A spindle-independent, myosin II–containing domain causes asymmetric cortical extension during anaphase of asymmetrically dividing Drosophila neuroblasts. Heterotrimeric G-proteins regulate the activity of the myosin II domain, and their loss leads to symmetric expansion and daughter cell size.
Abstract
The cytokinetic cleavage furrow is typically positioned symmetrically relative to the cortical cell boundaries, but it can also be asymmetric. The mechanisms that control furrow site specification have been intensively studied, but how polar cortex movements influence ultimate furrow position remains poorly understood. We measured the position of the apical and the basal cortex in asymmetrically dividing Drosophila neuroblasts and observed preferential displacement of the apical cortex that becomes the larger daughter cell during anaphase, effectively shifting the cleavage furrow toward the smaller daughter cell. Asymmetric cortical extension is correlated with the presence of cortical myosin II, which is polarized in neuroblasts. Loss of myosin II asymmetry by perturbing heterotrimeric G-protein signaling results in symmetric extension and equal-sized daughter cells. We propose a model in which contraction-driven asymmetric polar extension of the neuroblast cortex during anaphase contributes to asymmetric furrow position and daughter cell size.
INTRODUCTION
During development, asymmetric cell division is used repeatedly to generate daughter cells that differ in size and fate (Knoblich, 2008). Daughter cell size asymmetry, which may be important for maintaining progenitor growth potential (Jorgensen and Tyers, 2004), can result from asymmetric positioning of the cleavage furrow (Glotzer, 2004). The site on the cortex where the cleavage furrow forms can be specified by the mitotic spindle (Oliferenko et al., 2009; von Dassow, 2009). For example, in the Caenorhabditis elegans zygote the spindle is displaced posteriorally at the end of metaphase and the furrow forms accordingly, leading to large anterior and small posterior daughter cells (Albertson, 1984; Keating and White, 1998; Glotzer, 2004). However, the position of the furrow depends not only on the site of furrow selection, but also on the relationship between the site of spindle specification and its position relative to the poles of the cell. Thus it is possible that an asymmetric furrow could result from specification of a furrow site at the center of the cell followed by asymmetric movement of the cortex at the cell poles. The morphology changes during mitosis can be dramatic, prompting us to explore the role of polar cortical movements in furrow position.
The study of furrow positioning has focused on how the site on the cortex that will become the cleavage furrow is selected by the mitotic spindle (Glotzer, 2004; von Dassow, 2009). The spindle pathway for furrow site selection is initiated at the central spindle by the centralspindlin complex consisting of the kinesin Pavarotti (ZEN-4 in C. elegans), the RACGAP50 Tumbleweed (CYK-4 in C. elegans), and the RhoGEF Pebble (ECT-2 in C. elegans). At the cell cortex, centralspindlin activates a narrow band of GTP-loaded Rho GTPase (Bement et al., 2006), ultimately leading to recruitment and activation of actomyosin to initiate cleavage furrow constriction. Astral microtubules can sharpen the site of furrow selection by inhibiting activation of Rho at the poles.
In addition to the spindle-directed equatorial constriction that occurs during cleavage furrowing, other morphological changes can happen late in mitosis. Symmetrically dividing cells, such as cultured S2 cells, round up at the beginning of mitosis but elongate at the poles late in anaphase (Hickson et al., 2006; Kunda et al., 2008; Rosenblatt, 2008; Figure 1A). The elongation that results from polar extension (i.e., outward displacement of the cortex) allows the spindle to expand into the polar regions as anaphase progresses (Rosenblatt et al., 2004; Hickson et al., 2006). The degree to which the polar cortex extends in cells that divide asymmetrically has been less characterized. Here, we use neuroblast asymmetric cell division as a model system for investigating the role of polar extension in cleavage furrow position and daughter cell size.
