An in vivo structure–function analysis of the Drosophila homologue Mini spindles (Msps) identified novel domains that are necessary for the interplay between the conserved TOG domains and inter-TOG microtubule (MT) binding that underlies the ability of Msps to promote MT dynamic instability.
Abstract
Individual microtubules (MTs) exhibit dynamic instability, a behavior in which they cycle between phases of growth and shrinkage while the total amount of MT polymer remains constant. Dynamic instability is promoted by the conserved XMAP215/Dis1 family of microtubule-associated proteins (MAPs). In this study, we conducted an in vivo structure–function analysis of the Drosophila homologue Mini spindles (Msps). Msps exhibits EB1-dependent and spatially regulated MT localization, targeting to microtubule plus ends in the cell interior and decorating the lattice of growing and shrinking microtubules in the cell periphery. RNA interference rescue experiments revealed that the NH2-terminal four TOG domains of Msps function as paired units and were sufficient to promote microtubule dynamics and EB1 comet formation. We also identified TOG5 and novel inter-TOG linker motifs that are required for targeting Msps to the microtubule lattice. These novel microtubule contact sites are necessary for the interplay between the conserved TOG domains and inter-TOG MT binding that underlies the ability of Msps to promote MT dynamic instability.
INTRODUCTION
Microtubules are noncovalent polymers of the protein tubulin that perform essential transport and structural roles within eukaryotic cells (Lansbergen and Akhmanova, 2006; Rodriguez et al., 2003; Howard and Hyman, 2003). Microtubules exhibit a steady-state behavior, termed dynamic instability, in which individual microtubules randomly switch between phases of growth and shrinkage while the total level of microtubule polymer within the cell remains constant (Mitchison and Kirschner, 1984). Dynamic instability is crucial to microtubule function and allows cells to rapidly remodel the microtubule network in response to cell cycle cues or extracellular signals and to perform “search-and-capture” functions producing stable interactions between microtubules and organelles, kinetochores, or cortical binding sites. In many cell types, the slow-growing minus ends are attached to a microtubule-organizing center, whereas growth and shrinkage are caused by addition or loss of α- and β-tubulin heterodimers at the fast-growing plus ends. Microtubules prepared from purified tubulin do not exhibit the same parameters of dynamic instability as microtubules observed in extracts or in cells, underscoring the role of microtubule-associated proteins (MAPs) as key regulators of microtubule behavior (Kinoshita, 2001; Gardner et al., 2008; van der Vaart et al., 2009). MAPs can be categorized into two types: conventional MAPs, which bind along the microtubule lattice and stabilize, and microtubule tip–interacting proteins (+TIPs), which preferentially associate with plus ends to regulate growth, shrinkage, and attachment to other structures within the cell. Although many MAPs and +TIPs have been individually studied through genetic, biochemical, and cell biological approaches, it has become widely accepted that these molecules function in a complex hierarchy of interactions (Akhmanova and Steinmetz, 2008). For example, the +TIP EB1 has emerged as a key molecule necessary for recruitment of other +TIPs to growing plus ends (Vaughan, 2005). We lack a mechanistic understanding of how these interactions work in concert to regulate microtubule dynamics.
XMAP215 was the first protein identified that affected microtubule dynamics at their plus ends; since that time members of the XMAP215/Dis1 protein family have emerged as key regulators of dynamic instability (Kinoshita et al., 2002). XMAP215 was originally identified in Xenopus egg extracts as a factor that stimulated the polymerization rate of purified tubulin almost 10-fold in vitro (Gard and Kirschner, 1987). Conserved homologues have been found across all eukaryotic taxa, and, where tested functionally, they have all been implicated as promoters of microtubule dynamics and are required for proper assembly and function of meiotic and mitotic spindles (Cullen et al., 1999; Cullen and Ohkura, 2001; Gergely et al., 2003; Srayko et al., 2003; Gard et al., 2004). All XMAP215/Dis1 proteins exhibit a conserved domain structure characterized by the presence of NH2-terminal TOG domains. Tandem pairs of TOG domains bind to tubulin and can trigger robust microtubule nucleation in vitro when present as multiple pairs (Slep and Vale, 2007). In almost all multicellular eukaryotes, XMAP215 homologues possess an array of five TOG domains that can be classified into three types based on sequence similarity: TOG domains 1 and 3 are type A, TOG domains 2 and 4 are type B, and TOG domain 5 is classified as type C (Gard et al., 2004; Slep, 2009). This pattern suggests that animal XMAP215 homologues evolved by sequential duplications of their TOG domains, generating a pair of TOG domains that reduplicated to produce the modern-day tandem arrangement of five copies. Crystal structures of isolated TOG domains from several species (Al-Bassam et al., 2007; Slep and Vale, 2007) revealed that they are flat, “paddle”-shaped domains composed of six tandem HEAT repeats. HEAT repeats have a helix-loop-helix motif, with the highly conserved intrahelical loop regions serving as the proposed tubulin contact sites (Slep, 2009).
Although a role for XMAP215/Dis1 proteins as promoters of microtubule plus-end dynamics is well established in several systems, there is no unifying framework for how the domain structure of these proteins contributes to their localization and unique effects on microtubules in living cells. Two recent studies using high-resolution in vitro assays both demonstrated that XMAP215 acts as a microtubule polymerase, although they arrived at different conclusions as to the exact molecular mechanism of XMAP215's “enzymatic” action (Kerssemakers et al., 2006; Brouhard et al., 2008). In addition to the large body of work indicating that XMAP215/Dis1 family members affect microtubule growth, there is evidence that this family of proteins has a more complex role that also involves regulating microtubule shrinkage. Several in vitro studies have found potent effects on microtubule depolymerization in the presence of XMAP215 and other family members (van Breugel et al., 2003; Shirasu-Hiza et al., 2003; Brouhard et al., 2008). Perhaps most telling, depletion of Drosophila Mini spindles (Msps) in S2 cells resulted in nondynamic, paused microtubules (Brittle and Ohkura, 2005). On the basis of these data, the authors of that study suggested that XMAP215/Dis1 family members might act as “antipause” factors that exist to rapidly catalyze the transition from pause to either growth or shrinkage, thereby enhancing the dynamics of microtubules in addition to their ability to add monomers to the growing end.
In this study, we conducted an in vivo characterization of Drosophila Mini spindles (Msps) in cultured Drosophila S2 cells. We found that Msps exhibits EB1-dependent and spatially regulated localization to microtubules, localizing to microtubule plus ends in the cell interior and decorating the lattice of growing and shrinking microtubules in the cell periphery. RNA interference (RNAi) rescue experiments revealed that the NH2-terminal four TOG domains of Msps were sufficient to promote microtubule dynamics and EB1 comet formation and that the TOG domains function as paired units. We also identified TOG5 and novel inter-TOG linker motifs that are sufficient for binding to the microtubule lattice. These novel microtubule contact sites were necessary for Msps peripheral lattice association and allowed Msps to regulate dynamic instability. Thus Msps regulates microtubule dynamics through several mechanisms that involve its unique, multidomain structure.
RESULTS
Msps exhibits a differential localization to microtubule plus ends in the cell interior and to the microtubule lattice in the periphery of interphase S2 cells
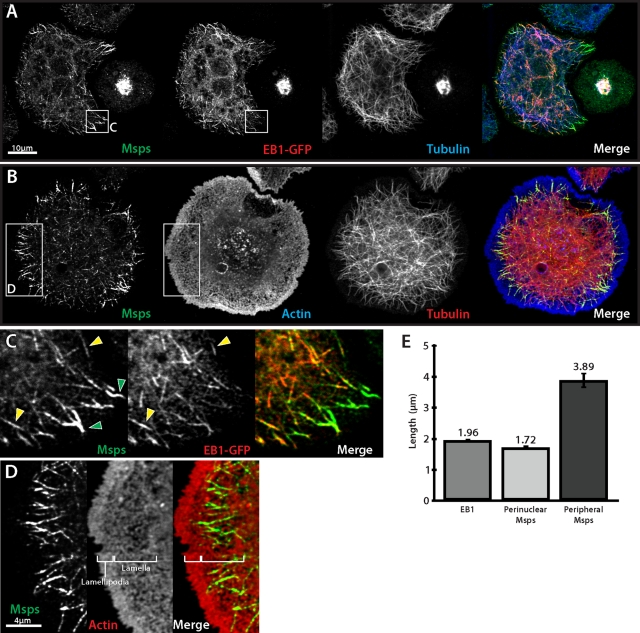
Brittle and Ohkura (2005) described the localization of Msps in S2 cells plated on concanavalin A (con A) and found it to be present on spindle microtubules and centrosomes in mitotic cells and as punctae along individual microtubules with prominent foci present at microtubule plus ends. We reexamined Msps localization using novel antibodies (Supplemental Figure S1A) raised against the second TOG domain (TOG2). Msps distribution in S2 cells was roughly similar in our hands; however, we observed more discrete patterns of localization along microtubules during interphase. Endogenous Msps accumulated at microtubule plus ends in the interior of the cell, labeling “comet”-like structures with a similar mean length to that of EB1 (Figure 1E). Triple labeling for Msps, microtubules, and EB1–green fluorescent protein (GFP; Figure 1A) revealed that Msps colocalized extensively with EB1 at these interior microtubule plus ends and confirmed prior observations that Msps behaves as a microtubule plus end–interacting protein (Lee et al., 2001). In the cellular periphery, however, in addition to plus-end localization, we found endogenous Msps decorated the microtubule lattice in a discontinuous pattern over several micrometers in length (Figure 1E). We also observed that some of the peripheral microtubule segments that exhibited Msps lattice association lacked EB1 (Figure 1C), suggesting that these microtubules had stopped growing just prior to fixation.
FIGURE 1:
Msps localizes to both microtubule plus ends and to the lattice of peripheral microtubules. (A) Interphase Drosophila S2 cell transfected with EB1::EB1-GFP (middle) and immunostained for Msps (left) and α-tubulin (right). (B) S2 cell immunostained for Msps (left), actin (middle), and α-tubulin (right). (C) Inset from A, Msps colocalizes with EB1 and peripheral microtubules that are EB1 negative. (D) Inset from B, Msps lattice accumulations (left) are coincident with actin-rich lamella (right). (E) Graph of microtubule decoration length for immunostained EB1 and interior-localized and peripheral Msps. Error bars, 95% CI.
The periphery of S2 cells plated on con A is made up of concentric actin-based subcompartments—the peripheral lamellipodium, the lamella, and the convergence zone—which are defined by local actin dynamics and are established by intracellular signaling cascades downstream of Rho-family GTPases (Iwasa and Mullins, 2007). Wittmann and Waterman-Storer (2005) demonstrated that the +TIP CLASP tracks with microtubule plus ends in the central cell body but associates with the microtubule lattice in the lamella and lamellipodium. To test the hypothesis that the compartment-based behavior of Msps resembled that of mammalian CLASP, we plated S2 cells on con A and stained them to visualize Msps, microtubules, and actin (Figure 1B). Whereas Msps staining rarely extended to the very peripheral lamellipodia in S2 cells, the pool of microtubule lattice–associated Msps coincided predominantly with the peripheral lamella (Figure 1D). This association did not depend on the actin cytoskeleton, as latrunculin treatment did not alter Msps localization to the microtubule lattice (unpublished data). These data indicate that Msps is able to interact with microtubules through at least two different modes—via an association with growing microtubule plus ends in a manner analogous to EB1, and to segments of the microtubule lattice similar to mammalian CLASP—and suggest that these two modes are spatially regulated in response to the localized cytoplasmic environment within the cell.
