3′ UTR; Furger et al., 1997; Hotz et al., 1997). This is especially crucial during development, as recently demonstrated for the 3′ UTR of BARP, which is sufficient to restrict expression of a green fluorescent protein (GFP) reporter exclusively to the salivary gland forms (Urwyler et al., 2007).
Consistent with posttranscriptional control, the T. brucei genome encodes for a large number of candidate RNA-binding proteins (De Gaudenzi et al., 2005). A few of them have been shown to participate in some differentiation processes (Hendriks et al., 2001; Paterou et al., 2006), but studies have been restricted to the culture bloodstream and procyclic forms. Hence, candidate proteins acting during development in the tsetse fly still need to be unraveled. We selected to investigate the contribution of ALBA (“acetylation lowers binding affinity”)–like proteins, a large family known for its eponymous ALBA domain (Pfam PF01918) with nucleic acid–binding ability (Aravind et al., 2003), since previous studies indicated a general role in the development program of several microorganisms. First, the ALBA-like protein Mdp2 is exclusively expressed during micronucleus to macronucleus development in the ciliate Stylonychia lemnae (Fetzer et al., 2002), a differentiation step characterized by massive DNA rearrangements also involving RNA molecules (Mochizuki et al., 2002). Second, in the apicomplexan parasite Plasmodium berghei, ALBA proteins are found concentrated in RNA-containing cytoplasmic foci, but exclusively in the female gametocyte (Mair et al., 2010). In Archaea, ALBA proteins are described as DNA binding in a histone-like manner, although they have been found to bind RNA in vivo (Forterre et al., 1999; She et al., 2001; Bell et al., 2002; Guo et al., 2003).
In this study, we investigate ALBA3 and ALBA4 and show that they are found in the cytoplasm of trypanosomes and aggregate in foci upon starvation stress, where they colocalize with the RNA-interacting protein DHH1 and with poly(A+) RNA. Inhibition of ALBA3/4 expression by RNA interference (RNAi) leads to multiple phenotypes that evoke the transition from the midgut to the proventricular stages. ALBA3/4 expression levels are dramatically reduced precisely at that stage of development in the tsetse fly, and forced expression of ALBA3 leads to perturbation in this crucial differentiation step.
RESULTS
The ALBA protein family in T. brucei
The genomes of T. brucei, Trypanosoma cruzi, and Trypanosoma vivax encode four proteins containing an ALBA domain, whereas only two are found in Trypanosoma congolense and in all Leishmania species. In T. brucei, two ALBA genes are found on chromosome 4: ALBA3 (Tb927.4.2040) and ALBA4 (Tb927.4.2030) and two on chromosome 11: ALBA1 (Tb11.02.2040) and ALBA2 (Tb11.02.2030; Supplemental Figure S1). ALBA1 and ALBA2 are small proteins of 12 and 14 kDa that contain only the ALBA domain (Pfam PF01918) and that show 53% identity on the protein level between each other. ALBA3 and ALBA4 are very divergent from ALBA1 and ALBA2, with which they share only 16% overall identity, restricted to the ALBA domain. ALBA3 and ALBA4 show high conservation between them, with 85% identity at the DNA level (Supplemental Figure S2A). The encoded proteins have a molecular weight of 21 and 25 kDa, respectively and contain, in addition to the ALBA domain, a C-terminal stretch of multiple RGG repeats that are believed to be important in nucleotide binding. This study focuses on the investigation of ALBA3 and ALBA4 since they are more likely to be true orthologues of ALBA proteins found in metazoa (Supplemental Figure S1B).
ALBA3/4 are cytosolic proteins that aggregate in mRNA-containing granules upon starvation
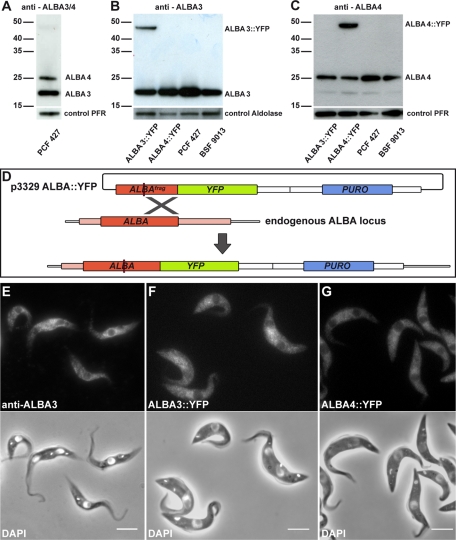
To investigate ALBA3 and ALBA4, both full-length proteins were expressed as glutathione S-transferase (GST) fusions and used to produce antisera in mice. Western blot analysis was carried out on total extracts of cultured procyclic trypanosomes of strain Lister 427 and confirmed that both proteins were expressed. They migrated at positions corresponding to their predicted size of 21 and 25 kDa for ALBA3 and ALBA4, respectively (Figure 1A). Of the 16 antisera produced, most of them recognized both bands with various efficiencies (see, e.g., Figure 1A), whereas one turned out to be more specific for ALBA3 (Figure 1B) and another one for ALBA4 (Figure 1C). Comparable levels of ALBA3 and ALBA4 proteins were found in the bloodstream form in culture (Figure 1, B and C). Next, cell lines expressing yellow fluorescent (YFP) fusion proteins with either ALBA3 or ALBA4 were generated in procyclic cells of Lister 427 and AnTat1.1 strains (this latter strain is fully competent to infect tsetse flies; see later discussion; Le Ray et al., 1977). The plasmids were linearized within the ALBA3 or ALBA4 coding sequence to target integration and subsequent expression of the fusion construct from the endogenous locus (Figure 1D). Protein expression was verified by Western blot using either the anti-ALBA3–specific antibody (Figure 1B) or the anti-ALBA4–specific antibody (Figure 1C). The YFP-tagged versions ran at positions in agreement with their molecular weights of 46 kDa (ALBA3::YFP; Figure 1B) and 50 kDa (ALBA4::YFP; Figure 1C), respectively. These results confirmed the specificity of the ALBA3 and ALBA4 antibodies. Moreover, as expected from the endogenous tagging procedure, the amount of untagged protein seemed reduced for both ALBA3 and ALBA4 (Figure 1, B and C).
FIGURE 1:
ALBA protein expression and localization in wild-type and ALBA::YFP–expressing cell lines. (A–C) Reactivity of different ALBA antibodies assessed by Western blot in the indicated cell lines (2 μg of total protein extracts per lane). (A) An antibody recognizing both ALBA3 (21 kDa) and ALBA4 (25 kDa) was used on protein extracts from procyclic form 427 (PCF) cells. (B) A specific anti-ALBA3 antibody and (C) an antibody mostly specific for ALBA4 were assessed in cell lines expressing either ALBA3::YFP (46 kDa) or ALBA4::YFP (50 kDa) fusion proteins in PCF wild-type Lister 427 and in bloodstream-form parasites (BSF) of the strain 9013 (Wirtz et al., 1999). (D) Schematic representation of the procedure used for endogenously tagging ALBA proteins. The plasmid p3329, containing a fragment of the ALBA gene upstream of the YFP sequence and a puromycin-resistance cassette, is linearized (red line) within the ALBA sequence before transfection. This allows homologous recombination with one of the endogenous ALBA copies to create YFP-tagged ALBA. (E) Cultured procyclic cells were analyzed by IFA using the anti-ALBA3 antibody (top) and counterstained with DAPI (white, bottom) merged with the phase contrast image. Cells of the Lister 427 strain expressing (F) endogenously ALBA3::YFP or (G) ALBA4::YFP were fixed with PFA and counterstained with DAPI (white, bottom). Scale bar, 5 μm.
Ten anti-ALBA antibodies were then used by immunofluorescence analysis (IFA) to evaluate the localization of ALBA3/4 proteins. After paraformaldehyde (PFA) fixation, all antibodies tested produced a clear signal evenly distributed in the cytoplasm of the cultured procyclic form (Figure 1E). ALBA3/4 were absent from the nucleus and the kinetoplast at all steps of the cell cycle. In contrast, signal was lost upon methanol fixation, suggesting that ALBA3/4 are cytosolic proteins that are solubilized upon membrane removal. Finally, observation of the ALBA::YFP direct fluorescence in live cells confirmed the exclusively cytoplasmic location and the absence of visible changes in the staining pattern during the cell cycle (Figure 1, F and G).
ALBA3 and ALBA4 are therefore unlikely to be involved in DNA binding, but as they possess RGG repeats, they could interact with RNA. In trypanosomes, several RNA-interacting proteins accumulate into cytoplasmic foci that harbor mRNA and are formed in starvation conditions or upon heat shock (Cassola et al., 2007; Kramer et al., 2008). The localization of ALBA3/4 was investigated in these two stress conditions, first by IFA with the anti-ALBA3 and an antibody against DHH1, a recognized marker of these granules (Kramer et al., 2008). In contrast to DHH1, the ALBA signal did not show discrete foci upon heat shock (Figure 2A). However, the ALBA signal was clearly concentrated in foci after starvation stress, where it colocalized with DHH1 (Figure 2B).