FIGURE 1:
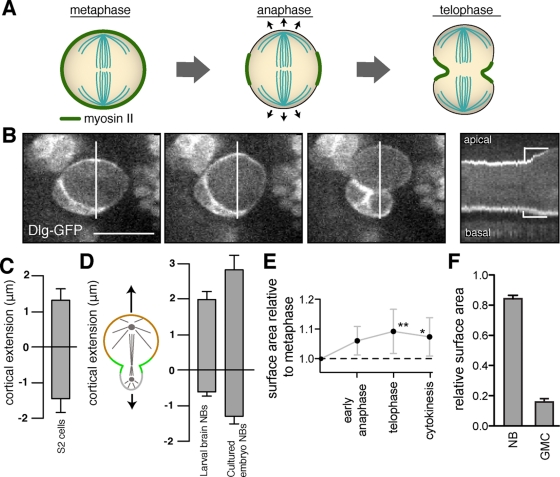
Drosophila neuroblasts undergo asymmetric polar elongation during anaphase. (A) Schematic of a symmetric division in which cortical extension is equal at both poles during anaphase. Myosin II is localized uniformly early in mitosis but becomes restricted to the equatorial region during anaphase. (B) Neuroblast cortical dynamics during mitosis using Dlg-GFP as a cortical marker. Selected frames from the movie are shown along with a kymograph of the entire division at 6-s intervals. The lines in the movie frames denote the section of the frame used for the kymograph. Cortical extension during anaphase is denoted by yellow brackets in the kymograph. The signal is enriched at the basal cortex because of contact with GMCs from previous divisions that also express GFP-Dlg (Supplemental Figure S1). (C) Mean anaphase polar extension in cultured Drosophila S2 cells transiently expressing Cherry-Zeus. The edge of the cell was marked at the point at which cytoplasmic fluorescence was no longer observed. Error bars, 1 SD. (D) Quantification of anaphase cortical extension in wild-type neuroblasts. The mean cortical extension from metaphase to the end of anaphase is shown for the apical (top) and basal (bottom) cortexes (NB, neuroblast). (E) The surface area of dividing neuroblasts measured using three-dimensional reconstruction normalized to that at the end of metaphase. The time points for measurements were early anaphase (completion of cortical extension), telophase (initiation of furrowing), and cytokinesis (completion of furrowing). (F) Mean relative surface areas of the daughter neuroblast (NB) and GMC that results from a neuroblast asymmetric cell division measured as in E at the completion of furrowing.
Drosophila neuroblasts are progenitors of the CNS, dividing to generate a larger apical cell that retains the neuroblast fate and a smaller basal ganglion mother cell (GMC) that assumes a differentiated fate (Doe, 2008; Knoblich, 2008). Neuroblasts divide rapidly, and daughter cell size asymmetry may be a mechanism for retaining sufficient resources to allow neuroblast self-renewal. The difference in fate of the two cells results from the polarization of fate determinants into separate apical and basal cortical domains that are precisely separated by the cleavage furrow (Knoblich, 2008). For example, the protein Miranda is localized to the basal cortex in metaphase and becomes segregated into the basal daughter cell as part of the machinery that confers GMC fate (Rolls et al., 2003; Lee et al., 2006; Atwood et al., 2007).
In addition to the spindle-directed pathway that controls targeting of furrow components such as centralspindlin to the equatorial cortex, neuroblasts possess a spindle-independent pathway that targets furrow components to the basal cortex (Cabernard et al., 2010). Shortly before the spindle directs recruitment to the equator, the spindle-independent pathway initiates contraction of the basal cortex (which becomes the GMC following abscission). A similar polar domain containing myosin II has recently been identified in C. elegans Q neuroblasts (Ou et al., 2010), which also divide to generate unequal-sized progeny, suggesting that the domain could be part of a common mechanism for daughter cell size asymmetry. Although the “asymmetric contraction” pathway is active in both Drosophila and C. elegans, and thus may be widely used in other systems, little is known about its mechanism. In particular, we know little about the cortical properties of the myosin-enriched (basal) and myosin-depleted (apical) neuroblast cortex.
RESULTS
Neuroblasts elongate asymmetrically during anaphase
We imaged larval brain neuroblasts expressing a green fluorescent protein (GFP) fusion to the cortical marker Discs Large (Dlg-GFP) or myosin II regulatory light chain (Spaghetti squash in Drosophila; Sqh-GFP) as a cortical marker to measure polar extension during mitosis (where “polar extension” refers to displacement of the cortex at the poles, without regard to the underlying mechanism). In contrast to the equal polar extension observed in symmetrically dividing cells (Rosenblatt et al., 2004; Hickson et al., 2006), neuroblast cortical extension is highly asymmetric (Figure 1, B and C). The neuroblast apical cortex (associated with the larger daughter cell that retains the neuroblast fate) extends significantly during anaphase, whereas the basal cortex (associated with the smaller differentiated cell) undergoes very little extension (Figure 1, B and D, and Supplemental Movie S1). On average, the apical pole extends over threefold more than the basal pole (2.0 vs. 0.6 μm). Thus polar extension in neuroblasts in intact tissue is highly asymmetric.