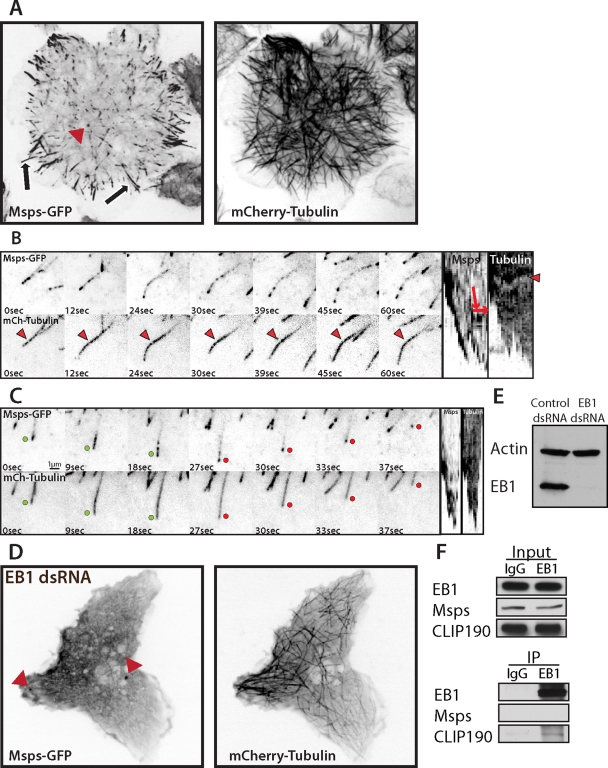
We performed simultaneous time-lapse imaging of Msps-GFP and mCherry-tubulin in S2 cells; Msps-GFP localization was identical to that of the endogenous protein observed by immunofluorescence (Figure 2A and Supplemental Movie S1). In the interior of the cell, Msps-GFP localized to growing microtubule plus ends. As microtubules grew into the periphery of the cell, Msps-GFP at the plus ends of individual microtubules frequently converted to a lattice-bound population that extended along the microtubule lattice in a discontinuous, punctate pattern (Figure 2A, arrows, and B). Peripheral, lattice-associated Msps-GFP punctae were dynamic and exhibited short, bidirectional movements along microtubules (Figure 2B and Supplemental Movie S2), often coalescing onto the end of a depolymerizing microtubule. In addition, foci of Msps-GFP often remained associated with the plus ends of microtubules after catastrophe and could track with the tips as they depolymerized (Figure 2C and Supplemental Movie S3), consistent with a recent in vitro report of family member, XMAP215 (Brouhard et al., 2008). In addition to microtubule localization, we also observed Msps-GFP associating with nonmotile cytoplasmic punctae (Figure 2A, arrowhead); double labeling with antibodies to Pericentrin-like protein (PLP) revealed these structures to be centrioles (unpublished data). From these observations we conclude that Msps is able to track microtubule plus ends during phases of growth or shrinkage in living cells and that it also interacts with the microtubule lattice in peripheral regions of the cell.
FIGURE 2:
Msps-GFP dynamics in S2 cells is EB1 dependent. (A) (Supplemental Movie S1) Interphase S2 cell expressing mCherry α-tubulin (right) and Msps-GFP (left) on microtubule plus ends in the interior, peripheral lattice accumulations (arrows), and centrioles (red arrowhead). (B) (Supplemental Movie S2) Msps-GFP dynamically moving along peripheral microtubules, represented as a kymograph (right). Red arrows on the kymograph (far right) indicate the direction of movement, and red arrowheads represent a fiduciary fluorescent mark along the microtubule that indicates microtubule translocation (kymograph, right). (C) (Supplemental Movie S3) Times series and kymograph of S2 cells expressing Msps-GFP (top) and mCherry α-tubulin (bottom). Growing ends are indicated by green dots and shrinking ends by red. (D) EB1-depleted S2 cell expressing Msps-GFP (left) and mCherry α-tubulin (right). (E) Western blot demonstrating EB1 knockdown from D over 6 d of RNAi. (F) Immunoprecipitation of EB1 and Western blot for EB1 (right, top), Msps (right, middle), and EB1-binding protein CLIP190 (right, bottom).
S2 cells exhibit two distinct populations of microtubule plus ends based on their origin and location between the cell interior and periphery
We next examined how Msps affects microtubule dynamics in living cells. We began by characterizing the behavior of microtubule plus ends in interphase S2 cells using either EB1-GFP or GFP–α-tubulin. EB1-GFP allowed us to specifically characterize the growth of microtubules in the cell interior, which is challenging due to the absence of an interphase microtubule-organizing center in Drosophila cells (Rogers et al., 2008), making the cell interior a complex meshwork of overlapping microtubules with indistinguishable plus and minus ends. The cell periphery, however, was amenable to analysis by GFP– α-tubulin, which allowed us to employ an automated microtubule-tracking algorithm (see Materials and Methods and Supplemental Figure S4; Zhang et al., 2011) to analyze the various parameters of microtubule dynamic instability.
Observation of EB1-GFP dynamics in S2 cells revealed that the majority of plus-end EB1 comets originated in the cell interior and translocated persistently through the cell with a velocity of 11.91 μm/min (±0.57, 95% confidence interval [CI]) toward the cell periphery, so that the majority of plus ends were oriented radially at the cell periphery (Supplemental Figure S1B, green tracks). When these interior EB1-GFP comets reached the cell cortex, they would disappear or stall momentarily before disappearing. We also observed a second population of EB1-GFP comets that specifically arose in the cell periphery, a region we defined to be coincidental with the cell lamella or roughly 3 μm from the cell edge (Iwasa and Mullins, 2007). These peripheral EB1-GFP comets likely represented either rescue events, growth of paused microtubules, or de novo nucleation of new microtubules originating in the cell periphery. Most surprisingly, these peripheral EB1-GFP comets polymerized at a slower velocity of 6.11 μm/min (±0.6, 95% CI; Supplemental Figure S1B, red tracks), suggesting that they perhaps experience different physical forces, such as actin retrograde flow, that influence their polymerization or that they have a different molecular complement at their plus ends than those in the cell interior.
In a series of complementary experiments, we observed peripheral microtubule dynamics by imaging GFP-tubulin. Microtubules formed a fairly stable polymer mass in the cell interior (Supplemental Figure S4A), whereas the peripheral microtubule ends were highly dynamic, best characterized by single growth and shrinkage events that spanned several micrometers with intermittent pauses and transitions to other dynamic states (Supplemental Figure S4C). The growth rate of these peripheral GFP-tubulin microtubules (6.134 μm/min; Table 1) also correlated with the slower population of peripheral EB1-GFP we had previously observed (Supplemental Figure S1B). Thus interphase S2 cells possess two populations of microtubule plus ends that exhibit different dynamics: 1) fast-growing, interior plus ends that grow persistently with little pause or catastrophe, and 2) slower, peripheral microtubules that are extremely dynamic, undergoing large growth and shrinkage events interspersed with pause.
TABLE 1:
Microtubule dynamics parameters rescued with NH2-terminal TOG-domain constructs.
| Events/minute | Velocity,b μm/min | |||||||
|---|---|---|---|---|---|---|---|---|
| Constructa | G → S | G → P | S→ G | S → P | P → G | P → S | Growth | Shortening |
| Control dsRNA (5, 2230) | 1.057 | 6.137 | 1.114 | 6.223 | 3.037 | 2.813 | 6.13 (±4.21, 3527) | 6.076 (±5.11, 2606) |
| Msps dsRNA (3, 1008) | 1.161 | 6.871 p = 0.004**c | 0.740 p = 0.039* | 6.728 p = 0.035* | 1.930 p = 0.007* | 4.057 p = 0.001** | 6.406 (±4.62, 897) | 6.727 p = 0.017* (±5.91, 1289) |
| Control dsRNA +TOG1-4 (3, 990) | 0.974 | 6.284 | 0.985 | 6.547 | 3.208 | 3.206 | 6.087 (±3.96, 1084) | 6.813 p = 0.0071* (±5.18, 1095) |
| Msps dsRNA +TOG1-4 (4, 440) | 0.945 | 7.0362 p = 0.0076* | 1.116 | 6.460 | 2.786 | 3.602 | 6.062 (±3.74, 471) | 6.229 (±4.63, 552) |
| Control dsRNA +TOG1-5 (6, 2011) | 1.08 | 6.757 p = 0.0018** | 1.127 | 6.800 p = 0.0037** | 3.114 | 3.116 | 6.191 (±4.0, 2361) | 7.551**** (±6.07, 2119) |
| Msps dsRNA +TOG1-5 (5, 2753) | 1.08 | 6.383 | 1.051 | 6.413 | 3.298 | 3.240 | 6.183 (±4.08, 3179) | 6.534 p = 0.043* (±5.38, 3350) |
aNumbers in parentheses are the number of cells analyzed and the analyzed events for each condition, respectively.
bStandard deviations and number of measurements are indicated in parentheses below each value.
cValues statistically different from control dsRNA where p < 0.05.
*p < 0.05; **p < 0.005; ****p < 0.00005. Statistical significance for transition rates was determined using a two-tailed permutation test with 10,000 resamples. For growth and shrinkage rates, a two-sample t test was used to determine statistical significance.
Msps and EB1 are mutually dependent for their normal dynamics at microtubule plus ends
We next tested the hypothesis that Msps interacts with other +TIPs for its localization and ability to regulate dynamic instability. We focused specifically on EB1 since both Msps and EB1 localize to microtubule plus ends throughout the cell cycle, and both proteins act to promote dynamic instability (Rogers et al., 2002; Brittle and Ohkura, 2005). Given these similarities, we explored functional interactions between Msps and EB1 by depleting one protein using RNAi and observing the other. Depletion of EB1 after 7 d of RNAi treatment (Figure 2E) completely abolished the accumulation of Msps-GFP at the plus end and lattice of microtubules (Figure 2D), and this loss of Msps-GFP from microtubules seemed to enhance its localization to centrioles during interphase (Figure 2D, arrowheads, and Supplemental Movie S4). In control experiments, immunofluorescence of the endogenous Msps in EB1 RNAi conditions exactly matched the localization of the exogenous transgene (Supplemental Figure S2, A and B). In the reciprocal experiment, depletion of endogenous Msps dramatically reduced the accumulation and pattern of endogenous EB1 (Supplemental Figure S2C) or EB1-GFP at microtubule plus ends in most cells (Figure 3, D and E) and completely eliminated it in a minority of the population (∼10% of cells; unpublished data). Under these conditions, EB1-GFP did not resemble typical EB1 “comets” observed in control cells (Figure 3, A and B), but instead formed spot-like punctae on microtubule ends in the cell interior, which exhibited short, back-and-forth displacements (Figure 3F) moving at a velocity of 3.72 μm/min (±0.21, 95% CI), as opposed to the persistent, track-like vectors seen in controls (Figure 3C), which moved at a rate of 11.91 μm/min (±0.57, 95% CI). Similar observations were recently reported in Drosophila sensory neurons (Stone et al., 2008), as well as with temperature-sensitive alleles of the Arabidopsis XMAP215 homologue, MOR1 (Kawamura and Wasteneys, 2008). To see whether these proteins interact, we performed coimmunoprecipitation using antibodies against EB1. Although we were able to pull down endogenous EB1 and a known EB1-binding protein, CLIP190, we were unable to coprecipitate endogenous Msps (Figure 2F), consistent with previous attempts to demonstrate binding in interphase extracts (Niethammer et al., 2007). This suggests that the mutual dependence for normal dynamics between EB1 and Msps may be due to synergistic effects on microtubule dynamics or may be mediated indirectly through another protein. Together, these data demonstrate that EB1 relies on Msps-dependent microtubule polymerization for normal dynamics, whereas proper localization of Msps is entirely dependent on EB1, although the two proteins may not interact directly.