FIGURE 2:
Localization of ALBA proteins in stress conditions and colocalization studies. Wild-type procyclic trypanosomes of the Lister 427 strain were subjected to different stresses: either (A) heat shock (42°C for 2 h) or (B) starvation (incubation in PBS for 2 h), fixed with PFA, and analyzed by IFA using the anti-ALBA3 antibody (left, green in merge) together with the anti-DHH1 antibody (middle left, red in merge) and counterstained with DAPI (blue in the phase contrast, right). (C) Cells expressing either ALBA3 or (D) ALBA4 as YFP fusions (left, green in merge) together with the DHH1 protein fused to mCherry (middle left, red in merge) were starved in PBS for 2 h, fixed with PFA. and counterstained with DAPI (blue in phase contrast, right). (E) Cells of the 427 strain starved in PBS for 2 h were analyzed by FISH-RNA to detect the poly(A+) RNA content in the cell (left, green in merge) coupled to IFA using the anti-ALBA3 antibody (middle left, red in merge); right, DNA content (DAPI staining). In E, image acquisition was carried out by confocal microscopy, and one z-plane (200 nm) for all is shown. Scale bar, 5 μm.
Because the access of the antibody to the antigen in the foci could be limited, these results were confirmed by analyzing the cell lines expressing endogenous ALBA3::YFP and ALBA4::YFP fusion proteins. In starvation conditions, both YFP fusion proteins started to aggregate into cytoplasmic foci as soon as after 10 min and reached a maximum accumulation after 2 h (unpublished data), confirming the IFA results. To further define ALBA localization, a mCherry::DHH1 construct (Kelly et al., 2007) was added to these cell lines, revealing that both proteins colocalized in cytoplasmic granules after 2 h of nutritional stress (Figure 2, C and D).
The colocalization of ALBA with a described RNA-binding protein in cytoplasmic foci prompted the investigation of poly(A+) RNA, which was shown to colocalize with the DHH1 protein upon starvation stress (Cassola et al., 2007). Therefore, the RNA–fluorescence in situ hybridization (FISH) method to detect poly(A+) RNA was coupled to IFA using the anti-ALBA3 antibody (Figure 2E). Whereas conventional epifluorescence microscopy suggested perfect colocalization, the analysis of Z-stacks by confocal microscopy revealed three types of foci in starved parasites: 1) those containing poly(A+) RNA and ALBA protein displaying equal signal intensity, 2) those containing poly(A+) RNA but no detectable ALBA protein, and 3) those showing a signal for ALBA protein but no detectable poly(A+) RNA signal. In conclusion, both ALBA proteins formed foci upon starvation stress conditions, colocalized with the DHH1 protein, and showed partial colocalization with poly(A+) RNA.
Depletion of ALBA3/4 blocks cell growth and induces morphological modifications
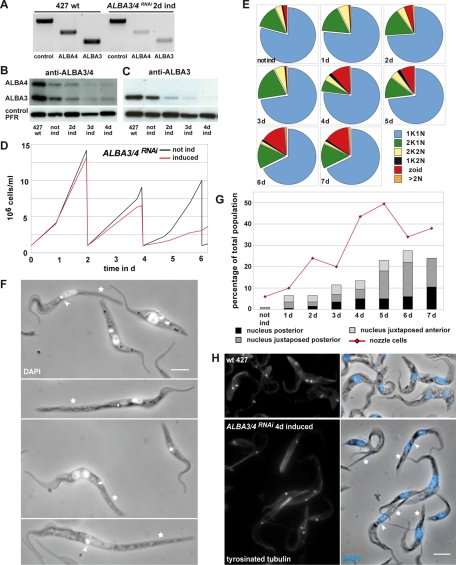
To evaluate the importance of ALBA3 and ALBA4 for trypanosomes, their expression was depleted either simultaneously or individually using RNAi in 29-13 procyclic parasites in culture. The first 400 base pairs of the ALBA3 and ALBA4 coding region show 94% identity (Supplemental Figure S2A) and were targeted for simultaneous knockdown of ALBA3 and ALBA4. The alignment of all ALBA DNA sequences (including the region of highest conservation; Supplemental Figure S2B) shows that the nucleotide identity is so low (maximum stretch of identity is limited to four consecutive nucleotides) that the possibility of cross-RNAi against ALBA1 or ALBA2 can be formally ruled out. The silencing efficiency of ALBA3/4 was assessed by semiquantitative reverse transcription-PCR (RT-PCR) (Figure 3A) and by Western blot using the antibody recognizing both ALBA3 and ALBA4 (Figure 3B) or an antibody recognizing exclusively ALBA3 (Figure 3C). The mRNA levels of both ALBA3 and ALBA4 were significantly reduced after 2 d of induction (Figure 3A). ALBA protein amounts dropped significantly starting from day 2 (Figure 3, B and C) and remained low after up to 7 d of continuous induction of RNAi (unpublished data). The anti-ALBA3 antibody was used in IFA to show protein down-regulation at the single-cell level (Supplemental Figure S3, B–D). The number of ALBA-positive cells decreased rapidly until day 3, when almost 100% of the cells were ALBA negative (Supplemental Figure S3B). Once it was proven that the ALBA proteins were efficiently knocked down in the ALBA3/4RNAi cell line, the growth in culture was monitored with and without tetracycline (Figure 3D). After a comparable proliferation in the first 2 d, the induced cells showed slightly slowed down growth after the first dilution step, but almost ceased to duplicate after the second dilution step, in contrast to the noninduced control, which subsequently needed dilution every 2 d (Figure 3D).
FIGURE 3:
ALBA3/4RNAi cells exhibit defects in cell growth and morphogenesis. (A) Semiquantitative RT-PCR on total RNA of the control 427 strain or the ALBA3/4RNAi cell line induced for 2 d, using specific primers for ALBA3 or ALBA4. (B, C) Western blot with 2 μg of total protein extracts per lane of either PCF Lister 427 wild type or of the ALBA3/4RNAi cell line either not induced or induced for 2, 3, and 4 d using (B) an antibody against ALBA3 and ALBA4 or (C) an anti-ALBA3–specific antibody. Anti-PFR antibody was used as a control. (D) Growth curve of the ALBA3/4RNAi cell line under noninduced (black) or induced (red) conditions during a 6-d period. (E) Proportion of cells in different cell cycle steps and DNA configuration during the time course of induction of the ALBA3/4RNAi cell line (n = 200 cells per time point). (F) Parasites of the ALBA3/4RNAi cell line induced for 4 and 5 d fixed with PFA and counterstained with DAPI (white). Scale bar, 5 μm; stars, cells with an elongated posterior end (nozzle); arrowheads, cells with a kinetoplast that is positioned anterior to the nucleus. (G) Percentage of aberrant cells in the ALBA3/4RNAi cell line during the time course of induction. Red curve represents the cells showing an elongated posterior end (nozzle phenotype), and the histogram represents the percentage of cells showing aberrant nucleus positioning (black, clearly posterior nucleus; dark gray, nucleus juxtaposed posterior to the kinetoplast; light gray, anterior juxtaposed nucleus). n = 200 cells per time point. (H) Cells of the 427 strain and the ALBA3/4RNAi cell line induced for 4 d were fixed in methanol and stained with the YL1/2 antibody to detect tyrosinated tubulin (left) and counterstained with DAPI (right, blue in the phase contrast). RNAi kinetic experiments were reproduced three times and always produced similar results (one representative experiment is shown). Scale bar, 5 μm; stars, cells with an elongated posterior end (nozzle); arrowheads, a kinetoplast that is positioned anterior to the nucleus.
Trypanosomes in culture divide by binary fission, and the cell cycle phases are recognized by number and position of DNA-containing organelles: G1/S (1 kinetoplast [K] and 1 nucleus [N]), G2/M (2K1N), and postmitotic cells (2K2N) (Figure 3E). Aberrant cell types were also recorded: 1K2N parasites, cells without nucleus called zoids (1K0N; Robinson et al., 1995), and cells with multiple nuclei (>2N). The noninduced state was considered as the control situation in which the population was split into 79% 1K1N, 14% 2K1N, and 4% 2K2N cells and a minority of various aberrant cell types (<3%). No major changes in these proportions were observed up to 3 d of induction, in agreement with the growth curve. However, after the fourth day of RNAi induction, the proportion of 2K1N cells decreased, whereas the percentage of zoids went up, reaching 15% after 6 d, indicating cell cycle defects (Ploubidou et al., 1999). In contrast to many RNAi mutants in T. brucei, we did not observe the emergence of multinucleated cells (Figure 3E).
To further investigate the consequences of the ALBA3/4 knockdown, cells induced at various time points were fixed and stained by IFA with the anti-ALBA3 antibody. The cells devoid of ALBA staining exhibited two main, striking phenotypes (Figure 3F). First, they showed increased length up to 40 μm (noninduced cell length in the 1K1N state is ∼25 μm) (Figure 3F, star). As early as 2 d after the induction of protein knockdown, 24% of the cells showed an elongated posterior end, reaching up to 50% after 5 d (Figure 3G, graph). This phenotype was previously reported in some mutants and termed “nozzle” (Hendriks et al., 2001; Li et al., 2003; Hammarton et al., 2004). However, a second phenotype specific to ALBA3/4RNAi was observed: the nucleus frequently appeared in a posterior position relative to the kinetoplast (Figure 3F, arrowhead). The percentage of such cells increased constantly during the course of RNAi induction (Figure 3G, histogram). ALBA3/4RNAi cells induced for 4 d were analyzed by scanning electron microscopy after extraction of the membrane, allowing direct visualization of the microtubule cytoskeleton corset and revealing the elongated posterior end with apparently correctly assembled microtubules (Supplemental Figure S3, E and F). In addition, we were able to have an inside view of a nucleus more posterior to the two kinetoplasts, combining the two phenotypes observed by light microscopy (Supplemental Figure S3F).