To ensure that asymmetric cortical extension was not a consequence of physical constraints imposed by the surrounding tissue, we measured cortical extension in cultured embryonic neuroblasts. These cells are dissociated from their surrounding tissue but continue to undergo asymmetric divisions (Siegrist and Doe, 2006). We found that cultured embryonic neuroblasts underwent asymmetric polar extension similar to their counterparts in the larval brain (Figure 1D and Supplemental Movie S2). We conclude that neuroblast asymmetric extension is an intrinsic property of the asymmetric cell division. Thus anaphase cortical extension differs significantly between symmetrically dividing cells and asymmetrically dividing neuroblasts. Whereas symmetrically dividing cells expand equally at both poles, neuroblast asymmetric divisions preferentially expand at the pole that becomes the larger daughter cell.
Asymmetric cortical extension is not caused solely by membrane synthesis
Neuroblast asymmetric cortical extension could occur by the preferential creation of new membrane at the apical surface. In this model the total surface area of the two daughter cells should be significantly larger than the surface area of the neuroblast before asymmetric cortical extension. To test this model, we measured the total surface area of the cell using three-dimensional reconstruction as a function of the cell cycle. We observed that the total surface area of the two daughter cells at the completion of cytokinesis is only ∼10% larger than the surface area of the metaphase neuroblast (Figure 1E). In contrast, the difference in surface area of the two daughter cells is much more extreme (Figure 1F). For example, the surface area of a representative neuroblast at metaphase was 923 μm2, and upon completion of cytokinesis the resulting neuroblast and GMC had surface areas of 828 and 163 μm2, respectively. Thus preferential membrane synthesis at the apical cortex is not sufficient to explain the asymmetric cortical extension that we observe, although it could contribute to the effect. Note that our measurements do not rule out a role for biased membrane flux.
Asymmetric cortical extension is independent of astral mictrotubules
To identify the cellular components responsible for cell-intrinsic asymmetric polar extension, we first focused on a possible role for the mitotic spindle, as it controls many of the morphological changes that occur during mitosis (von Dassow, 2009). At the poles, astral microtubules contact the cortex and could be responsible for controlling the asymmetric polar extension observed in neuroblasts. For example, asymmetric growth of the apical spindle could “push” the apical cortex outward. We examined whether astral mictrotubules are required for the difference in polar extension seen at the apical and basal neuroblast cortex by examining sas4 mutants, which lack astral microtubules (Basto et al., 2006). As shown in Figure 2A, the cortical extension in sas4 occurs predominantly at the apical pole, similar to wild-type neuroblasts. We conclude that astral microtubules are not required for asymmetric cortical extension.
FIGURE 2:
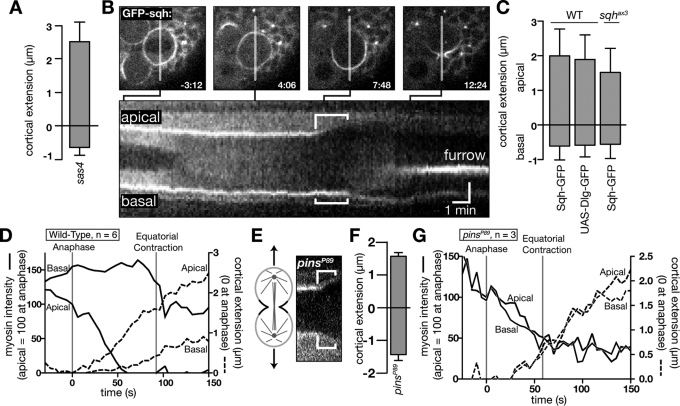
Basal myosin II is required for asymmetric polar elongation. (A) Mean anaphase polar extension in sas4 mutant neuroblasts. Error bars, 1 SD. (B) Kymograph of myosin II (Sqh-GFP) in a wild-type larval neuroblast imaged at 6-s intervals. Top, selected frames with time relative to nuclear envelope breakdown. A line marks the section of the frame used for the kymograph. Anaphase cortical extension is denoted by brackets in the kymograph. (C) Quantification of apical and basal cortical extension in sqhax3; sqh-GFP (larval neuroblasts), sqh-GFP, and worniu-Gal4; UAS-Dlg-GFP neuroblasts. Error bars, 1 SD. Anaphase onset was determined using spindle (Jupiter-cherry) or chromosome (His2A-mRFP) markers. (D) Time dependence of cortical myosin signal and cortical position for wild-type neuroblasts. Dashed lines indicate the cortical position at each pole relative to the position at anaphase start. Solid lines indicate the intensity at each pole relative to the apical cortical intensity at anaphase start (determined as in C). Equatorial contraction indicates the time point at which the initiation of furrow ingression was observed. (E) Kymograph of Sqh-GFP in pinsP89 mutant neuroblasts. Brackets denote polar extension during anaphase. (F) Quantification of anaphase cortical extension in pins mutant neuroblasts. Error bars, 1 SD. (G) Time dependence of cortical myosin signal and cortical position for pinsP89 mutant neuroblasts. Annotations as in D.