FIGURE 3:
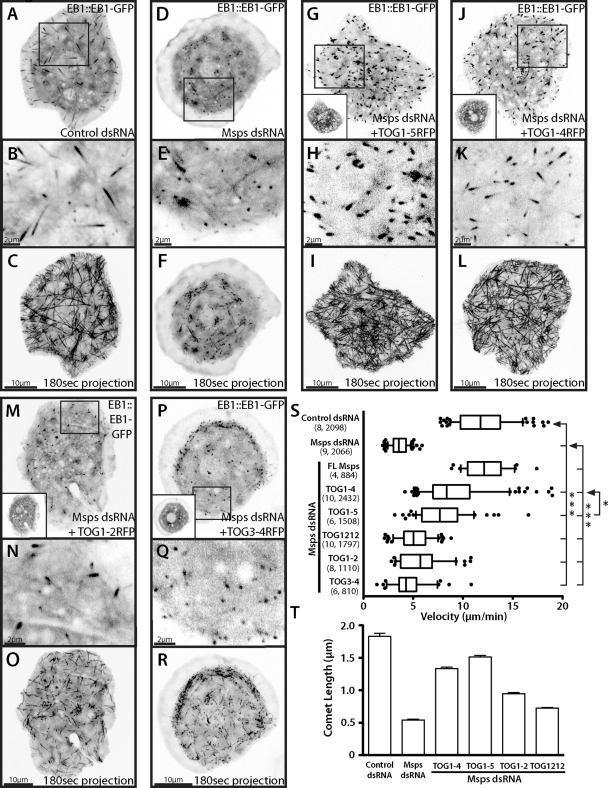
Expression of Msps TOG domains is sufficient to partially rescue Msps microtubule polymerization and EB1 dynamics. (A) (Supplemental Movie S4) Interphase S2 cell expressing EB1::EB1-GFP with wild-type localization (inset, B) and velocity visualized using a 180-s maximum projection (C). Localization and velocity are altered when cells are Msps dsRNA treated (D–F) but can be partially rescued by addition of TOG1-5 TagRFP (G–I), TOG1-4 TagRFP (J–L), TOG1-2 (M–O, and Supplemental Movie S5), and a very small amount by TOG3-4 (P–R). Insets in G, J, M, and P show TagRFP expression. (S) Distribution of EB1 velocities. Dots indicate velocities outside the 10th and 90th percentiles. Numbers in parentheses indicate cells analyzed and number of velocities, respectively. (T) Endogenous EB1 immunofluorescent comet length in cells treated with control or Msps dsRNA. Error bars, 95% CI. Asterisks indicate p value calculated using an unpaired t test between two conditions (*p < 0.05; ***p < 0.0005).
Msps TOG domains are sufficient to promote microtubule polymerization and EB1 dynamics
If Msps functions as a microtubule polymerase in living cells as it does in vitro, we hypothesized that the nondynamic microtubules produced by Msps depletion could not accommodate normal EB1 comet motility. Because TOG domains are sufficient to potently stimulate microtubule polymerization in vitro (Slep and Vale, 2007), we asked whether expression of the NH2-terminal domains of Msps was sufficient to rescue microtubule growth and, concomitantly, EB1 behavior. We depleted endogenous Msps using RNAi targeting either the 5′ untranslated region (5′ UTR) or the non–TOG domain–containing COOH-terminus (Supplemental Figure S1A) and then expressed versions of the gene tagged at its COOH-terminus with TagRFP and observed EB1-GFP dynamics specifically in the cell interior, where we could observe persistent and fast plus-end tracks in wild-type cells. Cells treated with Msps double-stranded (ds) RNA and transfected with TagRFP alone exactly recapitulated the aberrant EB1 tip localization pattern described earlier (unpublished data). Cells depleted of endogenous Msps and transfected with full-length Msps-TagRFP, however, exhibited EB1 comets of normal size that moved with an average velocity of 12.24 μm/min (±0.72, 95% CI), a rate that was not statistically different from EB1 movements in cells treated with control dsRNA (11.91 μm/min) (Figure 3S). We next prepared an expression construct embodying all five TOG domains (TOG1-5: residues 1–1428), transfected together with EB1-GFP, into cells depleted of endogenous Msps. Consistent with our hypothesis, TOG1-5 was able to rescue persistent EB1 plus-end tracking (Figure 3, G–I, and Supplemental Movie S5) to velocities of 8.12 μm/min (±0.74, 95% CI) (Figure 3S). To exclude the possibility that rescue of microtubule dynamics was due to overexpression of EB1-GFP, we performed the rescue without EB1-GFP and examined the localization of endogenous EB1 by immunofluorescence. We found that expression of TOG1-5 was sufficient to partially restore the localization pattern and length of endogenous EB1 on microtubule tips (Figure 3T), which has been shown to correlate with the measured velocity of EB1 comets in cells (Bieling et al., 2008). These data demonstrate that the TOG domains of Msps are sufficient to promote microtubule growth and indirectly support EB1 plus-end tracking in living cells.
TOG domains function in paired functional units during microtubule growth in vivo
We previously demonstrated that a tandem construct of Msps TOG1-2 formed a complex with tubulin by gel filtration chromatography but did not promote microtubule polymerization in vitro unless artificially homodimerized or expressed as an arrayed construct (TOG1212; Slep and Vale, 2007). These data suggested that the TOG domains of Msps functionally interact with tubulin as alternating paired arrays of A-type and B-type TOG domains. To test this hypothesis in vivo using the Msps RNAi/EB1-GFP rescue as an assay, we prepared an expression construct encoding the first four TOG domains (TOG1–4; residues 1–1080) fused to TagRFP and used this to replace endogenous Msps. As predicted by the model, TOG1-4 was also sufficient to partially rescue persistent EB1-GFP tip tracking to 9.42 μm/min (±0.66, 95% CI; Figure 3, J–L). This rescue of EB1 velocities was approximately equivalent to that of TOG1-5, suggesting that the first four TOG domains of Msps comprise the functional microtubule polymerase portion of the protein.
We next addressed whether individual pairs of TOG domains could functionally substitute for endogenous Msps using constructs encoding TOG1-2 (residues 1–498) or TOG3-4 (residues 583-–1080). It was striking that TOG1-2 exhibited a small rescue of EB1-GFP velocities within Msps-depleted cells (Figure 3, M–O, and Supplemental Movie S6). EB1-GFP comets in TOG1-2–expressing cells were smaller than in control RNAi or TOG1-4–expressing cells and exhibited slower velocities of 6.0 μm/min (±0.7, 95% CI). Although comet movement was less processive as compared with control or TOG1-4–rescued cells, EB1-GFP movement usually persisted in a single direction, albeit more slowly, with short phases of pause, unlike the saltatory, back and forth movement in untransfected Msps RNAi cells (Figure 3O and Supplemental Figure S3). This EB1 movement produced by TOG1-2 was never observed under any other condition. Expression of TOG3-4, however, failed to produce any qualitative rescue of EB1 velocity (Figure 3, P–R), although the measured velocity of 4.6 μm/min (±0.5, 95% CI) was statistically different from that of Msps-depleted cells. These data suggest that Msps requires two paired arrays of TOG domains to act cooperatively in vivo to promote microtubule polymerization. Our results also suggest that the Msps TOG domains are not functionally equivalent and may have differential affinities for tubulin. To further test this, we used a previously described construct (Slep and Vale, 2007) that has the first two TOG domains arrayed twice in a single polypeptide with the normal linkers preserved (linker1 and linker2) between the TOG domains (TOG1212 tRFP). Surprisingly, this construct was not able to significantly rescue EB1 velocities (5.2 μm/min ±0.4, 95% CI; Figure 3S) compared with TOG1-4, suggesting that the unique combination of TOG domains 1–4 is required to effectively act as a microtubule polymerase.
In addition to measuring EB1-GFP velocities, we also wanted to determine whether the same TOG-domain constructs could rescue other aspects of microtubule dynamics, specifically the highly dynamic peripheral microtubules we previously observed (Supplemental Figure S4C). Using our automated microtubule-tracking algorithm, we measured the dynamics of visible microtubule ends in the cell periphery after 7 d of either control or Msps dsRNA. After Msps depletion, microtubules in the periphery no longer exhibited the large growth and shrinkage events that characterized the wild-type peripheral microtubules. Although the average velocity of growth was unchanged between control and Msps dsRNA treatment (Table 1), as previously observed (Brittle and Ohkura, 2005), the amplitude of these growth and shrinkage events was severely diminished, with small spurts of growth and shrinkage interrupted by pause or a small rescue/catastrophe transition (Supplemental Figure S4C). In addition, the primary characteristic of this treatment was the significant increase in the frequencies of pause (Table 1), in agreement with a previous study (Brittle and Ohkura, 2005). These data are consistent with a role for Msps as an important enhancer of microtubule dynamicity. Expression of either TOG1-4 or TOG1-5 was able to partially restore most of the parameters of dynamic instability, although these conditions maintained higher frequencies of transitions from growth to pause, suggesting that they could suppress catastrophe but were not able to fully restore the large persistent growth events seen in control cells. Of interest, when these constructs were expressed in control or Msps dsRNA conditions, both TOG1-4 and TOG1-5 showed an increase in the microtubule shortening velocity over control cells, and in the case of TOG1-5/control dsRNA (7.55 μm/min), significantly greater than that of untransfected, Msps-depleted cells (6.894 μm/min). This suggested to us that in addition to promoting microtubule polymerization, these constructs also influence microtubule disassembly. The measured parameters of dynamic instability are summarized in Table 1. Overall these data reinforce the conclusion that the NH2-terminus of Msps is capable of partially rescuing both EB1 velocities and dynamic instability.