ALBA3/4RNAi-induced cells were stained with the YL1/2 antibody, which recognizes newly assembled tyrosinated α-tubulin (Kilmartin et al., 1982; Sherwin and Gull, 1989; Figure 3H), to evaluate whether the elongation in nozzle cells was due to an active polymerization of microtubules at the posterior pole. The staining was found at the basal bodies and the daughter flagellum, as well as at the posterior end in both control and ALBA3/4RNAi-induced cells, but ALBA3/4RNAi cells with an elongated posterior end displayed by far the brightest signal (Figure 3H, stars). In T. brucei, the length of the single flagellum that extends along the cell body controls cell size (Kohl et al., 2003). It was therefore measured in the ALBA3/4RNAi cells to exclude a contribution to cell elongation. Regardless of the cell length and the RNAi induction time point (days 4–6), the length of the flagellum remained in the normal range (unpublished data). In contrast, the distance from the kinetoplast to the posterior end correlated in a linear relationship with cell length, which can be linked to the exhaustive microtubule elongation at the posterior pole (unpublished data).
To assess whether these phenotypes were the result of silencing of ALBA3 or ALBA4 alone or of the combination of ALBA3 and ALBA4, constructs allowing expression of double-stranded (ds) RNA targeting specifically ALBA3 or ALBA4 (Supplemental Figure S3A) were produced and transfected in trypanosomes. Potent silencing of ALBA4 was achieved, but this did not result in a visible phenotype (unpublished data). In contrast, generation of a cell line silencing ALBA3 was very difficult either because of significant expression of ALBA3 dsRNA even before the addition of tetracycline or because of the emergence of cells that did not silence ALBA3 but proliferated rapidly (unpublished data). This was reported previously for several RNAi experiments (Chen et al., 2003) and prevented full investigation of the cell line. These data suggest that ALBA3 could be essential for procyclic trypanosomes. In summary, procyclic cells depleted of ALBA3/4 proteins elongated by active polymerization of microtubules at their posterior end relocated their nucleus to the posterior side of the kinetoplast and became arrested in the cell cycle.
ALBA3/4 expression profile during trypanosome development in the tsetse fly
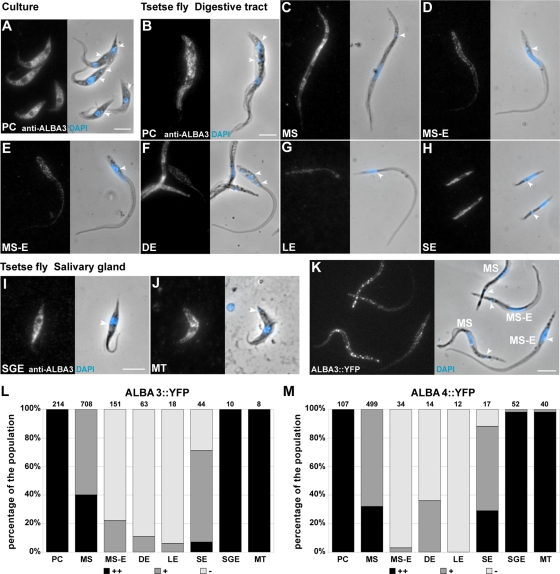
All the foregoing observations suggest that ALBA3/4 knockdown mimics several typical changes happening in cells that undergo a trypomastigote-to-epimastigote differentiation during parasite development in the anterior midgut of a tsetse fly. However, this differentiation step is not reproducible in the laboratory. To assess ALBA protein levels in vivo during the parasite cycle, tsetse flies were fed with procyclic parasites of the AnTat 1.1 strain. Flies were dissected at various time points of infection, and the obtained parasites were fixed in PFA to perform IFA with the selected anti-ALBA3 antibody. The procyclic form (PC) obtained from the fly midgut showed a cytoplasmic ALBA staining (Figure 4B) equivalent to that observed with the cultured form (Figure 4A). As the cell elongated to differentiate into the nonproliferative mesocyclic trypomastigote form (MS), no visible changes in ALBA signal intensity or profile were detected (Figure 4C). The following stage in the parasite cycle, the mesocyclic to epimastigote form (MS-E), is found mainly in the proventriculus of the fly and does not proliferate but is characterized by an elongated nucleus migrating toward the posterior end of the cell (Vickerman et al., 1988; Sharma et al., 2008; Rotureau et al., 2011). The MS-E parasites showed strongly reduced ALBA level (Figure 3, D and E, and Supplemental Figure S4). The ALBA level remained very low in cells that had adopted the epimastigote configuration, including after kinetoplast duplication and mitosis in the asymmetrically dividing epimastigote form (DE) (Figure 4F). After division, the short form (SE) reacquired ALBA signal, whereas the long one (LE) remained negative (Figure 4, G and H). Finally, all the forms in the salivary glands were positive for ALBA: the salivary gland epimastigotes (SGEs) and the infective metacyclic trypomastigote parasites (MTs) (Figure 4, I and J).
FIGURE 4:
ALBA3/4 expression level during parasite development in the tsetse fly. (A–J) Evolution of ALBA3/4 expression level assessed by IFA with the anti-ALBA3 antibody (left) and counterstained with DAPI (blue in the phase contrast image). ALBA signal in (A) Lister 427 procyclic cells in culture, (B–H) AnTat1.1 parasites derived from the digestive tract, and (I, J) salivary glands of the tsetse fly (normalized fluorescent signal). (K) Trypanosomes of the strain AnTat1.1 expressing endogenous ALBA3::YFP, derived from the tsetse proventriculus, fixed and stained with DAPI. Scale bars, 5 μm; arrowheads, the localization of the kinetoplast (A–K). (L, M) Percentage of strong (black), positive fluorescent (dark gray), and negative cells (light gray) at the different stages of the parasite cycle of AnTat1.1 parasites expressing endogenously tagged ALBA3::YFP (L) and ALBA4::YFP (M) obtained upon tsetse fly dissection. For ALBA3::YFP, data are the sum of two separate infection experiments (total 168 flies; 68 were dissected), whereas 93 flies (36 dissected) were used for ALBA4::YFP in one infection experiment. Analysis was performed on live cells as described in the text; the number of counted cells is indicated on top of the bars. Cells are shown in the order of appearance during the infection cycle in the tsetse fly: procyclic (PC), mesocyclic trypomastigote (MS), parasites in transition from mesocyclic trypomastigote to epimastigote (MS-E), and asymmetrically dividing epimastigote (DE) into a long (LE) and a short epimastigote (SE); in the salivary glands (I, J), salivary gland epimastigote (SGE) and metacyclic (MT).
These data indicate a precise control of ALBA3 and ALBA4 during trypanosome development in the fly. However, quantification of the results is challenging since these stages are present in low numbers and difficult to access, and, moreover, the necessary use of PFA fixation means that even fewer parasites are available for analysis (cells fixed in these conditions do not adhere well on microscope slides). We therefore chose to work with live cells expressing YFP fusion proteins, which also avoids limitations resulting from fixation approaches. Hence, tsetse flies were fed with either one of the ALBA::YFP AnTat1.1 cell lines and dissected at different time points of infection. For both strains, the rates of infection in the midgut (MG) and salivary glands (SG) were comparable with those for the parental wild-type AnTat1.1 strain (control MG, 40%, SG 10.7%; ALBA3::YFP MG, 54%, SG 10.3%; ALBA4::YFP, MG 33%, SG 11%). Figure 4K shows a typical example of ALBA3::YFP cells released from the proventriculus: although mesocyclic parasites showed strong ALBA signal, the mesocyclic to epimastigote forms seemed almost negative. In this experiment, parasites were released from fly tissues into phosphate-buffered saline (PBS), demonstrating that the formation of cytoplasmic starvation granules also occurs in parasites issued from an infection (Figure 4K).
Because endogenous ALBA fusion protein expression reflected the results obtained by IFA, ALBA expression profile was monitored by live videomicroscopy. However, parasites can only be seen upon dissection and are short lived after fly sacrifice, restricting time available for analysis. We selected to acquire images by analogue video recording. This is not a digital system, and absolute quantification cannot be performed, but three categories of cells can be clearly defined: negative (−), positive (+), and bright (++). By this mean, it was possible to analyze a sufficiently large number of cells (1216 for ALBA3::YFP and 775 for ALBA4::YFP) not reachable otherwise. The differentiation stage was determined by phase contrast imaging (for morphology) coupled with DNA staining by 4′,6-diamidino-2-phenylindole (DAPI), a dye that penetrates easily in live trypanosomes (Figure 4, L and M). Supplemental Movie S1 shows representative cells for each ALBA3::YFP parasite stage found in the fly, and Supplemental Movie S2 shows fields of cells of the same strain to illustrate how relative fluorescence intensity was evaluated.
All ALBA3::YFP cells showed a bright fluorescent signal at the PC stage, whereas the MS parasites were more heterogeneous (40% bright and 60% positive green cells; Figure 4L). The ALBA3 fluorescence intensity dropped further during the transition to the epimastigote form, with almost 80% of the parasites being negative. This was even more pronounced in the DEs (>90% of ALBA3::YFP negative cells). The ALBA signal was recovered in the SEs, whereas LEs remained negative, confirming the IFA data (Figure 4L). Finally, all trypanosome forms found in the salivary glands were strongly positive for ALBA3 (Figure 4L).
The expression profile of ALBA4::YFP turned out to be very similar to that of ALBA3 (Figure 4M and Supplemental Movie S3), with the exception of the DE, with 35% of positive fluorescent cells in ALBA4 versus 10% in ALBA3. This could be explained by a faster reemergence of ALBA4 protein in the future SE before cytokinesis is completed, going along with the higher percentage of SE-positive cells for ALBA4 versus ALBA3 (Figure 4M).