Cortical extension occurs at the onset of apical myosin II depletion
We examined the localization of the cortical factor myosin II to determine whether it could be important for polar extension in neuroblasts. In symmetrically dividing cells, myosin II is uniformly cortical in prophase but becomes confined to the equatorial region by late anaphase (Rosenblatt et al., 2004). Neuroblasts exhibit a similar pattern of myosin II localization, except that myosin II is retained at the basal cortex during anaphase along with Anillin and Pav, which are normally restricted to the furrowing region (Cabernard et al., 2010). In examining the localization of myosin II, we noticed a striking correlation with the loss of Sqh-GFP signal at the apical cortex and the onset of cortical extension, whereas myosin II remained at the basal cortex where extension was limited (Figure 2, B–D). Preferential apical clearing is not observed for the control proteins Dlg-GFP and Moe-RFP (Supplemental Figure S2). Furthermore, asymmetric cortical extension is not dependent on Sqh overexpression, as it also occurs in sqh mutants expressing Sqh-GFP (Figure 2C). The retention of myosin II on the basal cortex, along with the limited cortical extension at this location, prompted us to hypothesize that the basal domain containing furrow components inhibits cortical extension, limiting cortical extension to the apical cortex.
Asymmetric cortical extension requires a G-protein/Partner of Inscuteable–regulated basal furrow domain
As the onset of cortical extension is highly correlated with the loss of myosin II, we hypothesized that the “basal furrow domain” containing myosin II, Anillin, and Pavarotti, might be responsible for the preferential extension of the apical cortex. This domain is not regulated by the spindle but is instead controlled by cortical polarity factors such as Partner of Inscuteable (Pins; Cabernard et al., 2010). We examined cortical extension in symmetrically dividing pins mutants to determine how loss of the basal myosin domain influences extension. We confirmed that neuroblasts lacking Pins fail to form the basal myosin domain (Cabernard et al., 2010; Figure 2E). We find that these neuroblasts extend equally at both the apical and basal poles during anaphase (Figure 2, F and G). We conclude that Pins, which is required for the basal furrow domain, is also required for asymmetric polar extension. The known role of Pins in regulating asymmetric daughter cell size (Yu et al., 2000) suggests that these three processes—the basal furrow domain, asymmetric polar elongation, and daughter cell size asymmetry—are tightly coupled.
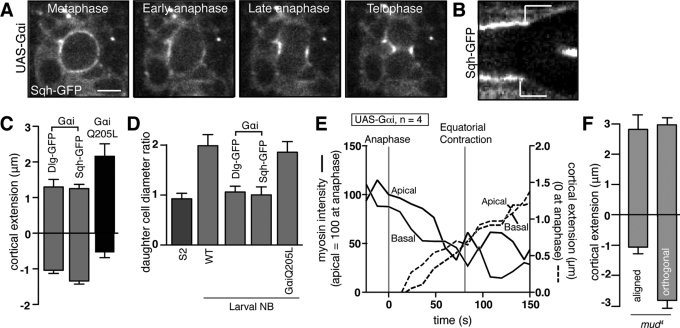
To further test the relationship between the basal furrow domain, asymmetric cortical elongation, and daughter cell size, we examined another genetic background in which neuroblasts divide symmetrically. Overexpression of the Pins-binding Gαi protein induces a high rate of symmetric divisions (Yu et al., 2000; Schaefer et al., 2001; Nipper et al., 2007). We imaged larval brain neuroblasts expressing Gαi under the control of the neuroblast-specific worniu-GAL4 driver to determine whether elevated Gαi levels concurrently alter the basal furrow domain and cortical extension prior to symmetric cell division. We found that the basal furrow domain failed to form in approximately half the neuroblasts, and neuroblasts lacking the domain expanded equally at both poles during anaphase (Figure 3, A–C and E, and Supplemental Movie S3). To determine whether the effect on the basal furrow domain and cortical extension is due to a greater pool of activated Gαi, we examined cortical extension in neuroblasts expressing the “activated” Q205L Gαi mutant (UAS-GαiQ205L driven with worniu-GAL4), which does not bind Pins or the Gβγ subunits (Schaefer et al., 2001). These neuroblasts divided normally, suggesting that the ability to bind Pins is required for Gαi-induced symmetric cell division (Figure 3, C and D, and Supplemental Movie S4). Thus heterotrimeric G-proteins and Pins are required for the basal furrow domain and asymmetric polar extension and daughter cell size.