Msps and XMAP/Dis1 family members are considered to be essential regulators of the mitotic spindle. Depletion of Msps by RNAi results in a range of spindle morphologies that can generally be classified as monopolar, multipolar, or small, “mini” spindles (Cullen et al., 1999). Because Msps TOG constructs could rescue some aspects of microtubule velocity and dynamics, we wanted to assess whether any of these constructs could rescue mitotic defects caused by Msps RNAi. S2 cells were transfected with Msps fragments tagged at their COOH-terminus with either TagRFP or enhanced GFP and treated with Msps dsRNA and in addition Cdc27 dsRNA during days 3–7 to enrich for mitotic cells (Goshima et al., 2007). Depletion of Msps alone gave an approximately equal distribution of monopolar, multipolar, and mini spindles (Supplemental Figure S5, A, top left, and B), with roughly 10.5% wild-type spindles, compared with untreated cells, which contained 90.4% wild-type bipolar spindles (Figure 5A, bottom, and Supplemental S5B). This phenotype was rescued by expression of the full-length Msps-GFP (85.2%; Supplemental Figure S5B). When either TOG1-4 or TOG1-5 was introduced, the distribution of spindle architecture shifted toward normal bipolar spindles (40.2 and 42.5%) or large monopolar spindles (53.9 and 54.1%; Supplemental Figure S5A, top right). These data suggest that the polymerase activity of Msps TOG domains can rescue spindle length defects caused by Msps RNAi depletion but cannot rescue spindle bipolarity.
FIGURE 5:
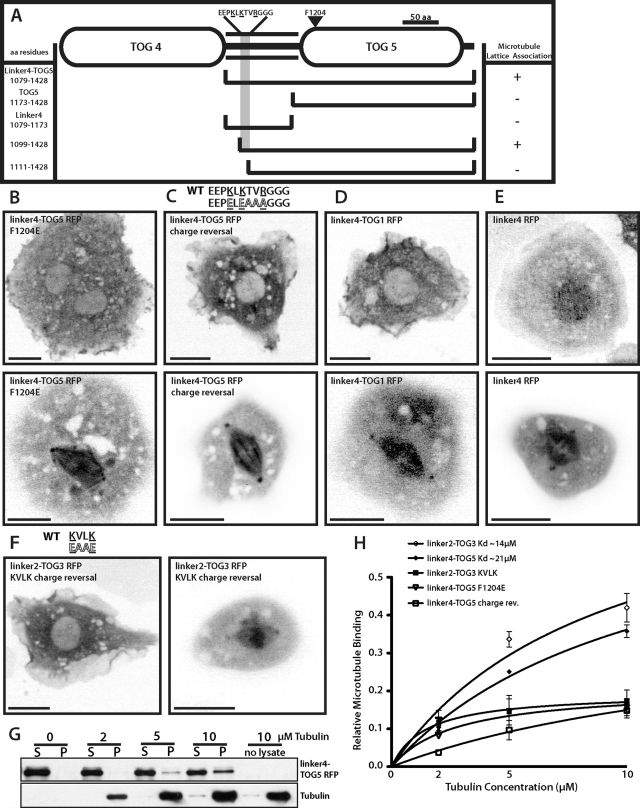
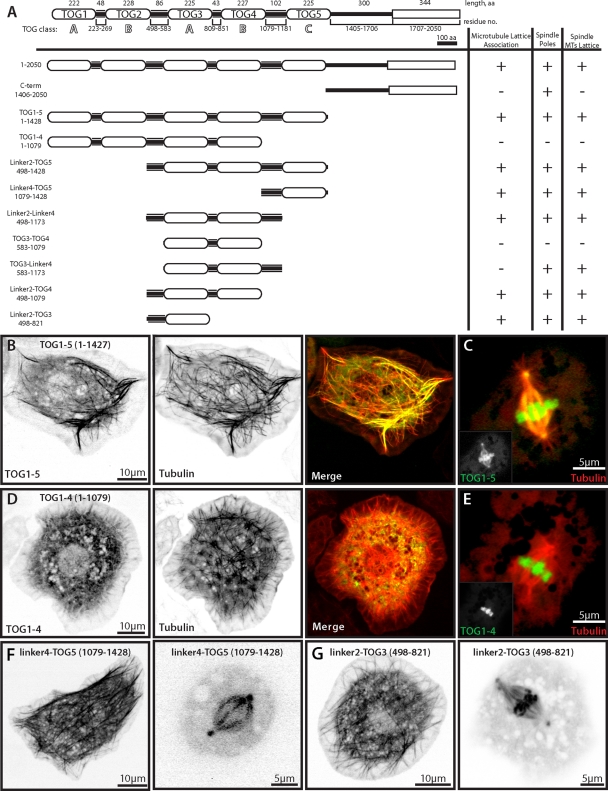
Msps has a novel microtubule lattice–binding site that spans the linker region between TOGs 4 and 5 and TOG5 itself. (A) Detailed domain structure of TOG4–TOG5 (821–1428). Interphase (top) and mitotic (bottom) localization of linker4-TOG5 F1204E TagRFP (B), linker4-TOG5 charge reversal TagRFP (residues 1102–1107) (C), chimeric linker4-TOG1 TagRFP (D), and linker4 TagRFP (residues 1079–1173) (E). (F) linker2-TOG3 TagRFP with KVLK charge-reversal mutation in interphase (left) and mitosis (right). Scale bars, 10 μm. (G) Representative Western blot from one in vitro microtubule cosedimentation assay replicate. (H) Binding curves summarizing in vitro cosedimentation assays for various Msps constructs. Dissociation constants are indicated where applicable. Error bars, 95% CI.
Msps has a novel microtubule lattice–binding site that spans the linker region between TOGs 4 and 5 and TOG5 itself
Having identified a function for the NH2-terminal TOG domains in regulating microtubule behavior, we wondered how other uncharacterized domains might also contribute to Msps' function in cells. Because full-length Msps exhibited dual modes of microtubule interaction, we wanted to identify the domain(s) that allowed the protein to interact with either the lattice or to the plus end. When transfected into S2 cells, TOG1-5 exhibited a very robust localization along the lengths of microtubules during interphase and mitosis (Figure 4, B and C), as well as spindle poles and condensed chromosomes in mitosis (Figure 4C). This pattern differed from the localization of full-length Msps, which was recruited to plus ends and to peripheral microtubule segments, as described earlier. Transfected cells exhibited relatively low cytoplasmic pools of TOG1-5–TagRFP, suggesting that this construct interacted with microtubules with a relatively high affinity. In contrast, TOG1-4, was predominantly soluble in transfected cells (Figure 4D). TOG1-4 did not localize to mitotic spindle microtubules or poles but did accumulate at condensed chromosomes in a manner similar to TOG1-5 (Figure 4E). These data led us to hypothesize that Msps possesses a microtubule lattice–binding site in the region of the TOG5 domain. To test this possibility, we generated a series of additional Msps truncations fused to TagRFP, cotransfected them into S2 cells together with GFP-tubulin, and scored them for their ability to localize to the interphase microtubule lattice and to the mitotic spindle and spindle poles (Figure 4A).
FIGURE 4:
Structure–function analysis of Mini spindles reveals two microtubule lattice–binding sites. (A) Domain structure of Drosophila Dis1/XMAP215 homologue Mini spindles. COOH-terminal TACC interaction domain is indicated as a rectangle. All Msps constructs were COOH-terminally tagged with TagRFP and transfected with GFP–α-tubulin to observe localization to distinct microtubule structures. (B) Msps TOG1-5 TagRFP (left) colocalizes with GFP–α-tubulin (middle) in interphase and spindle poles, spindle microtubules, and condensed chromosomes in mitosis (C). (D) Msps TOG1-4 TagRFP (left) does not colocalize with GFP–α-tubulin (middle) in interphase or mitosis but does localize to condensed chromosomes in mitosis (E). Insets represent single TagRFP channel from merge. (F) Linker4-TOG5 TagRFP in interphase (left) or mitotic (right) S2 cells. (G) Linker2-TOG3 in interphase (left) and mitotic (right) S2 cells.
Using the portion that comprised the difference between TOG1-4 and TOG1-5-TagRFP, we designed an expression construct starting from the linker region between TOG4 and TOG5 extending to TOG5 (linker4-TOG5, residues 1079–1428). When transfected into cells, linker4-TOG5-TagRFP associated strongly with microtubules, spindles, and poles (Figure 4F), indicating the presence of microtubule-binding activity in this region of Msps.
Multiple TOG domains are believed to have evolved through duplication events, such that linker2-TOG3 and linker4-TOG5 are similar class TOG domains and linkers of similar size with an overall positive charge. Therefore we postulated that linker2-TOG3 might be a second microtubule lattice–binding domain similar to that of linker4-TOG5. To assess this, we generated an equivalent construct that included the linker between TOG2 and TOG3 extending to the end of TOG3 (linker2-TOG3-TagRFP; residues 498–821). We found that linker2-TOG3 did exhibit microtubule- and spindle-binding activities, as well as the localization to condensed chromosomes as seen with TOG1-4 and TOG1-5 constructs (Figure 4G). The fact that this microtubule-binding activity is only apparent when the individual domain is expressed, as opposed to a larger region such as TOG1-4, suggests that it is either regulated or somehow masked in the context of full-length Msps or TOG1-4.
Finally, we examined the localization of the COOH-terminal domain (residues 1407–2050) of Msps fused to TagRFP. This construct did not associate with the lattice of interphase microtubules but was recruited to mitotic spindle poles (unpublished data), consistent with published observations that the COOH-terminus of XMAP215 targets it to the centrosome in Xenopus (Popov et al., 2001).
Msps possesses conserved microtubule-binding motifs within the inter-TOG linker regions
Our next objective was to define the minimal structural components in Msps linker4-TOG5 that were required for association with the microtubule lattice. Given the well-documented interactions between other TOG domains and tubulin, we tested the hypothesis that TOG5 would be sufficient to mediate this interaction. We subdivided the construct into linker4 (residues 1079–1173) or TOG5 (residues 1173–1427), fused these fragments to TagRFP, and expressed them in S2 cells (Figure 5A). Neither fragment associated with microtubules in vivo alone, but rather exhibited a diffuse localization throughout the cytoplasm (unpublished data), demonstrating that elements in both domains were required for microtubule lattice association. The orientation of the TOG domain and linker was also important, as TOG4-linker4 was not sufficient to bind microtubules in S2 cells (unpublished data).
To test the hypothesis that TOG5 interacts with the microtubule lattice in a similar manner to other TOG domains in vitro via its intra-HEAT repeat loops (Slep and Vale, 2007), we mutated phenylalanine to glutamic acid in linker4-TOG5. We previously found that a conserved nonpolar residue on the first HEAT repeat loop is absolutely required for tubulin binding in vitro (Slep and Vale, 2007). This mutation (F1204E) prevented linker4-TOG5 localization to microtubules in interphase cells (Figure 5B), suggesting that TOG5 is able to bind microtubules using the same surface used by other TOG domains to bind to tubulin. This point mutation did not prevent linker4-TOG5 from localizing to the spindle or spindle poles in mitotic cells, however, indicating that there is either an enhanced affinity or an indirect association with the spindle during mitosis (Figure 5B, bottom).