Taken together, the combination of IFA and live videomicroscopy analysis showed a defined expression profile of ALBA3 and ALBA4 proteins during the parasite cycle, marked by a significant drop at the trypomastigote to epimastigote transition.
Constitutive overexpression of ALBA3 proteins impairs differentiation
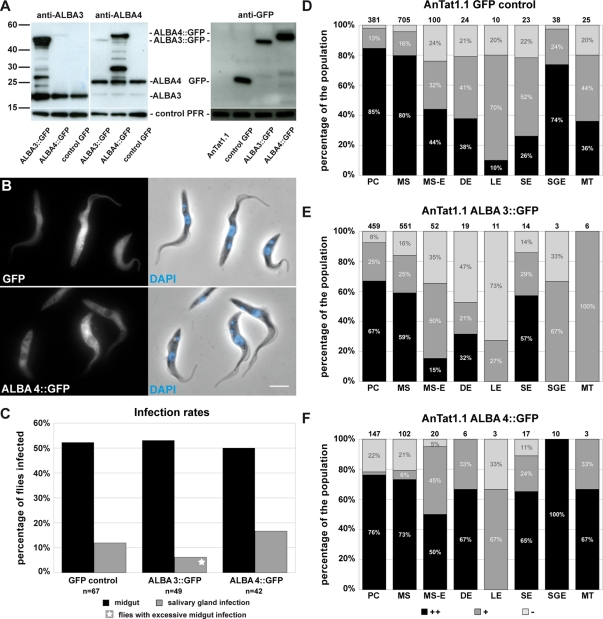
To understand the significance of the down-regulation of ALBA3/4 in the proventricular stages, we sought to overexpress them. However, regulatory elements required for expression at these particular stages are unknown. Hence, we selected to use the pHD67E vector, which had been reported to confer GFP reporter expression throughout the parasite cycle (Bingle et al., 2001). This construct was targeted to the rDNA locus, and the reporter GFP protein was expressed under the control of the EP procyclin promoter. Either ALBA3 or ALBA4 coding region was fused upstream of the GFP gene into this vector. The expression as GFP fusion was necessary to control the level of overexpression in individual cells during parasite development in the fly. Procyclic trypanosomes of the AnTat1.1 strain were transfected with the plasmid pHD67E as control or with pHD67EALBA3 and pHD67EALBA4. Protein expression levels were first analyzed by Western blot in cultured procyclic cells using the specific anti-ALBA3 or anti-ALBA4 antibodies (Figure 5A). Fusion proteins exhibited the expected electrophoretic mobility, and their abundance appeared to be equivalent to that of the endogenous ALBA3 or ALBA4, meaning that these cells were expressing double the amount of ALBA3 or ALBA4 (Figure 5A). This exogenous expression of ALBA::GFP did not lead to a down-regulation of the endogenous ALBA protein, but additional protein bands of intermediate size were detected with both the anti-ALBA3 and anti-ALBA4 antibodies (Figure 5A), possibly corresponding to degradation products of the fusion protein, a feature not seen during the endogenous tagging experiments (Figure 1, B and C). To compare the expression levels between cell lines, protein samples were probed with an anti-GFP antibody (Figure 5A). ALBA4::GFP was expressed to a level comparable to that of the control GFP, whereas ALBA3::GFP turned out to be less abundant, despite the use of the same expression system. In the GFP control cells, direct observation of fluorescence in live or fixed cells (Figure 5B) revealed that GFP expression was heterogeneous from one cell to the other and that the signal was detected in the cytoplasm as well as in the flagellum and the nucleus (Figure 5B, top). In contrast, ALBA3::GFP (unpublished data) and ALBA4::GFP fusion proteins (Figure 5B, bottom) were found exclusively in the cytoplasm. All three cell lines grew normally in culture, and cells did not exhibit any particular phenotype.
FIGURE 5:
Overexpression of ALBA3::GFP or ALBA4::GFP and consequences for fly infection. (A) Western blots with 2 μg of total protein samples of AnTat1.1 WT cells carrying a GFP gene (control GFP) or an extra copy of either ALBA3::GFP or ALBA4::GFP. Blots were incubated with a specific anti-ALBA3 antibody to detect the endogenous ALBA3 (expected molecular weight 21 kDa) and the ALBA3::GFP (46 kDa) protein (left), with a specific anti-ALBA4 antibody to detect the endogenous ALBA4 (25 kDa) and the ALBA4::GFP (50 kDa) protein (middle), or an anti-GFP antibody to reveal the control GFP protein (right) and the ALBA::GFP fusion proteins. A weak cross-reactivity of the anti-ALBA4 antibody with ALBA3 is visible. Anti-PFR antibody was used as a control. (B) Procyclic cells of the strain AnTat1.1 in culture expressing either GFP or exogenous ALBA4::GFP were fixed in PFA and counterstained with DAPI (left, direct fluorescence; right, phase contrast image and DNA in blue; scale bar, 5 μm). (C) Percentage of flies infected (in the midgut or the salivary glands) when challenged with AnTat1.1 cells expressing GFP control (sum of seven infection experiments; 214 flies, 67 dissected), ALBA3::GFP (sum of six infection experiments; 155 flies, 49 dissected), or ALBA4::GFP (sum of three infection experiments; 143 flies, 42 dissected) (n = total number of dissected flies). Infections with the ALBA3::GFP cell line in the salivary gland were only observed in flies with a highly infected midgut (star). (D–F) Percentage of bright (black), positive fluorescent (dark gray), and negative cells (light gray) in the populations of each stage of AnTat1.1 parasites expressing GFP alone (D), ALBA3::GFP (E), or ALBA4::GFP (F) during the course of infection. Analysis was performed on live trypanosomes directly upon fly dissection as described in the text; the number of counted cells is indicated on top of the bars. To reach the total numbers of parasites, data from all independent infection experiments were grouped for analysis as indicated in C. Cells are shown in the order of appearance during the infection cycle in the tsetse fly: procyclic (PC), mesocyclic trypomastigote (MS), parasites in transition from mesocyclic trypomastigote to epimastigote (MS-E), and asymmetrically dividing epimastigote (DE) into a long (LE) and a short epimastigote (SE); in the salivary glands, salivary gland epimastigote (SGE) and metacyclic (MT).
Several groups of flies were infected with each of the strains, and dissection was undertaken at late time points (days 25–30), when control infections should have reached maturity in the salivary glands. Infection rates were obtained by grouping data of nine independent experiments (Figure 5C). For the GFP control strain, 52% of the flies showed midgut infections, and salivary gland infections were detected in 12% of the flies (Figure 5C), which correspond to the levels observed in our laboratory for the AnTat1.1 wild-type strain (Rotureau et al., 2011). One-third of the established infections in the midgut led to a mature salivary gland infection. The ALBA3::GFP strain turned out to be as efficient as the control in developing midgut infections (53%). The level of salivary gland infections was slightly lower (6%; meaning that only 10% of the established infections in the midgut led to a mature infection in the fly saliva). It should be noted that these flies showed an unusually high number of midgut parasites in comparison to other midgut infections observed for the same cell line. For the ALBA4::GFP strain, 50% of the flies were infected in the midgut, and 17% showed parasites in the salivary glands, values that are very close to the control situation, with one-third of midgut infections leading to a mature development.
Live trypanosomes from all three strains were investigated by videomicroscopy as described earlier (Supplemental Movie S4). Starting with a high proportion of green fluorescent procyclic parasites (78–98%), the green signal evolved differently during progression in the parasite cycle (Figure 5, D–F). In the GFP control cell line, 80% of the cells were strongly positive at the midgut PC or MS stages, and negative parasites were rare (Figure 5D). The general trend in all proventricular stages was a drop in the proportion of strongly positive parasites, but the abundance of GFP-negative parasites remained constantly low, ∼20%. Cells in the salivary gland were positive in most cases (SGE and MT). In summary, the reporter GFP protein was present in 80 to nearly 100% of the cells in all stages of the parasite cycle, although its abundance was variable (Figure 5D). In contrast to the endogenous tagging experiments, different results were obtained for cells overexpressing ALBA3::GFP (Figure 5E) or ALBA4::GFP (Figure 5F). Infections with the ALBA4::GFP strain were characterized by a majority of cells expressing a high level of the fusion protein, whereas negative parasites were a minority in all stages investigated, with the exception of long epimastigotes (Figure 5F). This profile is similar to the expression of the control GFP alone. In contrast, proventricular parasites resulting from ALBA3::GFP infections showed a higher proportion of negative cells: 35% for the MS-E and up to 47% for the DE, whereas the percentage of positive SE was comparable to the results obtained for ALBA4::GFP. Only a few cells could be analyzed in the salivary glands due to reduced parasite load, but all turned out to be negative or weakly positive (Figure 5E), in contrast to the other two cell lines (Figure 5, D and F). These results show that overexpression of the ALBA4 protein as GFP fusion is maintained in each stage during progression of the parasite cycle without impairing the rate of fly infectivity. In contrast, although the same expression system was used, the ALBA3::GFP protein was down-regulated at the transition from trypomastigote to epimastigote stage, like the endogenous ALBA3 protein. Moreover, parasites expressing large amounts of ALBA3::GFP proteins were not observed in the salivary glands, suggesting that strong overexpressers cannot complete the final part of the parasite cycle.