FIGURE 3:
G-protein signaling regulates the basal furrow domain. (A) Myosin II (Sqh-GFP) localization in larval brain neuroblasts expressing Gαi using worniu-Gal4; UAS-Gαi. Images shown were taken 12 s apart. Scale bar, 5 μm. (B) Kymograph of Sqh-GFP signal across the poles from movie in A. Cortical extension is marked by the white lines. (C) Mean polar elongation for neuroblasts expressing Gαi or Gαi Q205L, a constitutively active variant that does not bind Gβγ or Pins. Cortical extension for Gαi is shown with two different cortical markers (Sqh-GFP or Dlg-GFP). Error bars, 1 SD. (D) Comparison of daughter cell size ratio for various cell types examined here. For asymmetrically dividing cells, this ratio was determined by dividing the diameter of the apical cell by the diameter of the basal cell. (E) Time dependence of cortical myosin signal and cortical position for neuroblasts expressing Gαi. Dashed lines indicate the cortical extension at each pole, whereas solid lines indicate the normalized intensity at each pole (as in Figure 2D). (F) Anaphase cortical extension for mud4 mutants where the spindle was aligned with basal myosin domain or orthogonal to it.
Asymmetric cortical extension requires alignment of the spindle with the basal furrow domain
We examined cortical extension in mud mutants to examine the consequence of decoupling the spindle-directed and basal furrows on polar extension. Mud orients the spindle with the polarity axis, and loss of mud function leads to randomization of spindle position (Guan et al., 2000; Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). In mud mutant neuroblasts with misaligned spindles, structures resembling polar bodies form at the basal furrow domain prior to equatorial contraction from the spindle-directed furrow (Cabernard et al., 2010). We examined cortical extension in this context to determine whether the basal furrow domain must be aligned with the spindle for asymmetric cortical extension. We found that although the cortex opposite the basal furrow domain begins to expand, subsequent spindle-induced equatorial furrowing overcomes this extension and ultimately leads to symmetric extension at the spindle poles and daughter cell size (Figure 3F). Thus coupling of both spindle-independent and spindle-dependent contractile pathways is required for asymmetric polar extension.
Asymmetric cortical extension does not require spindle-directed equatorial contraction
Our results suggest that the basal furrow domain is required for asymmetric cortical extension. The basal domain could bias cortical extension toward the apical pole in at least two possible ways. The basal domain could increase the rigidity of the basal cortex, thereby focusing cortical extension induced by spindle-induced equatorial contraction to the apical cortex. Alternatively, the basal domain itself could be responsible for extension at the apical cortex. To distinguish between these models, we treated neuroblasts with Colcemid to depolymerize microtubules, thereby inhibiting formation of the spindle and the spindle-directed furrow (Brinkley et al., 1967; Cabernard et al., 2010). To allow these neuroblasts treated with Colcemid to pass through the spindle checkpoint without a spindle, the Colcemid treatments were done in a rod background (Basto et al., 2000; Chan et al., 2000; Savoian et al., 2000; Cabernard et al., 2010). The presence of the basal furrow domain alone is sufficient for asymmetric cortical extension, as anaphase cortical extension is indistinguishable from wild-type neuroblasts (Figure 4, A and B). Thus we conclude that spindle-directed equatorial contraction is not required to produce asymmetric cortical extension.
FIGURE 4:
Asymmetric cortical extension does not require spindle-induced equatorial contraction. (A) Sequence of a Colcemid treated rod mutant neuroblast expressing Jupiter-cherry to ensure the absence of a spindle. Scale bar, 10 μm. (B) Quantification of cortical extension in Colcemid-treated rod mutants. Error bars, 1 SD. (C) Model for the role of the basal furrow domain in daughter cell size asymmetry.