To identify the residues in linker4 that are necessary for microtubule association, we made a series of NH2-terminal truncations of the linker4-TOG5 fragment. A construct composed of residues 1099–1427 associated with microtubules, but a construct spanning residues 1111–1427 did not (Figure 5A), suggesting that the 12 residues (EEPKLKTVRGGG) spanning amino acids 1099–1111 were important for microtubule lattice binding. Because microtubule binding is often mediated through electrostatic interactions between positively charged residues on MAPs and the overall negatively charged surface of tubulin, we hypothesized that reversing the amino acid charge of residues within this necessary region of linker4 would eliminate microtubule association. Mutation of lysines and an arginine in this region of linker4-TOG5-TagRFP to either glutamic acid or alanine was sufficient to eliminate microtubule association in interphase (Figure 5B). This construct exhibited a weak, albeit very distinct, localization to the spindle and spindle poles as cells entered mitosis, similar to linker4-TOG5 F1204E. These data indicate that this positively charged patch of residues in the linker4-TOG5 sequence is required for its association with microtubules during interphase.
We also tested whether TOG5 was unique in its ability to cooperatively bind microtubules with linker4 or whether other TOG domains would be sufficient for recruitment to microtubules by preparing a chimeric protein consisting of linker4 fused to TOG1. Linker4-TOG1 did not associate with microtubules in interphase; however, it did localize to the spindle and spindle poles during mitosis (Figure 5D), suggesting that the linker is able to recruit Msps to the mitotic spindle. GFP fusions of either linker2 (unpublished data) or linker4 localized to the cytoplasm during interphase and to the spindle and spindle poles in dividing cells (Figure 5E). Taken together, these data identify a region in linker4 required for binding of linker4-TOG5 to interphase microtubules and further suggest that there is some specificity for TOG5 for this association. They also suggest that a second mechanism exists for recruiting Msps to the mitotic spindle via the inter-TOG linkers.
We next examined the sequences of linker2 or linker4 to determine whether there were additional microtubule-binding motifs present in these regions of the protein. Although many Dis1/XMAP215 family members retain a high degree of sequence similarity within TOG domains, the linker regions connecting them share less sequence similarity. Generally, the linker2 and linker4 among higher animal and plant XMAP215 family members share similar lengths (∼100 amino acids) and an overall higher percentage of positively charged residues relative to linkers 1 and 3. By using the previously described region within linker4 (1099–1111), we were able to distinguish three unique motifs that occurred from NH2-terminal to COOH-terminal within the linker2 or 4 region of Dis1/XMAP215 family members (Supplemental Figure S5C). The most conserved feature of these motifs was a sequence exactly matching or similar to KVLK within the third linker motif. Although the exact EEPKLKTVRGGG sequence was absent from Msps linker2, this domain does possess a stretch of residues rich in basic and nonpolar amino acids that also contains this KVLK motif. To test whether this region of homology could represent another cryptic microtubule-binding site, we mutated the KVLK motif to EAAE within linker2-TOG3 (residues 498–583) and found that this completely abrogated the interphase microtubule-binding activity but not mitotic spindle localization of linker2-TOG3 (Figure 5F). These data suggest the presence of a second microtubule interaction site in the second inter-TOG linker that contains at least one highly conserved binding motif.
To biochemically verify the Msps fragment binding to microtubules, we tested their ability to cosediment with microtubules in vitro. We translated fragments of Msps as TagRFP fusions in vitro, incubated the proteins with Taxol-stabilized microtubules, and centrifuged them through a glycerol cushion. The supernatant and microtubule pellet were then collected and immunoblotted with antibodies against TagRFP (Figure 5G). Both linker4-TOG5 and linker2-TOG3 cosedimented with microtubules; we were able to extrapolate an apparent Kd by varying the concentration of tubulin (Figure 5H). Linker2-TOG3 and linker4-TOG5 had similar apparent Kd of 14 and 21 μM, respectively. Linker-TOG constructs containing mutations that abrogated microtubule association in vivo only minimally cosedimented with tubulin at levels approaching background detection. These domains bind tubulin at lower affinities than what was been observed for other full-length XMAP/Dis1 family members (Spittle et al., 2000; Al-Bassam et al., 2010). This lower affinity may be due to cooperative interactions within the full-length protein between both microtubule-binding sites and the added interactions of TOGs 1, 2, and 4 with tubulin.
The linker regions are necessary for Msps peripheral lattice association and influence microtubule dynamics in the cell periphery
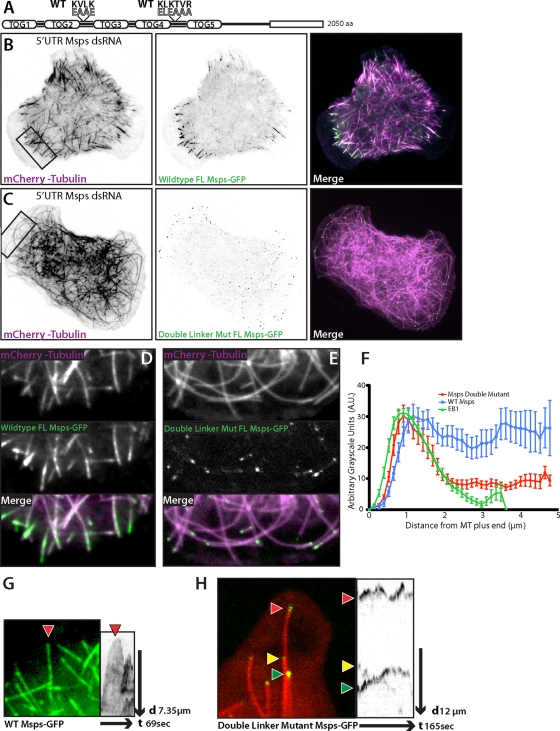
To determine the contributions of linker2-TOG3 and linker4-TOG5 to Msps dynamics and function in cells, we created full-length constructs that contained the linker2 KVLK charge reversal (residues 498–583), the linker4 charge reversal (residues 1099–1111), or both mutations (Figure 6A). The dynamics of either single mutant in cells depleted of endogenous Msps appeared qualitatively similar (unpublished data) to that of wild-type Msps (Figure 6B). This suggests that in context of the full-length molecule, the inter-TOG linker microtubule-binding sites act redundantly or can compensate for loss of the other. However, the double-mutant Msps (Double Mut Msps) displayed significantly altered localization and dynamics compared with wild-type Msps when expressed in Msps-depleted cells (Figure 6C). First, in the peripheral lamella, the microtubule lattice association was greatly reduced (Figure 6E and Supplemental Movie S7) compared with wild type (Figure 6D). Double Mut Msps–GFP displayed no discernible association along the length of microtubules and no longer decorated the lattice as a discontinuous patch of GFP fluorescence (Figure 6D). Instead the Double Mut Msps localized primarily to small, comet-like structures on the plus end of lamella microtubules (Figure 6E). Use of fluorescence intensity line scans showed that this reduction in the peripheral lattice binding was readily apparent compared with both wild-type Msps-GFP and EB1-GFP (Figure 6F). Second, expression of Double Mut Msps also affected the morphology and dynamics of microtubules in the cell periphery. In control cells, microtubules that enter the cell lamella are often highly dynamic, growing straight until they encounter the cell cortex at a perpendicular angle before undergoing a catastrophe that depolymerizes the microtubule out of the peripheral region (Figure 6, B and D). In cells expressing only Double Mut Msps, however, long microtubules were often observed to curl away from the periphery (Figure 6, C and E, and Supplemental Movie S7); we postulated this was caused by continued microtubule growth after contact with the cell cortex. Our hypothesis was confirmed by time-lapse images in which microtubule plus ends in the cell periphery grew and stalled at the cell edge or began to curve and grow parallel to the cell lamellipodia. These peripheral microtubules were decorated with small comet/dots of Double Mut Msps and seemed to exhibit shorter durations of depolymerization before recovering and growing again (Figure 6H and Supplemental Movie S8) compared with wild-type Msps (Figure 6G and Supplemental Movie S8).
FIGURE 6:
Mutation to the linker regions of Msps abrogates interaction with the lattice of peripheral microtubules. (A) Cartoon schematic of full-length double-mutant Msps (Double Mut Msps) that contains the linker2 KVLK charge reversal and linker4 charge reversal. (B) Interphase S2 cell depleted of endogenous Msps using 5′ UTR dsRNA, expressing wild-type Msps-GFP (B) (Supplemental Movie S6) or Double Mut Msps-GFP (C). (D–E) Inset of B (left) and C (right) with Msps-GFP (top left) or Msps Double Linker Mut (top right), mCherry-Tubulin (middle), and the merge (bottom). (F) Line scans across plus ends of S2 cells expressing EB1::EB1-GFP, full-length wild-type Msps-GFP, or full-length double-mutant Msps-GFP (Double Mut Msps). Line scans for Msps are of peripheral microtubules at equal exposures and time frames; error bars, SD. (G) (Supplemental Movie S7) Peripheral microtubule decorated with wild-type Msps-GFP and the associated kymograph. (H) (Supplemental Movie S8) Peripheral microtubule end with Double Mut Msps-GFP and kymograph.
To examine the dynamics of these peripheral microtubules, we first established a baseline of microtubule dynamics within cells expressing exogenous full-length (FL) Msps-GFP. Expression of FL Msps-GFP in addition to endogenous Msps (control-treated cells) closely replicated untransfected control cells, with the exception of increased growth and shrinkage rates (Table 2). Rescue of Msps 5′ UTR dsRNA-treated cells with FL Msps-GFP caused an increase in microtubule growth and shrinkage, suppressed catastrophe, and increased microtubule transitions out of pause. Expression of a single linker2-mutated, full-length Msps-GFP elevated growth and shrinkage rates but had no affect on the dynamic transitions of control-treated cells and was fully competent to rescue Msps 5′ UTR depletion.
TABLE 2:
Microtubule dynamics parameters with full-length Msps transgenes.
| Events/minute | Velocity,b μm/min | |||||||
|---|---|---|---|---|---|---|---|---|
| Constructa | G → S | G→ P | S →G | S → P | P → G | P → S | Growth | Shortening |
| Control dsRNA (5, 2230) | 1.057 | 6.137 | 1.114 | 6.223 | 3.037 | 2.813 | 6.134 (±4.21, 3527) | 6.077 (±5.11, 2606) |
| Control dsRNA + FL-Msps (3, 545) | 1.286 | 6.415 | 1.149 | 6.107 | 2.446 | 3.219 | 6.568 p = 0.0029**c (±5.19, 1308) | 7.283 ****c (±6.24, 1322) |
| Msps dsRNA (3, 1008) | 1.161 | 6.871 p = 0.004**c | 0.740 p = 0.039*c | 6.728 p = 0.007*c | 1.930 p = 0.035*c | 4.057 p = 0.001**c | 6.406 (±4.62, 897) | 6.727 p = 0.017*c (±5.91, 1289) |
| Control dsRNA + FL-Msps (3, 545) | 1.286 | 6.415 | 1.149 | 6.107 | 2.446 | 3.219 | 6.568 (±5.19, 1308) | 7.283 ****c (±6.24, 1322) |
| Msps dsRNA + FL-Msps (3, 1219) | 1.313 | 5.842 p = 0.011*d | 1.633 p = 0.0096*d | 5.577 p = 0.018*d | 3.275 p = 0.0023**d | 2.971 | 7.021 p = 0.011*d (±5.59, 3431) | 8.415 ****d (±7.48, 2803) |
| Control dsRNA + Linker2 Mut (3, 473) | 1.633 | 6.257 | 1.573 | 5.930 | 2.886 | 2.594 | 7.591 ****d (±7.28, 1045) | 9.092 ****d (±8.71, 1030) |
| Msps dsRNA + Linker2 Mut (3, 509) | 1.202 | 6.050 | 1.534 | 5.635 | 2.813 | 3.949 | 7.585 ****d (±6.248, 1183) | 9.063 ****d (±8.228, 1105) |
| Control dsRNA + Linker2&4 Mut (3, 665) | 1.266 | 6.233 | 1.238 | 5.976 | 2.306 | 2.373 p = 0.0076*d | 7.334 p = 0.0007**d (±6.40, 1414) | 9.795 ****d (±8.81, 1366) |
| Msps dsRNA + Linker2&4 Mut (3, 421) | 1.019 | 5.658 p = 0.0089*d | 1.2010 | 5.513 p = 0.0315*d | 1.751 p = 0.0159*d | 2.113 p = 0.0016**d | 5.516 ****d (±6.45, 1110) | 7.329 (±7.89, 949) |
aNumbers in parentheses are the number of cells analyzed and the analyzed events for each condition, respectively.
bStandard deviations and number of measurements are indicated in parentheses below each value.
cValues statistically different from control dsRNA where p < 0.05.
dValues statistically different from control dsRNA + FL-Msps where p < 0.05.