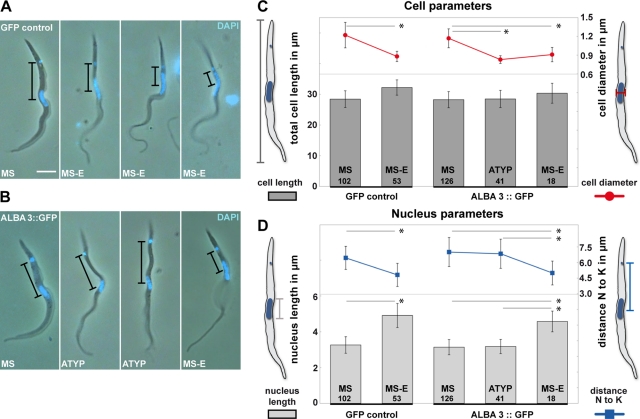
During the live video analysis of the ALBA3::GFP cells (but not of the GFP control or in the ALBA4::GFP strain) in the anterior midgut and proventriculus, our attention was attracted by a significant proportion of parasites that showed atypical cell diameter and DNA organelle positioning (Figure 6). Several morphometric parameters were measured: the length of the parasite (from the posterior end of the cell to the flagellum tip), the cell diameter, the distance between kinetoplast and nucleus centers, and the length of the nucleus. To avoid artifacts potentially caused by fixation methods, this analysis was performed on cells from the live movies of the GFP control and the ALBA3::GFP parasites (Figure 6). Two populations were identified in the GFP control: MS cells with a fairly large diameter (1.2 μm) and with a round nucleus positioned close to the center of the cell (102 of 155 analyzed cells; Figure 6A) and MS-E cells with a thinner diameter (0.9 μm) and an elongated, oval nucleus undergoing migration toward the posterior end of the cell (Figure 6, C and D, left). These two phenomena were always found to occur in parallel but never uncoupled. Although typical MS (126 of 185 analyzed cells) and MS-E (18 of 185) were present, an atypical cell type (ATYP) was also observed in the ALBA3::GFP cell line (41 of 185). These cells presented a pronounced thin cell diameter (0.83 ± 0.06 μm) typical for MS-E but, strikingly, despite having the cell diameter of MS-E, they exhibited a nonelongated nucleus (3.18 ± 0.4 vs. 4.58 ±0.58 μm in MS-E) whose position was more anterior than that observed in MS-E. The distance to the kinetoplast was 6.84 ± 1.40 μm instead of 5.0 ± 1.14 μm (Figure 6, B–D).
FIGURE 6:
Characterization of the transition from mesocyclic trypomastigote to epimastigote stage in the GFP control and the ALBA3::GFP–expressing strain. Still images extracted from movies of cells of the AnTat1.1 strain expressing GFP control (A) or the ALBA3::GFP (B) after fly dissection. Phase images were superimposed with the fluorescent DAPI signal in blue. Black bars indicate the distance between the nucleus and the kinetoplast. Scale bar, 5 μm. Cellular (C) and nuclear parameters (D) in mesocyclic (MS), parasites in transition from mesocyclic trypomastigote to epimastigote (MS-E), and atypical forms (ATYP, found in ALBA3::GFP infected flies only). Numbers of analyzed parasites are indicated in the bars. (C) Left axis, total cell length plotted as dark gray bars (μm ± SD); right axis, cell diameter shown as red curve (μm ± SD). (D) Left axis, nucleus length plotted as light gray bars (μm ± SD); right axis, distance between the nucleus and the kinetoplast centers shown as blue curve (μm ± SD). Analysis of variance tests were performed, and significant comparisons by Tukey ad hoc posttests are indicated on the histogram with *p < 0.0001.
In conclusion, this atypical form encountered in the ALBA3::GFP overexpressing strain displayed the morphology of an MS-E regarding the cell diameter, but the nucleus length and its distance to the kinetoplast were found to be comparable to the mesocyclic form. This suggests that the presence of the ALBA3::GFP protein delayed or inhibited migration of the nucleus toward the posterior end of the cell during the trypomastigote to epimastigote transition, further strengthening the hypothesis that ALBA3 is implicated in this differentiation process.
DISCUSSION
Very little is known about the way in which trypanosomes accomplish the different transitions that mark their development in the tsetse fly (Sharma et al., 2009). This is due to the failure to reproduce these steps in vitro and to the difficulty in obtaining sufficiently high cell numbers from infected flies for manipulation. We showed here that the combination of endogenous tagging and monitoring of fluorescent fusion proteins by live microscopy is a powerful approach to assess protein expression at the individual cell level in all parasite developmental stages in the tsetse fly, although the investigation remains tedious. Coupled to IFA detection, it revealed a drastic drop in the amount of both ALBA3/4 exclusively during differentiation from the mesocyclic trypomastigote stage to the epimastigote stage in the anterior midgut. A dilution effect due to changes in cell shape can be ruled out, as the cell volume remains constant during this transition (Rotureau et al., 2011). This step is a one-way decision: postmesocyclic parasites are committed to differentiation, in contrast to mesocyclic trypomastigotes, which, once issued from fly dissection and transferred to culture medium, are able to transform back to the proliferating procyclic form after 5 d (Van Den Abbeele et al., 1999).
We propose that this ALBA3/4 depletion that takes place during the transition from the mesocyclic to the epimastigote stage correlates with an involvement of these proteins in differentiation. Remarkably, ALBA3/4 RNAi knockdown at the procyclic stage in culture reproduces several aspects of this transition (cell cycle arrest, posterior elongation, nucleus migration at the posterior end), with the exception of the cell diameter, which does not shrink, suggesting that additional factors need to come into play. Conversely, forced expression of ALBA3 fused to GFP impaired this same differentiation step in the fly proventriculus, resulting in the presence of numerous individuals showing reduced cell diameter as expected, but where elongation and migration of the nucleus to the posterior end are either blocked or slowed down. This was accompanied by a reduction in the efficiency of salivary gland infections (only 10% of infected midguts led to salivary gland infection, compared with ∼33% in GFP or ALBA4::GFP controls), which only occurred in flies heavily infected in the midgut, which should facilitate the passage of the bottleneck leading to the salivary glands (Oberle et al., 2010). Moreover, the parasites that reached the salivary glands displayed reduced ALBA3::GFP amounts compared with their ALBA4::GFP and GFP-alone counterparts, suggesting that excessive expression of ALBA3 is not compatible with this step of the parasite life cycle.
In molecular terms, the observed phenotypes could reflect a direct role for ALBA3/4 proteins in cytoskeleton rearrangements or an indirect role if ALBA3/4 act on other molecules that in turn are controlling development. ALBA proteins were shown recently to bind to a specific element found in the 3′ UTR of the GPEET procyclin mRNA (Mani et al., 2011), which encodes one of the main trypanosome surface proteins (Urwyler et al., 2005). In the absence of ALBA, the expression of a reporter flanked by this 3′ UTR is down-regulated. This takes place at the protein level, suggesting that ALBA proteins could participate to the control of stability and/or translation of mRNA carrying this element (Mani et al., 2011). Of interest, the expression of the GPEET procyclin protein is reduced during the development stages in the anterior midgut (Urwyler et al., 2005), that is, exactly when ALBA protein levels are reduced. These data support the view that ALBA proteins could interact with mRNA, as suggested by the presence of multiple RGG domains, the formation of cytoplasmic granules upon starvation that colocalize with the mRNA-interacting protein DHH1 (Kramer et al., 2010), and the association with some, but not all, poly(A+) RNA. Recent data indicate that the four trypanosome ALBA proteins could make protein complexes, in which ALBA3 would be the core component (Mani et al., 2011). In that study, silencing of ALBA3 alone was sufficient to generate a growth phenotype similar to what was observed for joint silencing of ALBA3 and ALBA4. The knockdown of ALBA3 alone resulted in a reduction of the amount of ALBA1, ALBA2, and ALBA4 proteins consistent with the presence of ALBA3 at the core of the protein complex. In contrast, knocking down ALBA4 alone did not result in a visible phenotype (Mani et al., 2011). These results are coherent with our study that shows that cells did not tolerate the same overexpression level for ALBA3 as for ALBA4, that impaired transformation in the fly was observed only upon ALBA3 overexpression (although this expression was more modest than for ALBA4), and that silencing of ALBA4 alone did not produce a visible phenotype. In summary, this work provides the first molecular and cellular data on a candidate protein for the control of differentiation during an essential and limiting step of the parasite cycle (Oberle et al., 2010).
Besides trypanosomes, ALBA proteins are found in numerous eukaryotic organisms, where their function remains to be investigated. In the female gamete of Plasmodium berghei, ALBA proteins are present in cytoplasmic structures harboring mRNA (Mair et al., 2010). The directed storage of specific mRNA is indispensable for the developmental program after fertilization and thus for the ookinete development. In agreement with our findings in trypanosomes, ALBA proteins could be important in the developmental fine tuning during the differentiation of certain cell types in multiple organisms, including mammals, highlighting the interest for further investigation of this multifaceted protein family.
MATERIALS AND METHODS
Animal care
Animals were housed in the Institut Pasteur animal facilities, which are accredited by the French Ministry of Agriculture for performing experiments on live mice, in compliance with the French and European regulations on care and protection of laboratory animals (European Directive 86/609/UE). Protocols for immunization were approved by the veterinary staff of the Institut Pasteur animal facility and were performed in compliance with National Institutes of Health Animal Welfare Assurance A5476-01 issued on 7 February 2007.
Trypanosome culture, RNAi, and stress induction
The T. brucei brucei Lister 427 strain for in vitro studies or the pleomorphic AnTat1.1 strain (Le Ray et al., 1977) for experiments in the tsetse fly were used. For the RNAi experiments, the Lister 427 derivative 29-13 (Wirtz et al., 1999) was chosen, as it disposes of a tetracycline-inducible system for dsRNA expression by T7 RNA polymerase. All procyclic parasites were cultured at 27°C in SDM79 (Brun and Schönenberger, 1979) supplemented with 10% fetal calf serum (FCS). In the case of the pleomorphic AnTat1.1 strain, 20 mM glycerol was added to the culture medium. For induction of RNAi, tetracycline was added to the medium at each dilution step at a final concentration of 1 μg/ml.