DISCUSSION
Morphological changes that occur during mitosis involve both positive and negative signals that emanate from the mitotic spindle. For example, in C. elegans the first division yields unequal-sized daughter cells (Albertson, 1984; Keating and White, 1998; Glotzer, 2004). At the metaphase-to-anaphase transition the spindle midzone is displaced toward the posterior end of the zygote and is believed to positively influence furrow formations, whereas astral microtubules repress furrowing at the poles (Albertson, 1984; Keating and White, 1998; Glotzer, 2004). Thus asymmetric positioning of the spindle before anaphase ultimately leads to a displaced furrow and unequal-sized daughter cells. In neuroblasts, however, the spindle is symmetrically positioned before anaphase, suggesting that other mechanisms are responsible for neuroblast asymmetric division (Cai et al., 2003; Kaltschmidt et al., 2000; Siller and Doe, 2009). Recently, components normally restricted to the cleavage furrow were found to be localized to the neuroblast basal cortex (Cabernard et al., 2010). We found that this “basal furrow domain” restricts cortical extension that normally happens at both poles during the division of many small, symmetrically dividing cells. This restriction of cortical extension limits the size of the future basal cell while allowing the future apical cell to expand during anaphase (Figure 4C).
The contribution of asymmetric cortical extension to daughter cell size explains why asl and cnn mutants, which lack astral microtubules, divide asymmetrically although they have a symmetric spindle (Bonaccorsi et al., 1998; Basto et al., 2006). We argue that the asymmetric spindle poles (large apical pole, small basal pole) observed in wild-type neuroblasts late in the cell cycle (Fuse et al., 2003) are a consequence, not a cause, of asymmetric cortical extension. Lack of basal cortical extension prevents the spindle from growing at this pole, whereas it is free to grow at the apical pole. In fact, in S2 cells where cortical extension has been inhibited at both poles by RNA interference knockdown of Rho kinase (Hickson et al., 2006), spindles become bent and distorted during anaphase. We propose that asymmetric cortical extension in neuroblasts channels spindle growth into the apical pole, resulting in a highly symmetric spindle. The basal myosin domain does not passively halt the extension at the basal pole by acting against the forces produced by spindle-directed equatorial contraction, but instead is directly responsible for asymmetric cortical extension. Nevertheless, the basal and spindle-directed myosin domains must work together for asymmetric cortical extension to occur (Figure 3F).
How is the basal furrow domain regulated? Several components that function upstream of myosin II in the canonical furrow (e.g., Pav and Anillin) are also present in the basal furrow domain (Cabernard et al., 2010). Previous work showed that the polarity protein Pins is required to establish a basal myosin domain, whereas Gαi mutants do not perturb the domain (Cabernard et al., 2010). A linear model in which Gβγ promotes the basal myosin domain but is inhibited by Gαi, which is in turn inhibited by Pins, is consistent with these observations, and our data provide further support. Pins binds Gαi such that it is dissociated from Gβγ, so in cells that lack Pins, heterotrimeric G-protein complex assembly is favored and both Gα and Gβγ activity are reduced. As Gαi mutants have normal basal furrow domains (Cabernard et al., 2010), Gαi is not required for the basal furrow domain, suggesting that Gβγ is the relevant furrow domain regulator. The Gαi overexpression results presented here are also consistent with this model. We observed that expression of Gαi, which would inactivate any free Gβγ, leads to loss of the basal furrow domain. However, expression of constitutively active Gαi, which does not bind Gβγ but does bind downstream effectors, has no effect on the furrow domain. Thus we propose that Gβγ activity is essential for establishing the basal myosin domain, asymmetric cortical extension, and unequal daughter cell size. Future work will be directed toward rigorously testing this model and identifying the link between polarity proteins and myosin.
MATERIALS AND METHODS
Fly stocks
All mutant chromosomes were balanced over CyO actin:GFP, TM3 actin:GFP, Ser, e, or TM6B Tb. We used Oregon R as wild type and the following mutant chromosomes and fly strains: Sqh:GFP (Royou et al., 2002), worGal4 (Albertson and Doe, 2003), UAS-Cherry:Jupiter (Cabernard and Doe, 2009), worGal4 and UAS-Dlg:eGFP (Koh et al., 1999), UAS-Gαi and UAS-GαiQ205L (Schaefer et al., 2001; kindly provided by J.A. Knoblich), mud4 (Guan et al., 2000), pinsP89 (Yu et al., 2000), FRT82B Sas-4M (Basto et al., 2006), rodH4.8 (Basto et al., 2000), UAS-His2A-mRFP (Emery et al., 2005), UAS-moe-RFP (Schwabe et al., 2005), and UAS-Jupiter-GFP (Karpova et al., 2006). All crosses were performed at 25°C except UAS-Gαi crosses, which were performed at room temperature and transferred to 30°C approximately 8 h prior to imaging.