*p < 0.05; **p < 0.005; ****p < 0.00005. Statistical significance for transition rates was determined using a two-tailed permutation test with 10,000 resamples. For growth and shrinkage rates, a two-sample t test was used to determine statistical significance.
We next examined the effect of Double Mut Msps on peripheral microtubule dynamics. In control-treated cells, Double Mut Msps had little effect on microtubule dynamics, although it did result in elevated growth and shrinkage rates. However, when S2 cells were depleted of endogenous Msps by 5′ UTR dsRNA, Double Mut Msps had a notable effect on both the rates of growth and shrinkage and the dynamic transitions of microtubules. Double Mut Msps did not elevate growth rates in the way that other full-length constructs did and had a slightly negative effect on growth rates, unlike any other construct we had previously analyzed. More surprising was the affect on dynamic transitions. Rescue by Double Mut Msps-GFP resulted in a decrease in transitions from either growth or shrinkage into pause (G/S → P), as well as the inverse circumstance of paused microtubules transitioning into growing or shrinking microtubules (P → G/S; Table 2). If we broadly classify paused microtubules as nondynamic and growing or shrinking microtubules as dynamic, these data suggest that Double Mut Msps-GFP microtubules were more likely to stay locked in either dynamic or nondynamic states and were unable to transition normally between these states. This suggests that the interactions of Msps with the lattice of peripheral microtubules plays a key role in balancing dynamic instability in this population of plus ends.
DISCUSSION
Growing evidence from several model systems indicates that members of the XMAP215/Dis1 protein family enhance microtubule dynamic instability and mitotic spindle assembly (Kinoshita et al., 2002). These multidomain proteins are highly conserved across taxa, and there is particular interest in the human homologue, ch-TOG, as its overexpression has been documented in several cancer cell types (Charrasse et al., 1995, 1998). Despite the importance of these proteins, we know relatively little about their dynamics in living cells or about how their domain structure relates to their function in vivo. In this study, we used the Drosophila XMAP215 homologue Mini spindles (Msps) as a model to address these questions in living S2 cells. We found that Msps exhibits a complex and dynamic behavior in vivo. It localizes both to growing and shrinking microtubule plus ends. In the actin-rich cell periphery, Msps-GFP punctae exhibit a discontinuous and dynamic association with the microtubule lattice. Consistent with roles for both Msps and EB1 as key regulators of dynamic instability, we observed that depletion of either of these proteins leads to alteration of the other's dynamics at microtubule plus ends. Remarkably, microtubule dynamics and EB1 velocity could be partially restored by replacing endogenous Msps with the NH2-terminal four TOG domains TOG1-4. Our structure–function analysis also revealed the presence of novel microtubule lattice–binding domains, which include sequences in two of the inter-TOG linker domains. These linkers contain novel motifs responsible for this interaction and are important for the normal dynamics of Msps, as mutation of these motifs causes a loss of Msps from the lattice of peripheral microtubules and an alteration to the morphology and dynamics of the microtubule cytoskeleton. Thus our data demonstrate that Msps is able to interact with microtubules through at least two mechanisms and that this bimodal interaction is required for normal dynamic instability.
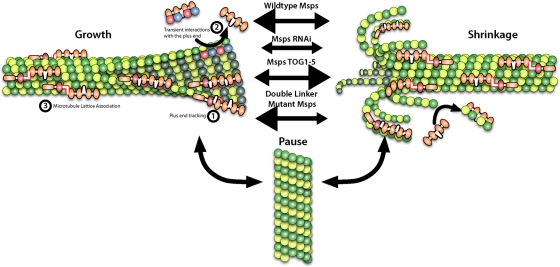
Our study, the first detailed examination of a Msps/Dis1 family member in a living animal cell, revealed that Msps exhibits a complex pattern of dynamics. Throughout the cell, Msps localizes to the plus ends of growing microtubules, consistent with other published descriptions of Msps behavior in early Drosophila embryos (Lee et al., 2001) and in vivo dynamics of homologues Stu2 (Saccharomyces cerevisiae; He et al., 2001; Kosco et al., 2001; van Breugel et al., 2003; Tanaka et al., 2005; Wolyniak et al., 2006), Alp14 (Schizosaccharomyces pombe; Sato et al., 2004; Garcia, 2001; Nakaseko et al., 2001), and AlpA (Aspergillus nidulans; Enke et al., 2007). Our observations in S2 cells are in line with family members acting as microtubule polymerases that promote the incorporation of free tubulin at the plus end. Msps also exhibits dynamics similar to the in vitro behavior of XMAP215 (Brouhard et al., 2008), as both proteins remain associated with microtubules during phases of growth and shrinkage. The biological significance of Msps association with shrinking microtubules is not understood; however, Stu2 and XMAP215 do destabilize microtubules under specific in vitro experimental conditions (Shirasu-Hiza et al., 2003; van Breugel et al., 2003; Brouhard et al., 2008). Persistent interaction with shrinking microtubules may reflect a physiologically important role for Msps during depolymerization in vivo. We favor a model based on this interaction with both growing and shrinking microtubules, first articulated by Shirasu-Hiza et al. (2003) and further elaborated by Brittle and Ohkura (2005), in which Msps acts not only as a polymerase but as an antipause factor that is capable of “catalyzing” the transition to either polymerization or depolymerization (Figure 7). Although it has primarily been shown to enhance the growth of microtubules, Msps is also necessary for their transition to disassembly in vitro and in vivo. These two seemingly opposing roles are mediated by 1) the TOG domains and plus-end localization, which enhance microtubule growth, and 2) microtubule lattice–binding sites within linker2-TOG3 and linker4-TOG5, which enhance the transition between dynamic and nondynamic states and also influence microtubule disassembly rates. Regulation and balance of these two domains maintain normal microtubule dynamics. In the future, it will be interesting to learn whether these in vivo dynamics also apply to XMAP215 or human ch-TOG, which remain largely uncharacterized in cells.
FIGURE 7:
Msps association with the microtubule lattice and plus end affect its function as both a microtubule polymerase and antipause factor. Msps is able to interact with microtubules through 1), plus-end association, 2), addition of tubulin subunits by transient interactions with microtubule end, and 3) association with the microtubule lattice. Msps is an antipause factor that enhances growth and shrinkage of microtubules, as RNAi of Msps leads to an increase in microtubule pause. Msps' ability to enhance either growth or shrinkage is due to its plus-end association and lattice-binding domain, respectively.
One of the central observations from this work was that the NH2-terminal TOG domains of Msps are the key determinants of the protein's activity in vivo. Several previous studies indicated a role for TOG domains as primary binding sites for tubulin in XMAP215/Dis1 proteins (Spittle et al., 2000; Popov et al., 2001; Gard et al., 2004; Al-Bassam et al., 2007; Slep and Vale, 2007). Biochemical analysis of the TOG domains from Msps revealed that TOG1-2 was the minimal construct that was able to bind tubulin (Slep and Vale, 2007). Moreover, a construct consisting of a tandem array of TOG1212 functioned as a potent microtubule nucleator in vitro, suggesting that multiple pairs of TOG domains act in concert to promote polymerization. We found that a fragment of Msps containing TOG1-4 was sufficient to rescue many aspects of microtubule dynamic instability in living S2 cells depleted of endogenous Msps, in terms of both interior EB1-GFP growth velocities and persistence and the frequencies of catastrophe and rescue. Consistent with the in vitro data, constructs embodying TOG1-2 imparted only a slight rescue of microtubule growth in S2 cells, whereas TOG3-4 did not. These data imply that individual pairs of TOG domains may contribute unequally to the activity of the protein but do function cooperatively to promote persistent microtubule growth. This is supported by recent work that found that TOG1 and 2, specifically, play a key role in XMAP215's polymerase activity in vitro (Widlund et al., 2011). The lack of rescue by TOG1212 also indicates that each pair of TOG domains is uniquely suited to fulfill the cooperative function of tubulin addition. Although TOG1-4 exerted a strong effect on tubulin incorporation and loss from the plus end as measured by GFP-tubulin and EB1-GFP growth rates, the TOG1-4 protein exhibited a predominantly cytoplasmic localization and did not accumulate at microtubule tips. These data suggest a mechanism in which TOG1-4 is able to associate with one or more tubulin heterodimers to transiently promote their addition or loss from the plus end (Figure 7, ①) without interacting in a processive manner (Figure 7, ②) or associating with the microtubule lattice (Figure 7, ③).
Another key observation is the identification of novel microtubule interaction sites spanning the TOG3 and TOG5 and their preceding inter-TOG linkers. These sites are necessary for association with the microtubule lattice and for the lattice-associated diffusive movements exhibited by full-length Msps. Our analyses identified the presence of conserved motifs in the inter-TOG regions that were enriched in basic amino acid residues, which we predict form an electrostatic interaction with the negative COOH-terminus of tubulin. The linkers between Msps' TOG domains are predicted to be disordered stretches without secondary structure that we found to cooperate with the highly structured TOG3 or TOG5 to mediate microtubule binding. The cooperation between ordered and disordered domains to mediate microtubule binding has become a common trend as more of the structural determinants of MAP–microtubule association have recently been elucidated (Guimaraes et al., 2008; Subramanian et al., 2010). These disordered regions seem to allow a diffusion-capable attachment to microtubules in addition to the ability to “titer” the interaction strength by charged residue addition or modification (i.e., phosphorylation; Kumar et al., 2009).