To induce nutritional stress, parasites were washed twice in PBS at room temperature and incubated 2 h at 27°C in an appropriate volume of PBS at cell densities between 5 and 10 million cells/ml (Cassola et al., 2007). To induce heat shock, parasites in their culture medium were transferred to 41°C for 2 h (Kramer et al., 2008).
Tsetse fly infection, maintenance, and dissection
Teneral males of Glossina morsitans morsitans from 8 to 96 h posteclosion were obtained from the Institut de Recherche pour le Développement (IRD), Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD), UMR 177 (Trypanosomes), Campus International de Baillarguet, Montpellier, France. Tsetse flies were infected with cultured parasites during their first meal through a silicone membrane (Rotureau et al., 2011). Freshly differentiated cultured procyclic AnTat1.1 were used at 10 million cells/ml in SDM79 medium supplemented with 10% FCS, 60 mM N-acetylglucosamine (Peacock et al., 2006), and 2.5% (wt/vol) bovine serum albumin (Kabayo et al., 1986). Tsetse flies were subsequently maintained in Roubaud cages at 27°C and 70% hygrometry and fed twice a week through a silicone membrane with fresh rabbit blood in heparin. Flies were starved for at least 48 h before being dissected 7–35 d postingestion. Salivary glands were immediately isolated upon dissection. Whole tsetse alimentary tracts, from the distal part of the foregut to the rectum, were dissected in a drop of PBS or SDM79 without FCS and hemin and arranged lengthways to screen for parasite presence. Foregut and proventriculus were then physically separated from the midgut in two distinct liquid drops. Parasites were released from tissues in the medium/PBS drop and were further treated no later than 15 min after dissection. For the AnTat1.1 ALBA::YFP experiments, 261 flies were infected. For the ALBA::GFP overexpression experiments, 517 flies were infected. Fly batches were usually split in two so as to have half of the flies infected with a control strain and half with the investigated cell line expressing various ALBA fusion proteins.
Protein expression and antibody production
For antibody production, ALBA3 and ALBA4 were expressed as recombinant GST fusion proteins in Escherichia coli. Full-length ALBA3 (Tb927.4.2040) and ALBA4 (Tb927.4.2030) were amplified from genomic DNA by PCR using Phusion polymerase (Finnzymes; Thermo Scientific, Waltham, MA) and primer pairs with 5′ BamHI and 3′ EcoRI flanking sequences. A common forward primer for ALBA3 and ALBA4 GCACAAGGGATCCATGCCTTCATATCCG (BamHI site underlined) was used in combination with either an ALBA3-specific reverse primer CAAATGAATTCTGATGCCTTACTCCTC (EcoRI site underlined) or an ALBA4-specific reverse primer GCTCGAATTCAGTTGCCTTCACCCAAATCG (EcoRI site underlined). PCR products were ligated into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. ALBA3/4 sequences were excised using EcoRI and BamHI enzymes and ligated into compatible sites of the pGEX B vector (GE Healthcare, Piscataway, NJ). Plasmids were sequenced and transformed into E. coli strain BL21. GST or GST-ALBA fusion protein expression was induced by addition of 1 mM isopropyl β-d-1-thiogalactopyranosid and analyzed by SDS–PAGE. GST-coupled ALBA proteins were purified as previously described (Smith and Johnson, 1988), and a total of 20 μg was subcutaneously injected four times (at 3-wk intervals) into BALB/c mice for immunization. After bleeding, sera were absorbed against GST. Sera from mice immunized with the GST protein alone were used as negative controls. For reasons of simplicity, the antibodies are referred to as anti-ALBA3, anti-ALBA4, and anti-ALBA3/4 as determined by Western blot. Note that the specificity might be different in IFA, as proteins are not denatured with SDS. For most IFA studies the anti-ALBA3 was used, as it showed the best signal-to-noise ratio.
Plasmid construction and trypanosome stable transformation
ALBA proteins were tagged using two different strategies. First, for endogenous tagging, the vector p3329 was used (Kelly et al., 2007; http://web.me.com/mc115/mclab/resources.html). It allows endogenous tagging with enhanced YFP at the C-terminal end of the protein and selection by puromycin. ALBA3 (520 base pairs) and ALBA4 (400 base pairs) C-terminal sequences were chemically synthesized by GeneCust Europe (Dudelange, Luxembourg) and subcloned into the p3329 vector after excision of the existing stuffer sequence by KpnI and BamHI. Before transfection into trypanosomes, the plasmids were linearized within the ALBA sequence by AfeI (ALBA3) and FspAI in (ALBA4). The plasmid for the endogenous expression of the mCherry-tagged version of DHH1 was linearized with NheI (Kramer et al., 2008).
For exogenous overexpression of ALBA proteins as GFP-tagged versions, the complete coding sequence of ALBA3 and ALBA4 was cloned in the plasmid pHD67E (Bingle et al., 2001) upstream of the GFP sequence using the HindIII restriction site. A common forward primer was used, GCACAATAAGCTTATGCCTTCATATCCG (HindIII site underlined), in combination with specific reverse primers, GCACTAAGCTTCGCTCCTCATTGCCACC (HindIII site underlined) for ALBA3 and GCACTAAGCTTCGGTTGCCTTCACCCAA (HindIII site underlined) for ALBA4. Before transfection, the plasmids were linearized at the unique NotI site in the rDNA intergenic targeting region.
For the generation of a cell line able to knock down ALBA3 and ALBA4 mRNA, the sequence for dsRNA expression was selected by the RNAit algorithm (Redmond et al., 2003). The common 400–base pair sequence of the ALBA3 and ALBA4 genes was amplified by PCR with the forward primer GAGACTCGAGATGCCTTCATATCCGTCGCA (XhoI site underlined) and the reverse primer AGAGAAGCTTTGTGGTTCACATCCAGCGGC (HindIII site underlined) and ligated into the corresponding sites of the pZJM vector (Wang et al., 2000). The ALBA insert is flanked by T7 promoters facing each other, allowing tetracycline-inducible dsRNA expression. For single RNAi, the sequences as shown in Supplemental Figure S3A were chemically synthesized by GeneCust Europe and subcloned into the pZJM vector after excision of the existing stuffer sequence by XhoI and HindIII. For RNAi against ALBA3 a fragment of 300 base pairs was used (the C-terminal 75 base pairs of the CDS and 227 base pairs of its 3′ UTR) and for ALBA4 a fragment of 350 base pairs (the C-terminal 77 base pairs of the CDS and 272 base pairs of its 3′ UTR). Before transfection into trypanosomes, all pZJM plasmids were linearized at the unique NotI site in the rDNA intergenic targeting region.
Trypanosomes were transfected with the plasmid constructs by Nucleofector technology (Lonza, Italy) as described (Burkard et al., 2007). Transfectants were selected by addition of the appropriate antibiotic concentration, and clonal populations were obtained by limited dilution.
Fluorescence analysis
Parasites expressing a fluorescent fusion protein were either analyzed directly by live video microscopy or after fixation for 30 min in a 4% paraformaldehyde (PFA) solution in PBS. In both cases, DNA was stained using DAPI (1 μg/ml).
For immunodetection, cultured parasites were washed twice in SDM79 medium without serum. Parasites obtained from the fly were spread directly on poly-l-lysine–coated slides before fixation. Two main methods of fixation were used. For methanol fixation, parasites were settled on poly-l-lysine–coated slides, air dried, and fixed in methanol at –20°C for 5 min, followed by a rehydration step for 10 min in PBS. For PFA fixation, parasites were incubated for 10–30 min at room temperature with a 4% PFA solution in PBS at pH 7 and left to settle on poly-l-lysine–coated slides. After a permeabilization step with 0.1% Nonidet P-40, samples were blocked for 1 h with 1% bovine serum albumin (BSA) in PBS. For immunodetection, washed slides were incubated with the appropriate dilution of the first antibody in 0.1% BSA in PBS for 1 h. After three 5-min washes, secondary antibodies coupled to a fluorochrome were used in the same conditions as the first antibodies. Cells were stained by DAPI and mounted with ProLong antifade reagent (Invitrogen). Antibodies used in PFA fixation were the anti-ALBA antibodies diluted 1/800 and the anti-DHH1 antibody diluted 1/1500 (Kramer et al., 2008, 2010). Antibodies used in methanol fixation are MAb25, recognizing an epitope found all along the trypanosome axoneme (Pradel et al., 2006; Absalon et al., 2007); L3B2, recognizing the FAZ filament (Kohl et al., 1999); and the YL1/2 antibody, recognizing tyrosinated α-tubulin (Kilmartin et al., 1982). Subclass-specific secondary antibodies coupled to Alexa 488 and Cy3 (1/400; Jackson ImmunoResearch Laboratories, West Grove, PA) were used for double labeling. Samples were observed either with a DMR microscope (Leica, Wetzlar, Germany) and images captured with a CoolSnap HQ camera (Roper Scientific, Tucson, AZ) or with a DMI4000 microscope (Leica) and images acquired with a Retiga-SRV camera (QImaging, Surrey, Canada). Pictures were analyzed and cell parameters were measured using IPLab Spectrum 3.9 software (Scanalytics, Rockville, MD, and BD Biosciences, San Diego, CA) and ImageJ 1.38X software (National Institutes of Health, Bethesda, MD). Images were merged and superimposed using Adobe Photoshop (CS2; San Jose, CA).