Cell culture
Drosophila S2 cells were maintained at room temperature in Schneider's medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (Sigma-Aldrich). Cells were transiently transfected with pMT-Zeus-Cherry with Effectene (Qiagen, Valencia, CA) according to the manufacturer's instructions, and expression was induced with 0.5 M copper sulfate 20 h prior to imaging. Immediately before imaging, cells were resuspended in Schneider's medium supplemented with 10% fetal bovine serum.
In vitro neuroblast culture
Primary cell cultures were made from embryos aged 6–8 h as previously described (Grosskortenhaus et al., 2005). They were then prepared for live imaging by resuspension in Chan and Gehrings balanced saline solution supplemented with 2% FBS.
Live imaging
Second or third larval brains were prepared for imaging as previously described (Siller et al., 2005). Five to nine Z steps were collected at 1- to 2-μm intervals every 6–12 s. Live imaging was performed using a spinning disk confocal microscope equipped with a Hamamatsu EM-CCD camera (Hamamatsu, Japan) using a 63 × 1.4 numerical aperture oil immersion objective. Pixel intensity measurements were performed using ImageJ (National Institutes of Health, Bethesda, MD). A linear photobleaching correction was added, using the intensity of a region outside the cell as a reference. Cortical extension was determined by measuring the position of the cell edges at the poles with a section from the middle of the cell as determined by examination of sections throughout the cell.
Colcemid treatment was performed on the strain +;worGal4, UAS-Cherry:Jupiter, Sqh:GFP; rodH4.8 (Cabernard et al., 2010), using a final concentration of 0.1 μm/ml, with live imaging beginning immediately after treatment.
Three-dimensional reconstructions for surface area analysis were done using the BoneJ plugin for ImageJ with a sampling value of 2 (Doube et al., 2010).
Supplementary Material
Acknowledgments
We thank Brad Nolen and members of the Prehoda lab for helpful comments. C.Q.D. is an Investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health Grant 068032 (K.E.P.) and the American Heart Association (C.C.).
Abbreviations used:
- Dlg
discs large
- GFP
green fluorescent protein
- GMC
ganglion mother cell
- moe
moesin
- mRFP
monomeric red fluorescent protein
- NB
neuroblast
- pins
partner of inscuteable
- rod
rough deal
- sqh
spaghetti squash (Drosophila myosin II)
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0173) on September 21, 2011.
REFERENCES
- Albertson DG. Formation of the first cleavage furrow in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5:166–170. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J Cell Sci. 2007;120:3200–3206. doi: 10.1242/jcs.014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Gomes R, Karess RE. Rough Deal and ZW10 are required for the metaphase checkpoint in Drosophila. Nat Cell Biol. 2000;2:939–943. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–86. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bement WM, Miller AL, von Dassow G. Rho GTPase activity zones and transient contractile arrays. Bioessays. 2006;28:983–993. doi: 10.1002/bies.20477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S, Giansanti MG, Gatti M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J Cell Biol. 1998;142:751–61. doi: 10.1083/jcb.142.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Neumüller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Stubblefield E, Hsu TC. The effect of Colcemid inhibition and reversal on the fine structure of the mitotic apparatus of Chinese hamster cells in vitro. J Ultrastruct Res. 1967;19(1) doi: 10.1016/s0022-5320(67)80057-1. [DOI] [PubMed] [Google Scholar]
- Cabernard C, Doe CQ. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Dev Cell. 2009;17:134–141. doi: 10.1016/j.devcel.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Cabernard C, Prehoda KE, Doe CQ. A spindle-independent cleavage furrow positioning pathway. Nature. 2010;467:91–94. doi: 10.1038/nature09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Yu F, Lin S, Chia W, Yang X. Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell. 2003;112:51–62. doi: 10.1016/s0092-8674(02)01170-4. [DOI] [PubMed] [Google Scholar]
- Chan GGT, Jablonskit SA, Starr DA, Goldberg ML, Yen TJ. Human ZW10 and Rod kinetochore proteins are novel components of the mitotic checkpoint. Nat Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Doube M, Kłosowski MM, Arganda-Carreras I, Cordelières F, Dougherty RP, Jackson J, Schmid B, Hutchinson JR, Shefelbine SJ. BoneJ: free and extensible bone image analysis in ImageJ. Bone. 2010;47:1076–1079. doi: 10.1016/j.bone.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitain MG, Knoblich JA. Asymmetric Rab 11 endosomes regulate delta cycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Fuse N, Hisata K, Katzen AL, Matsuzaki F. Heterotrimeric G proteins regulate daughter cell size asymmetry in Drosophila neuroblast divisions. Curr Biol. 2003;13:947–954. doi: 10.1016/s0960-9822(03)00334-8. [DOI] [PubMed] [Google Scholar]