Beyond the structural basis for microtubule lattice association, we observed that Msps uses this mechanism in a spatially restricted manner in the actin-rich lamella of S2 cells. In other cell types, microtubules that enter this subcellular compartment exhibit decreased catastrophe rates due to the small GTPase Rac1 regulating MAPs that influence microtubule dynamics (Wittmann et al., 2003, 2004). In this regard, the spatial transition from plus-end binding to MAP lattice association has also been observed in mammalian epithelial cells for the +TIP CLASP (Wittmann and Waterman-Storer, 2005). In our hands, the Drosophila CLASP homologue Orbit/MAST, when tagged with GFP, displayed plus-end dynamics similar to that of EB1 and did not differentially localize in the periphery of the cell (J.C and S.R., unpublished observations). One possibility is that Msps functions in an orthologous manner to mammalian CLASP, which could be due to their shared structure as TOG domain–containing proteins. In any case, we speculate that this transition from tip tracking to lattice binding reflects a regulated change in the conformation of Msps. Given the observation that TOG1-5 decorates the microtubule lattice constitutively, we also hypothesize that the COOH-terminus of the protein is involved in this regulation and acts to “gate” the microtubule-binding activity of linker4-TOG5 in the full-length protein. Future studies will address the basis of this regulation.
Ablating these microtubule lattice association sites in the full-length molecule had a dramatic effect on the dynamics and morphology of the microtubule cytoskeleton. Although lattice association is commonly a stabilizing property of most MAPs, we were surprised to find that without its normal lattice association, Msps Double Mutant produced very stable microtubules that exhibited fewer transitions and continuous growth after microtubules encountered the cell cortex. In contrast, although Msps dsRNA also produces very stable microtubules, these microtubules are seldom able to persistently grow against the actin retrograde flow in the peripheral lamella. These data, in conjunction with the increased shrinkage rates we observed when cells expressed TOGs1-4 or TOG1-5, lead us to propose that these lattice association sites may influence the catalysis between dynamic states and the shrinkage rates of peripheral microtubules. Microtubules in peripheral regions of S2 cells exhibit increased dynamic instability as compared with the interior of the cell, and our data suggest a mechanism through which Msps might regulate these peripheral behaviors through its lattice association.
We hypothesize that this influence on dynamics could be caused by three non–mutually exclusive mechanisms. First, Msps lattice association could act to “strip” Msps from the plus end, potentially taking heterodimers with it and lowering the concentration of this polymerase from the microtubule tip. Second, Msps' lattice-binding domains could act to slightly perturb lateral interactions between heterodimers along the decorated protofilament. This cascade of small perturbations could act to prime the microtubule lattice for depolymerization several micrometers distal to the plus end. Finally, lattice-bound Msps could be acting in concert with other +TIPs, such as kinesin-13 depolymerases, to influence catastrophe and shrinkage.
Msps' dependence on EB1 for its plus-end localization in interphase has recently become more clear, based on two recent studies identifying novel EB1 adaptor proteins, SLAIN/Sentin. In mammalian cells, SLAIN binds both ch-TOG and EB1 and is required for ch-TOG plus-end tracking (van der Vaart et al., 2011). In Drosophila, a similar protein, Sentin, was also described. Although it has not yet been shown to bind Msps, RNAi depletion of Sentin alters the plus-end localization of Msps (Li et al., 2011). In our hands, depletion of EB1 removed Msps from both the plus end and lattice of microtubules. Although we do know the mechanism for how Msps is lost from both plus end and lattice, this suggests that the ability of full-length Msps to recognize the microtubule lattice is in some way tied to its plus-end association. One possibility could be a whole-molecule conformational change that occurs when Msps interacts with EB1 at the growing end, which might be required to license Msps for association along the microtubule lattice. This most likely is concurrent with some other spatially regulated control that gives Msps a bimodal function between the cell interior and the periphery. EB1's plus-end localization does not require Msps, as a population of EB1 remains on microtubule tips following Msps depletion; however, its dynamics is drastically altered. Rescue of EB1 velocities using the TOG-domain region of Msps suggests that EB1 relies on the polymerase activity of Msps for its normal dynamics. This also seems true since Msps fragments that rescue EB1-GFP comet formation do not seem to localize to the microtubule plus end, although we cannot rule out the possibility that there may be transient pools of TOG-domain fragments at the plus end. Instead, we favor a model in which these TOG-domain fragments act en masse to chaperone tubulin onto the plus end, subtly influencing the on and off rates of heterodimer addition. This creates a plus-end structure with sufficient binding sites to support more normal EB1 comets.
On the basis of our data, Msps exhibits at least two modes of interaction with microtubules, both of which are essential to promote normal parameters of dynamic instability. We envision a cycle of interactions that begins upon association between soluble Msps and one or more tubulin heterodimers. The Msps–tubulin complex then recognizes the microtubule plus end and associates transiently. The molecular basis of this recognition is unknown but might reflect a conformation of tubulin at the plus end (e.g., a growing sheet), a chemical signature (e.g., a cap of GTP tubulin), the presence of another +TIP such as EB1, or some combination of these. On binding, Msps delivers its tubulin “cargo” to promote polymerization. In the cell interior, Msps then dissociates, thus behaving as a typical +TIP. In the cell periphery, however, following tubulin delivery, Msps receives a signal that causes it to engage its lattice-binding activity and to diffuse along the microtubule surface. Msps also associates with microtubule plus ends upon transition to catastrophe, perhaps working cooperatively with destabilizing factors such as kinesin-13 proteins and stathmin/OP18.
MATERIALS AND METHODS
Cell culture and RNA interference
Culture and RNAi of Drosophila S2 cells were performed as previously described (Rogers and Rogers, 2008). For RNAi, the T7 promoter sequence was appended to gene-specific primer sequences to generate dsRNA using T7 RiboMAX in vitro transcription (Promega, Madison, WI). Primer sequences used for ∼1.5 kb of the Msps 5' UTR are as follows: forward, 5′-CGCAACGACGCTGTTGG-3′, and reverse, 5′-TCGTGTTTCGTACGCTAC-3′. Msps COOH-terminal dsRNA was made against amino acids 1752–1927, forward, 5′-GCCGAAGTTTACAGACCTGC-3′, and reverse, 5′-TGTACTTGTGAAATGGGGCA-3′. EB1 dsRNA primers were as follows: forward, 5′-GAGAATGGCTGTAAACGTCTACTCCACAAATGTG-3′, and reverse, 5′-GAG ATGCCCGTGCTGTTGGCACAGGCGTTTA-3′. Cdc27 dsRNA primers were made according to previously used methods (Goshima et al., 2007).
Immunofluorescence microscopy
Methodology for the fixation of S2 cells was adapted from previously described protocols (Rogers and Rogers, 2008). S2 cells were seeded on coverslips coated with con A for 1 h, washed briefly in BRB80 buffer (80 mM 1,4-piperazinediethanesulfonic acid, pH 6.9; 1 mM magnesium sulfate; 1 mM ethylene glycol tetraacetic acid), and fixed for 10 min in methanol prechilled to −80°C. Antibodies used in this study for immunofluorescence were as follows: Msps (described later), 1:1000; actin (MAB1501; Millipore, Billerica, MA), 1:500; α-tubulin (DM1α; Sigma-Aldrich, St. Louis, MO), 1:1000; and anti-EB1 (Rogers et al., 2002), 1:500. Secondary antibodies Cy2, Rhodamine Red, and Cy5 (Jackson ImmunoResearch Laboratories, West Grove, PA) were used at a final concentration of 1:300. Cells were imaged using either an Eclipse Ti-E (Nikon, Melville, NY) or a TCS SP5 X (Leica, Wetzlar, Germany) laser-scanning confocal microscope.
Immunoblotting
Samples for immunoblots were prepared as described (Rogers et al., 2009) and resolved on 7–12% SDS–PAGE gels. RNAi efficacy was assayed by immunoblot, and the protein loads were normalized using α-actin antibody (MAB1501). Percentage depletion was determined by densitometry using scanned film images with ImageJ (National Institutes of Health, Bethesda, MD).
Immunoprecipitation
S2 cells grown under normal culture conditions (see earlier discussion) were pelleted at 8000 rpm for 2 min and resuspended in lysis buffer (150 mM NaCl, 1 mM dithiothreitol, 50 mM Tris, 0.5% Triton X-100, 2.5 mM phenylmethylsulfonyl fluoride, 0.5 mM EDTA, and Complete EDTA-Free Protease Inhibitor Cocktail [Roche, Indianapolis, IN]). Lysates were precleared by centrifugation and then diluted twofold with lysis buffer. Samples were removed for input controls before being incubated with either mouse immunoglobulin G (Sigma-Aldrich), or anti-EB1 (Rogers et al., 2002) at 4°C for 2 h, followed by incubation with Sepharose–protein A beads for another 2 h. Sepharose–protein A bead immunocomplexes were washed three times with lysis buffer and then resuspended in Laemmli sample buffer. Samples were run on SDS–PAGE and immunoblotted using anti-EB1 (Rogers et al., 2002), anti-Msps (this work), or anti-CLIP190 (Dzhindzhev et al., 2005).
Antibody production
Antibodies were raised in rabbits (Proteintech Group, Chicago, IL) against Drosophila Mini spindles TOG2 domain (residues 267–500; Slep and Vale, 2007). Antibodies were further affinity purified using either recombinant TOG2 or TOG1-TOG2 (residues 1–505; Slep and Vale, 2007).
Microtubule cosedimentation assay
Microtubule cosedimentation was performed using a variation of previously published work (Spittle et al., 2000; Campbell and Slep, 2011). Briefly, fragments of Msps were subcloned using Topo-D pEntry vectors (Invitrogen, Carlsbad, CA) with a COOH-terminal TagRFP and then recombined into the pDEST17 bacterial expression vector with 5′ T7 promoter sites. These plasmids were then used as templates for in vitro transcription/translation reactions (TNT; Promega). After a 90-min incubation, the lysates were diluted threefold in BRB80 buffer and clarified for 30 min at 100,000 × g. A 60-μl amount of the clarified lysate (representing 100–200 ng translated protein according to the manufacturer's estimates) was then added to varying concentrations of Taxol-stabilized microtubules for 20 min. These reactions were then spun through a glycerol cushion for 30 min at 100,000 × g, and samples were taken from the supernatant (60 μl of supernatant added to 60 μl of 2× Laemmli buffer) and washed pellet (60 μl of fresh BRB80 with 60 μl of Laemmli buffer). Equal volumes were loaded onto a SDS–PAGE gel from all reactions and were immunoblotted for TagRFP at 1:1000 (Evrogen, Moscow, Russia) and then 1:750 anti–rabbit horseradish peroxidase. The resulting film was scanned, and densitometry was performed to determine the relative amount of translated protein in the supernatant and microtubule pellet. Relative binding affinity was plotted as the percentage of total protein pelleted, that is, the pellet's intensity divided by the added intensity of both supernatant and pellet. These were plotted against the concentration of microtubules, and a curve was fit to the points using nonlinear regression of one-site binding (GraphPad Prism, La Jolla, CA). On the basis of the curve formula, an apparent Kd was calculated where exactly half of the translated protein would be expected to pellet with microtubules.