RNA-FISH coupled to IFA
For detection of total poly(A+) RNA by FISH, parasites were harvested, washed, allowed to adhere to poly-l-lysine–coated microscope slides, and fixed with 8% paraformaldehyde in PBS for 20 min. Slides were incubated for 10 min with a 25 mM solution of NH4Cl and washed with PBS. Cells were simultaneously permeabilized and blocked for 1 h in 0.5% saponin (Sigma-Aldrich, St. Louis, MO) and 2% BSA in PBS. Prehybridization was performed for 2 h at room temperature in hybridization solution: 2% BSA, 5× Denhardt, 4× SSC (0.6 M NaCl and 0.06 M sodium citrate), 5% dextran sulfate, 35% deionized formamide (Sigma-Aldrich), 0.5 mg/ml wheat germ tRNA (Sigma-Aldrich), and 10 U/ml RNasin (Promega, Madison, WI). Hybridization was performed overnight in the dark at room temperature in a humid chamber in the presence of 1 ng/ml Alexa 488–conjugated oligo(dT)30 probe in hybridization solution. Samples were protected from light sources during the rest of the procedure. Slides were washed at 5-min intervals: once in 4× SSC plus 35% deionized formamide, once in 4× SSC, once in 2× SSC, and finally in PBS for 10 min. Before performing IFA as described in the preceding paragraph, blocking was performed during 30 min with 10% fetal calf serum in PBS.
Slides were analyzed using a Zeiss inverted microscope (Axiovert 200) equipped with an oil immersion objective (63× with a 1.4 numerical aperture) and a spinning disk confocal head (CSU22; Yokogawa, Sugarland, TX). Images were acquired using Volocity software with an EMCCD camera (C-9100; Hamamatsu, Hamamatsu, Japan). For presentation purposes only, the contrast of the images in Figure 6B was optimized by the same settings for every panel in Adobe Photoshop according to editorial policies.
Live video microscopy
Parasites found in the fly were obtained as described earlier in a drop of medium without serum directly on microscope slides. The pieces of tissue were removed and 5 μl of a solution of DAPI at 1 μg/ml added to the drop. Cells were covered by a coverslip and observed with a DMI4000 microscope (Leica). Videos were acquired using a COHU 460LI camera (Pieper, Schwerte, Germany) coupled to a DVD recorder (RDR-HX725; Sony, Tokyo, Japan). This is an analogue system in which neither exposure time nor other parameters can be adapted. The different fluorescence signals were acquired by changing the filters directly on the microscope. Movies were processed using the software Mpeg Streamclip 1.8 (Squared 5, www.squared5.com/) and ImageJ 1.38X and were assembled with iMovie (Apple, Cupertino, CA).
Western blot
Cells were washed in PBS and boiled in Laemmli sample buffer before SDS–PAGE separation, loading 2 μg (∼0.2 million cells) of total cell protein per lane. Proteins were transferred to polyvinylidene fluoride membranes, blocked overnight with 5% skimmed milk in PBS, and incubated with primary antibodies diluted in 1% milk and 0.1% Tween 20 in PBS for 1 h. The anti-ALBA3–specific antibody was diluted 1/2000, and the anti-ALBA4–specific one was diluted 1/500. To detect GFP, we used an anti-GFP antibody (Invitrogen) diluted 1/500. As loading controls, antibodies anti-PFR (L13D6; Kohl et al., 1999) diluted 1/50 and anti-aldolase (a kind gift of Paul Michels, Brussels, Belgium) diluted 1/1000 were used. Membrane washes were performed with 0.2% Tween 20 in PBS. Species-specific secondary antibodies coupled to horseradish peroxidase (GE Healthcare) were diluted 1/20,000 in 1% milk and 0.1% Tween 20 in PBS and incubated with the membranes for 1 h. Final detection was carried out by using an enhanced chemiluminescence kit according to manufacturer's instructions (Amersham, Piscataway, NJ).
RT-PCR
Total RNA was extracted from cells grown with or without tetracycline for the indicated period of time and purified using TRIzol (Invitrogen). DNA was eliminated by DNase treatment, and RNA purity was confirmed by conventional PCR. After primer calibration and determination of optimal conditions, semiquantitative RT-PCR was performed as described (Durand-Dubief et al., 2003). At least one primer was selected to be different from those used for cloning the knockdown construct. As independent control, we performed RT-PCR in parallel with described primers for TbODA7 (Tb11.01.5550; Duquesnoy et al., 2009).
Scanning electron microscopy of detergent-extracted cells
For detergent-extracted cytoskeleton preparations, trypanosome cells were treated with Triton X-100 as previously described (Absalon et al., 2008).
Supplementary Material
Acknowledgments
We thank Bernadette Tchicaya and Jerome Janelle from the UMR 177 IRD-CIRAD team, headed by Gérard Cuny (Montpellier, France), for providing tsetse flies. We are grateful to Jan Van Den Abbeele for providing the trypanosome AnTat 1.1 cell line. We acknowledge Mark Carrington and Paul Michels for providing anti-DHH1 and anti-aldolase antibodies, respectively. We are grateful to Christophe Machu and Pascal Roux at the imaging platform (PFID) of the Institut Pasteur for supervision and help with confocal microscopy. We thank Susanne Kramer and Mark Carrington for their very useful instructions in designing stress experiments and their analysis, as well as for providing various plasmids for endogenous tagging. We acknowledge Gerald Späth and Mark Carrington for critical reading of the manuscript and Isabel Roditi and Jan Mani for sharing data prior to publication. This work was funded by the Centre National de la Recherche Scientifique, the Institut Pasteur, and a MIE, Maladies Infectieuses et leur environnement grant from the Agence Nationale de la Recherche. I.S. is funded by an Aides à la Formation-Recherche fellowship from the Fonds National de la Recherche, Luxembourg, and B.R. by a Roux postdoctoral fellowship from the Institut Pasteur.
Abbreviations used:
- ALBA
acetylation lowers binding affinity
- BARP
brucei alanine rich protein
- DE
dividing epimastigote
- GST
glutathione S-transferase
- IFA
immunofluorescence analysis
- LE
long epimastigote
- MG
midgut
- MS
mesocyclic trypomastigote form
- MS-E
mesocyclic trypomastigote to epimastigote transition
- MT
metacyclic trypomastigote
- PC
procyclic trypomastigote form
- PFA
paraformaldehyde
- PFR
paraflagellar rod
- SE
short epimastigote
- SG
salivary glands
- SGE
salivary gland epimastigote
- UTR
untranslated region
- WT
wild-type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0511) on September 30, 2011.
REFERENCES
- Absalon S, Blisnick T, Bonhivers M, Kohl L, Cayet N, Toutirais G, Buisson J, Robinson D, Bastin P. Flagellum elongation is required for correct structure, orientation and function of the flagellar pocket in Trypanosoma brucei. J Cell Sci. 2008;121:3704–3716. doi: 10.1242/jcs.035626. [DOI] [PubMed] [Google Scholar]
- Absalon S, Kohl L, Branche C, Blisnick T, Toutirais G, Rusconi F, Cosson J, Bonhivers M, Robinson D, Bastin P. Basal body positioning is controlled by flagellum formation in Trypanosoma brucei. PLoS One. 2007;2:e437. doi: 10.1371/journal.pone.0000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy S, Gibson WC, Lehane MJ. Interactions between tsetse and trypanosomes with implications for the control of trypanosomiasis. Adv Parasitol. 2003;53:1–83. doi: 10.1016/s0065-308x(03)53002-0. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Anantharaman V. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4:R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Besteiro S, Barrett MP, Riviere L, Bringaud F. Energy generation in insect stages of Trypanosoma brucei: metabolism in flux. Trends Parasitol. 2005;21:185–191. doi: 10.1016/j.pt.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Bingle LE, Eastlake JL, Bailey M, Gibson WC. A novel GFP approach for the analysis of genetic exchange in trypanosomes allowing the in situ detection of mating events. Microbiology. 2001;147:3231–3240. doi: 10.1099/00221287-147-12-3231. [DOI] [PubMed] [Google Scholar]
- Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2009;375:148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- Brun R, Schönenberger Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Burkard G, Fragoso CM, Roditi I. Highly efficient stable transformation of bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol. 2007;153:220–223. doi: 10.1016/j.molbiopara.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Cassola A, De Gaudenzi JG, Frasch AC. Recruitment of mRNAs to cytoplasmic ribonucleoprotein granules in trypanosomes. Mol Microbiol. 2007;65:655–670. doi: 10.1111/j.1365-2958.2007.05833.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hung CH, Burderer T, Lee GS. Development of RNA interference revertants in Trypanosoma brucei cell lines generated with a double stranded RNA expression construct driven by two opposing promoters. Mol Biochem Parasitol. 2003;126:275–279. doi: 10.1016/s0166-6851(02)00276-1. [DOI] [PubMed] [Google Scholar]
- Daniels JP, Gull K, Wickstead B. Cell biology of the trypanosome genome. Microbiol Mol Biol Rev. 2010;74:552–569. doi: 10.1128/MMBR.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gaudenzi J, Frasch AC, Clayton C. RNA-binding domain proteins in kinetoplastids: a comparative analysis. Eukaryot Cell. 2005;4:2106–2114. doi: 10.1128/EC.4.12.2106-2114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza W, Attias M, Rodrigues JC. Particularities of mitochondrial structure in parasitic protists (Apicomplexa and Kinetoplastida) Int J Biochem Cell Biol. 2009;41:2069–2080. doi: 10.1016/j.biocel.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Dean S, Marchetti R, Kirk K, Matthews KR. A surface transporter family conveys the trypanosome differentiation signal. Nature. 2009;459:213–217. doi: 10.1038/nature07997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquesnoy P, et al. Loss-of-function mutations in the human ortholog of Chlamydomonas reinhardtii ODA7 disrupt dynein arm assembly and cause primary ciliary dyskinesia. Am J Hum Genet. 2009;85:890–896. doi: 10.1016/j.ajhg.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Kohl L, Bastin P. Efficiency and specificity of RNA interference generated by intra- and intermolecular double stranded RNA in Trypanosoma brucei. Mol Biochem Parasitol. 2003;129:11–21. doi: 10.1016/s0166-6851(03)00071-9. [DOI] [PubMed] [Google Scholar]
- Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18:2798–2811. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Moya SM, Estévez AM. Posttranscriptional control and the role of RNA-binding proteins in gene regulation in trypanosomatid protozoan parasites. Wiley Interdiscip Rev RNA. 2010;1:34–46. doi: 10.1002/wrna.6. [DOI] [PubMed] [Google Scholar]
- Fetzer CP, Hogan DJ, Lipps HJ. A PIWI homolog is one of the proteins expressed exclusively during macronuclear development in the ciliate Stylonychia lemnae. Nucleic Acids Res. 2002;30:4380–4386. doi: 10.1093/nar/gkf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Confalonieri F, Knapp S. Identification of the gene encoding archeal-specific DNA-binding proteins of the Sac10b family. Mol Microbiol. 1999;32:669–670. doi: 10.1046/j.1365-2958.1999.01366.x. [DOI] [PubMed] [Google Scholar]
- Furger A, Schurch N, Kurath U, Roditi I. Elements in the 3′ untranslated region of procyclin mRNA regulate expression in insect forms of Trypanosoma brucei by modulating RNA stability and translation. Mol Cell Biol. 1997;17:4372–4380. doi: 10.1128/mcb.17.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Xue H, Huang L. Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo. Mol Microbiol. 2003;50:1605–1615. doi: 10.1046/j.1365-2958.2003.03793.x. [DOI] [PubMed] [Google Scholar]
- Hammarton TC, Engstler M, Mottram JC. The Trypanosoma brucei cyclin, CYC2, is required for cell cycle progression through G1 phase and for maintenance of procyclic form cell morphology. J Biol Chem. 2004;279:24757–24764. doi: 10.1074/jbc.M401276200. [DOI] [PubMed] [Google Scholar]
- Hendriks EF, Robinson DR, Hinkins M, Matthews KR. A novel CCCH protein which modulates differentiation of Trypanosoma brucei to its procyclic form. EMBO J. 2001;20:6700–6711. doi: 10.1093/emboj/20.23.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz HR, Hartmann C, Huober K, Hug M, Clayton C. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 1997;25:3017–3026. doi: 10.1093/nar/25.15.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabayo JP, DeLoach JR, Spates GE, Holman GM, Kapatsa GM. Studies on the biochemical basis of the nutritional quality of tsetse fly diets. Comp Biochem Physiol A Comp Physiol. 1986;83:133–139. doi: 10.1016/0300-9629(86)90100-3. [DOI] [PubMed] [Google Scholar]
- Kelly S, et al. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol. 2007;154:103–109. doi: 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L, Robinson D, Bastin P. Novel roles for the flagellum in cell morphogenesis and cytokinesis of trypanosomes. EMBO J. 2003;22:5336–5346. doi: 10.1093/emboj/cdg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L, Sherwin T, Gull K. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J Eukaryot Microbiol. 1999;46:105–109. doi: 10.1111/j.1550-7408.1999.tb04592.x. [DOI] [PubMed] [Google Scholar]
- Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Queiroz R, Ellis L, Hoheisel JD, Clayton C, Carrington M. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J Cell Sci. 2010;123:699–711. doi: 10.1242/jcs.058511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J Cell Sci. 2008;121:3002–3014. doi: 10.1242/jcs.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Ray D, Barry JD, Easton C, Vickerman K. First tsetse fly transmission of the “AnTat” serodeme of Trypanosoma brucei. Ann Soc Belg Med Trop. 1977;57:369–381. [PubMed] [Google Scholar]
- Li Y, Li Z, Wang CC. Differentiation of Trypanosoma brucei may be stage non-specific and does not require progression of cell cycle. Mol Microbiol. 2003;49:251–265. doi: 10.1046/j.1365-2958.2003.03575.x. [DOI] [PubMed] [Google Scholar]
- Mair GR, Lasonder E, Garver LS, Franke-Fayard BM, Carret CK, Wiegant JC, Dirks RW, Dimopoulos G, Janse CJ, Waters AP. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 2010;6:e1000767. doi: 10.1371/journal.ppat.1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani J, Guttinger A, Schimanski B, Heller M, Acosta-Serrano A, Pescher P, Spath G, Roditi I. Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One. 2011;6:e22463. doi: 10.1371/journal.pone.0022463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Calvillo S, Vizuet-de-Rueda JC, Florencio-Martinez LE, Manning-Cela RG, Figueroa-Angulo EE. Gene expression in trypanosomatid parasites. J Biomed Biotechnol. 2010;2010:525241. doi: 10.1155/2010/525241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001037. doi: 10.1371/journal.ppat.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberle M, Balmer O, Brun R, Roditi I. Bottlenecks and the maintenance of minor genotypes during the life cycle of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001023. doi: 10.1371/journal.ppat.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterou A, Walrad P, Craddy P, Fenn K, Matthews K. Identification and stage-specific association with the translational apparatus of TbZFP3, a CCCH protein that promotes trypanosome life-cycle development. J Biol Chem. 2006;281:39002–39013. doi: 10.1074/jbc.M604280200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock L, Ferris V, Bailey M, Gibson W. Multiple effects of the lectin-inhibitory sugars D-glucosamine and N-acetyl-glucosamine on tsetse-trypanosome interactions. Parasitology. 2006;132:651–658. doi: 10.1017/S0031182005009571. [DOI] [PubMed] [Google Scholar]
- Ploubidou A, Robinson DR, Docherty RC, Ogbadoyi EO, Gull K. Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J Cell Sci. 1999;112:4641–4650. doi: 10.1242/jcs.112.24.4641. [DOI] [PubMed] [Google Scholar]
- Pradel LC, Bonhivers M, Landrein N, Robinson DR. NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J Cell Sci. 2006;119:1852–1863. doi: 10.1242/jcs.02900. [DOI] [PubMed] [Google Scholar]
- Redmond S, Vadivelu J, Field MC. RNAit: an automated Web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol Biochem Parasitol. 2003;128:115–118. doi: 10.1016/s0166-6851(03)00045-8. [DOI] [PubMed] [Google Scholar]
- Reuner B, Vassella E, Yutzy B, Boshart M. Cell density triggers slender to stumpy differentiation of Trypanosoma brucei bloodstream forms in culture. Mol Biochem Parasitol. 1997;90:269–280. doi: 10.1016/s0166-6851(97)00160-6. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K. Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J Cell Biol. 1995;128:1163–1172. doi: 10.1083/jcb.128.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotureau B, Subota I, Bastin P. Molecular bases of cytoskeleton plasticity during the Trypanosoma brucei parasite cycle. Cell Microbiol. 2011;13:705–716. doi: 10.1111/j.1462-5822.2010.01566.x. [DOI] [PubMed] [Google Scholar]
- Sharma R, Gluenz E, Peacock L, Gibson W, Gull K, Carrington M. The heart of darkness: growth and form of Trypanosoma brucei in the tsetse fly. Trends Parasitol. 2009;25:517–524. doi: 10.1016/j.pt.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Peacock L, Gluenz E, Gull K, Gibson W, Carrington M. Asymmetric cell division as a route to reduction in cell length and change in cell morphology in trypanosomes. Protist. 2008;159:137–151. doi: 10.1016/j.protis.2007.07.004. [DOI] [PubMed] [Google Scholar]
- She Q, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T, Gull K. Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell. 1989;57:211–221. doi: 10.1016/0092-8674(89)90959-8. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–1076. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Szoor B, Wilson J, McElhinney H, Tabernero L, Matthews KR. Protein tyrosine phosphatase TbPTP1: a molecular switch controlling life cycle differentiation in trypanosomes. J Cell Biol. 2006;175:293–303. doi: 10.1083/jcb.200605090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S, Studer E, Renggli CK, Roditi I. A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Mol Microbiol. 2007;63:218–228. doi: 10.1111/j.1365-2958.2006.05492.x. [DOI] [PubMed] [Google Scholar]
- Urwyler S, Vassella E, Van Den Abbeele J, Renggli CK, Blundell P, Barry JD, Roditi I. Expression of procyclin mRNAs during cyclical transmission of Trypanosoma brucei. PLoS Pathog. 2005;1:e22. doi: 10.1371/journal.ppat.0010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Abbeele J, Claes Y, van Bockstaele D, Le Ray D, Coosemans M. Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology. 1999;118((Pt 5),):469–478. doi: 10.1017/s0031182099004217. [DOI] [PubMed] [Google Scholar]
- Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci. 1997;110((Pt 21),):2661–2671. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- Vickerman K, Tetley L, Hendry KA, Turner CM. Biology of African trypanosomes in the tsetse fly. Biol Cell. 1988;64:109–119. doi: 10.1016/0248-4900(88)90070-6. [DOI] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Wright JR, Siegel TN, Cross GA. Histone H3 trimethylated at lysine 4 is enriched at probable transcription start sites in Trypanosoma brucei. Mol Biochem Parasitol. 2010;172:141–144. doi: 10.1016/j.molbiopara.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.