- Glotzer M. Cleavage furrow positioning. J Cell Biol. 2004;164:347–351. doi: 10.1083/jcb.200310112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskortenhaus R, Pearson B, Marusich A, Doe CQ. The neuroblast gene expression “clock” is regulated by cytokinesis-dependent and cell cycle-independent mechanisms. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Guan Z, Prado A, Melzig J, Heisenberg M, Nash HA, Raabe T. Mushroom body defect, a gene involved in the control of neuroblast proliferation in Drosophila, encodes a coiled-coil protein. Proc Natl Acad Sci USA. 2000;97:8122–8127. doi: 10.1073/pnas.97.14.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GRX, Echard A, O'Farrell PH. Rho kinase controls cell shape changes during cytokinesis. Curr Biol. 2006;16:359–370. doi: 10.1016/j.cub.2005.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutterer A, Glotzer M, Mishima M. Clustering of centralspindlin is essential for its accumulation to the central spindle and the midbody. Curr Biol. 2009;19:2043–2049. doi: 10.1016/j.cub.2009.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–R1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt JA, Davidson CM, Brown NH, Brand AH. Rotation and asymmetry of the mitotic spindle direct asymmetric cell divisions in the developing central nervous system. Nat Cell Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- Karpova N, Bobinnec Y, Fouix S, Huitorel P, Debec A. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil Cytoskeleton. 2006;63:301–312. doi: 10.1002/cm.20124. [DOI] [PubMed] [Google Scholar]
- Keating HH, White JG. Centrosome dynamics in early embryos of Caenorhabditis elegans. J Cell Sci. 1998;111:3027–3033. doi: 10.1242/jcs.111.20.3027. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Koh YH, Popova E, Thomas U, Griffith LC, Budnik V. Regulation of DLG localization at synapses by CaMKII-dependent phosphorylation. Cell. 1999;98:353–363. doi: 10.1016/s0092-8674(00)81964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Pelling AE, Liu T, Baum B. Moesin controls the cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol. 2008;18:91–101. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ. Lgl, Pins, and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE. Galphai generates multiple Pins activation states to link cortical polarity and spindle orientation in Drosophila neuroblasts. Proc Natl Acad Sci USA. 2007;104:14306–14311. doi: 10.1073/pnas.0701812104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliferenko S, Chew TG, Balasubramanian MK. Genes Dev. 2009;23:660–674. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- Ou G, Stuurman N, D'Ambrosio M, Vale RD. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science. 2010;300:677–680. doi: 10.1126/science.1196112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J. Mitosis: moesin and the importance of being round. Curr Biol. 2008;18:91–101. doi: 10.1016/j.cub.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J, Cramer LP, Baum B, McGee KM. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- Royou A, Sullivan W, Karess R. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J Cell Biol. 2002;158:127–137. doi: 10.1083/jcb.200203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian MS, Goldberg ML, Reider CL. The rate of poleward chromosome movement is attenuated in Drosophila zw10 and rod mutants. Nat Cell Biol. 2000;2:948–952. doi: 10.1038/35046605. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Schwabe T, Bainton RJ, Fetter RD, Heberlein U, Gaul U. GPCR signaling is required for blood-brain barrier formation in Drosophila. Cell. 2005;123(1):133–144. doi: 10.1016/j.cell.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 2006;133:529–536. doi: 10.1242/dev.02211. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMa-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Siller KH, Serr M, Steward R, Hays TS, Doe CQ. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/Dynactin in spindle assembly and mitotic checkpoint control. Mol Biol Cell. 2005;16:5127–5140. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G. Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 2009;19:165–173. doi: 10.1016/j.tcb.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of Partner of Inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in Inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.