Molecular biology and transfection
EB1::EB1-GFP was constructed by subcloning ∼1.5 kb of genomic DNA sequence 5′ of the EB1 gene in front of the 5′ start of EB1-GFP (Rogers et al., 2002). All fragments of Mini spindles were subcloned into a metallothionein promoter, pMT A vector backbone (Invitrogen) that contained a COOH-terminal fusion of TagRFP (a gift from Roger Tsien; Shaner et al., 2008). A previously described Msps construct (Lee et al., 2001) was used as a cDNA template, and all constructs were amplified by either Pfu or KOD polymerase (Novagen, Gibbstown, NJ). Full-length Msps-GFP was subcloned using the Gateway TopoD pEntr system (Invitrogen) into a final zeocin-selectable pIZ backbone (Invitrogen) that had both the metallothionein promoter and the Gateway (Invitrogen) LR recombination sites inserted into the multicloning site. Transfections were performed using the Amaxa Nucleofector II transfection system (Lonza, Basel, Switzerland) according to manufacturer's protocols. Constructs were induced 24 h after transfection with 40 μM copper sulfate for ∼12–18 h before imaging.
Live-cell microscopy
S2 cells were seeded onto Con A–coated glass-bottom dishes (MatTek, Ashland, MA) in Schneider's growth medium at room temperature (∼25°C) 1 h prior to imaging. Time-lapse images were acquired with a 100×, numerical aperture 1.45, Plan Apochromat objective using a Yokogawa (PerkinElmer, Waltham, MA) or VT-Hawk (VisiTech, Sunderland, United Kingdom) confocal system, captured with an Orca-ER or Orca-R2 camera (Hamamatsu, Hamamatsu, Japan), respectively. MetaMorph (Molecular Devices, Sunnyvale, CA) and VisiTech Vox software were used to control the respective confocal systems and acquire images. Images were acquired at 3-s intervals over periods of 3–10 min.
EB1-GFP comet tracking
EB1-GFP velocities were acquired from time-lapse movies (described earlier, Yokogawa spinning disk) using the ImageJ Manual Tracking plugin (Fabrice Cordelieres, Institut Curie, Orsay, France). Single comets were manually tracked for their full lifetime, and the mean comet velocity was calculated using the instantaneous frame-to-frame velocity. The collection of mean comet velocities were then plotted by a box-and-whisker plot, and statistical comparisons were made between two conditions using an unpaired t test (GraphPad Prism).
Automated microtubule tracking
Definition of the tracking region.
For each cell, we tracked microtubules in a hand-selected region-of-interest (ROI). It consists of an outer (the cortex; Supplemental Figure S4A) and an inner (to avoid tracking the crowded interior) boundary. In all calculations, the data from cells of the same treatment (control dsRNA, Msps dsRNA + TOG1-4, etc.) were pooled.
Definition of growth and shrinkage.
To define growth and shortening behavior, we calculated the angle α between the direction of the displacement of the microtubule tip and the direction of the microtubule itself. A value of cos α > 0.2 (α < ∼78°) indicated probable growth, and cos α < −0.2 (α > ∼101°) indicated probable shortening. For a microtubule to be counted as growing or shortening, we required that it displayed the same behavior (either growing or shortening) for at least two consecutive frames and that the overall displacement in a run of growth or shrinkage be at least 3 pixels.
Calculating state populations.
State populations were calculated as the total amount of time observed for a state (growing, shortening, or paused) divided by the total amount of time observed in the trajectories. Confidence intervals were estimated by a BCa-corrected bootstrap procedure that resampled the trajectories included in this statistic 1000 times.
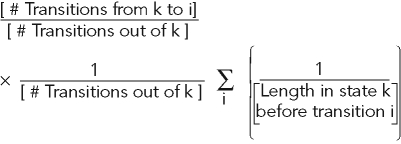
Calculating transition rates
To calculate transition rates, each frame of a microtubule trajectory was marked as grow, shorten, or pause. Grow and shorten states were defined as previously described. Pause was defined as neither growing nor shortening. The lifetimes of states that began and ended in the middle of a trajectory (not on a boundary), as well as the state to which they transitioned, were collected. The rate of transition between an initial state k (k = grow, shrink, or pause) and a final state l (l ≠ k) is given by
 |
We used a bootstrap approximation to construct 95% confidence intervals of each transition, with 1000 resamples of all lifetimes. Statistical significance was established using a two-sided permutation test with 10,000 resamples of the statistic, rcontrol,i –rtreatment,i, where control,i and rtreatment,i correspond to the rates calculated in resample i of the control vector and the treatment vector, respectively (e.g., Msps dsRNA, Msps dsRNA + TOG1-4, etc.).
Supplementary Material
Acknowledgments
We thank Mark Peifer and members of the Rogers, Peifer, and Slep labs for helpful advice and H. O. Lee for critically reading the manuscript. We also thank Hiro Ohkura, Jordan Raff, and Roger Tsien for sharing reagents. S.S. and A.M. are supported by National Institutes of Health Grant R01GM086536 to A.M. This work was supported by grants from the National Institutes of Health (RO1GM081645) and Beckman Foundation to S.L.R.
Abbreviations used:
- con A
concanavalin A
- MAP
microtubule-associated protein
- Msps
Mini spindles
- +TIP
microtubule plus end–tracking protein
- 5′ UTR
5′ untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0520) on September 30, 2011.
REFERENCES
- Akhmanova A, Steinmetz M. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008 doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal structure of a TOG domain: conserved features of XMAP215/Dis1-family TOG domains and implications for tubulin binding. Structure. 2007;15:355–362. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, Surrey T. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol. 2008;183:1223–1233. doi: 10.1083/jcb.200809190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittle AL, Ohkura H. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. EMBO J. 2005;24:1387–1396. doi: 10.1038/sj.emboj.7600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Slep KC. αβ-Tubulin and microtubule-binding assays. Methods Mol Biol. 2011;777:87–97. doi: 10.1007/978-1-61779-252-6_6. [DOI] [PubMed] [Google Scholar]
- Charrasse S, Mazel M, Taviaux S, Berta P, Chow T, Larroque C. Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. Eur J Biochem. 1995;234:406–413. doi: 10.1111/j.1432-1033.1995.406_b.x. [DOI] [PubMed] [Google Scholar]
- Charrasse S, Schroeder M, Gauthier-Rouviere C, Ango F, Cassimeris L, Gard DL, Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J Cell Sci. 1998;111(Pt 10):1371–1383. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- Cullen CF, Deák P, Glover DM, Ohkura H. mini spindles: a gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Ohkura H. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat Cell Biol. 2001;3:637–642. doi: 10.1038/35083025. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Rogers SL, Vale RD, Ohkura H. Distinct mechanisms govern the localisation of Drosophila CLIP-190 to unattached kinetochores and microtubule plus-ends. J Cell Sci. 2005;118:3781–3790. doi: 10.1242/jcs.02504. [DOI] [PubMed] [Google Scholar]
- Enke C, Zekert N, Veith D, Schaaf C, Konzack S, Fischer R. Aspergillus nidulans Dis1/XMAP215 protein AlpA localizes to spindle pole bodies and microtubule plus ends and contributes to growth directionality. Eukaryotic Cell. 2007;6:555–562. doi: 10.1128/EC.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Becker BE, Josh Romney S. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int Rev Cytol. 2004;239:179–272. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Hunt AJ, Goodson HV, Odde DJ. Microtubule assembly dynamics: new insights at the nanoscale. Curr Opin Cell Biol. 2008;20:64–70. doi: 10.1016/j.ceb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–341. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Howard J, Hyman AA. Dynamics and mechanics of the microtubule plus end. Nature. 2003;422:753–758. doi: 10.1038/nature01600. [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E, Wasteneys GO. MOR1, the Arabidopsis thaliana homologue of Xenopus MAP215, promotes rapid growth and shrinkage, and suppresses the pausing of microtubules in vivo. J Cell Sci. 2008;121:4114–4123. doi: 10.1242/jcs.039065. [DOI] [PubMed] [Google Scholar]
- Kerssemakers JWJ, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Kinoshita K. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–1343. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Habermann B, Hyman AA. XMAP215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 2002;12:267–273. doi: 10.1016/s0962-8924(02)02295-x. [DOI] [PubMed] [Google Scholar]
- Kosco KA, Pearson CG, Maddox PS, Wang PJ, Adams IR, Salmon ED, Bloom K, Huffaker TC. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol Biol Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Lyle KS, Gierke S, Matov A, Danuser G, Wittmann T. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J Cell Biol. 2009;184:895–908. doi: 10.1083/jcb.200901042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Akhmanova A. Microtubule plus end: a hub of cellular activities. Traffic. 2006;7:499–507. doi: 10.1111/j.1600-0854.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nature Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- Li W, Miki T, Watanabe T, Kakeno M, Sugiyama I, Kaibuchi K, Goshima G. EB1 promotes microtubule dynamics by recruiting Sentin in Drosophila cells. J Cell Biol. 2011;193:973–983. doi: 10.1083/jcb.201101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M. M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol. 2001;11:537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Kronja I, Kandels-Lewis S, Rybina S, Bastiaens P, Karsenti E. Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol. 2007;5:e29. doi: 10.1371/journal.pbio.0050029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov AV, Pozniakovsky A, Arnal I, Antony C, Ashford AJ, Kinoshita K, Tournebize R, Hyman AA, Karsenti E. XMAP215 regulates microtubule dynamics through two distinct domains. EMBO J. 2001;20:397–410. doi: 10.1093/emboj/20.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Peifer M, Rogers SL. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell. 2008;19:3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- Sato M, Vardy L, Angel Garcia M, Koonrugsa N, Toda T. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol Biol Cell. 2004;15:1609–1622. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu-Hiza M, Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. J Cell Biol. 2003;161:349–358. doi: 10.1083/jcb.200211095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC. The role of TOG domains in microtubule plus end dynamics. Biochem Soc Trans. 2009;37:1002–1006. doi: 10.1042/BST0371002. [DOI] [PubMed] [Google Scholar]
- Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittle C, Charrasse S, Larroque C, Cassimeris L. The interaction of TOGp with microtubules and tubulin. J Biol Chem. 2000;275:20748–20753. doi: 10.1074/jbc.M002597200. [DOI] [PubMed] [Google Scholar]
- Srayko M, Quintin S, Schwager A, Hyman AA. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr Biol. 2003;13:1506–1511. doi: 10.1016/s0960-9822(03)00597-9. [DOI] [PubMed] [Google Scholar]
- Stone MC, Roegiers F, Rolls MM. Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol Biol Cell. 2008;19:4122–4129. doi: 10.1091/mbc.E07-10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142:433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- van Breugel M, Drechsel D, Hyman A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J Cell Biol. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B, Akhmanova A, Straube A. Regulation of microtubule dynamic instability. Biochem Soc Trans. 2009;37:1007–1013. doi: 10.1042/BST0371007. [DOI] [PubMed] [Google Scholar]
- van der Vaart B, et al. SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J Cell Biol. 2011;193:1083–1099. doi: 10.1083/jcb.201012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT. TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, Howard J. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci USA. 2011;108:2741–2746. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J Cell Biol. 2003;161:845–851. doi: 10.1083/jcb.200303082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Bokoch GM, Waterman-Storer CM. Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J Biol Chem. 2004;279:6196–6203. doi: 10.1074/jbc.M307261200. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak MJ, Blake-Hodek K, Kosco K, Hwang E, You L, Huffaker TC. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol Biol Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, et al. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat Cell Biol. 2011;13:361–370. doi: 10.1038/ncb2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.