Abstract
Bachground
Hepatitis C virus (HCV) infection is a well-documented etiological factor for hepatocellular carcinoma (HCC). As HCV shows remarkable genetic diversity, an interesting and important issue is whether such a high viral genetic diversity plays a role in the incidence of HCC. Prior data on this subject are conflicting.
Objectives
Potential association between HCV genetic mutations or strain variability and HCC incidence has been examined through a comparative genetic analysis merely focused on a single HCV subtype (genotype 4a) in a single country (Egypt).
Study design
The study focused on three HCV sequence datasets with explicit sampling dates and disease patterns. An overlapping HCV Core/E1 domain from three datasets was used as the target for comparative analysis through genetic and phylogenetic approaches.
Results
Based on partial Core/E1 domain (387 bp), genetic and phylogenetic analysis did not identify any HCC-specific viral mutations and strains, respectively.
Conclusions
The Core/E1 domain of HCV genotype 4a in Egypt does not contain HCC-specific mutations or strains. Additionally, sequence errors resulting from the polymerase chain reaction, together with a strong evolutionary pressure on HCV in patients with end-stage liver disease, have significant potential to bias data generation and interpretation.
Keywords: hepatitis C virus, hepatocellular carcinoma, cirrhosis, RT-PCR, Egypt
1. Background
The causal relationship between hepatitis C virus (HCV) infection and hepatocellular carcinoma (HCC) is well documented.1,2 About 1–3% patients with chronic HCV infection will develop HCC in the United States.3 HCV-related tumorigenesis has been studied extensively and almost all HCV-encoded viral proteins, especially Core protein, can cause cellular transformation through multiple mechanisms.4 As a positive, single-strand RNA virus, a remarkable feature of HCV genome is the high genetic diversity with at least six major genotypes and more than 100 subtypes.5 Based on underlying mechanisms in HCV-related HCC, it is important to know whether such a high viral genetic diversity plays a differential role in the incidence of HCC. In other words, is the incidence of HCC preferentially associated with specific HCV genotype(s)/subtype(s)/strain(s) or particular mutations in the HCV genome? Due to the lack of appropriate small animal models supporting the HCV life cycle, these issues have been studied mostly in clinical settings.6–17 However, published reports have yielded conflicting data concerning these questions.6–17 The development of HCC is a long-term, multi-step process affected by many factors from both the host and the virus. To assess the role of viral genetic diversity in the incidence of HCC, it is therefore necessary to have a well-designed experimental strategy that minimizes the interference from other factors contributing to carcinogenesis.
2. Objectives
In the present study, the potential association between HCV genetic mutations or strain variability and HCC incidence has been examined through a comparative genetic analysis merely focused on a single HCV subtype (genotype 4a) in a single country (Egypt).
3. Study design
3.1. HCV sequence data collection
Three HCV sequence datasets were included in this study. The first HCV dataset was derived from a nationwide epidemiological study designed to evaluate the prevalence of HCV in Egyptian blood donors.18 The dataset consists of 49 HCV genotype 4a E1/Core sequences with assigned GenBank accession numbers from AF271825 to AF271873, representing a subset of blood donors from 15 geographically diverse governorates in Egypt.19 The second dataset was generated in our laboratory in a study to investigate the role of HCV genotype in end-stage liver disease in Egypt.20 This dataset includes a total of 146 HCV E1/Core sequences corresponding to 97 patients with HCC, 43 patients with cirrhosis and 6 individuals without end-stage liver disease (GenBank accession numbers HQ615723 to HQ615868).20 The final dataset includes 36 HCV E1/Core sequences from a study that investigated familial transmission of HCV in an Egyptian village.21 A summary for three datasets is presented in Table 1.
Table 1.
Summary of the three datasets used in this study. CHC, chronic hepatitis C infection
| Data | Subjects | Number | Sampling date (year) | Sampling pattern | Country |
|---|---|---|---|---|---|
| 1 | Blood donors | 49 | 1993 | Nationwide | Egypt |
| 2 | HCC | 97 | 2003 | Nationwide | Egypt |
| Cirrhosis | 43 | ||||
| CHC | 6 | ||||
| 3 | CHC | 36 | 2002 | A single village | Egypt |
3.2. Control experiment to estimate data quality
The three datasets were generated from three different laboratories. It is thus possible that some nucleotide differences may simply result from differences in experimental protocols. In each laboratory, the sequences were obtained from serum samples through direct sequencing of reverse transcription-polymerase chain reaction (RT-PCR) product. Thus, the use of different primers in the RT-PCR protocols is a potential concern.22 We determined the HCV Core/E1 sequences for five randomly selected samples using primer sets from all three laboratories (Table 2). Sequences were compared for the estimation of potential influence by primer selection. In our experimental protocol, RT and PCR were respectively conducted with M-MLV reverse transcriptase (Promega) and AmpliTaq DNA polymerase (Applied Biosystems) as we described previously.22
Table 2.
The primers used for the generation of the three HCV Core/E1 datasets. The numbering of primer positions is according to HCV H77 strain (GenBank accession number AF009606). Degenerate bases are matched with standard International Union of Pure and Applied Chemistry (IUPAC) codes. RT, reverse transcription
| Dataset | Primers | Polarity | Sequence (5′-3′) | Position | Application | Product size |
|---|---|---|---|---|---|---|
| 1 | 987R_H77 | Anti-sense | cgtaggggaccagttcatcatcat | 1305–1328 | RT/1st round PCR | 474 bp |
| 493S_H77 | Sense | gcaacagggaaccttcctggttgctc | 834–859 | 1st round PCR | ||
| 502S_H77 | Sense | aaccttcctggttgctctttctctat | 843–868 | 2nd round PCR | ||
| 975R_H77 | Anti-sense | gttcatcatcatatcccatgccat | 1293–1316 | |||
| 2 | CER1414 | Anti-sense | cccrcsaggachccccagtg | 1395–1414 | RT/1st round PCR | 596 bp |
| CEF696 | Sense | tgggtaaggtcatcgatacc | 697–716 | 1st round PCR | ||
| CEF723 | Sense | gcggcttcgccgacctcatg | 724–743 | 2nd round PCR | ||
| CER1318 | Anti-sense | ccagttcatcatcatrtccca | 1299–1319 | |||
| 3 | E4R | Anti-sense | gttggrctccarttcatcatc | 1307–1327 | RT/1st round PCR | 469 bp |
| Core823 | Sense | ggatcaaytaygcaacaggg | 823–842 | 1st round PCR | ||
| Core850 | Sense | ccggttgctcyttytctatc | 850–869 | 2nd round PCR | ||
| E2R | Anti-sense | cagttcatcatcatrtccct | 1299–1318 |
3.3. Genetic analysis
The genetic analysis was performed between datasets 1 and 2, while dataset 3 was used as a reference control. The target domain for comparative analysis was an overlapping region among these datasets, 387 bp in length from nucleotide position 873 through 1259 (all position numbering in the study is based on HCV strain H77, GenBank accession number AF009606). A consensus sequence corresponding to this target domain was first generated from 41 unrelated HCV genotype 4a sequences deposited in the Los Alamos HCV database.23 Nucleotide (387 sites) and amino acid (129 sites) frequencies were calculated against the consensus sequence at each site, followed by Chi-square test. Next, we evaluated intra-group mutation patterns and selection pressure by both Tajima’s D test24 and the calculation of genetic diversity parameters, including genetic distance (d), the number of synonymous substitutions per synonymous site (dS), the number of non-synonymous substitutions per non-synonymous site (dN) and dN/dS values. Tajima’s D test (coding region) was done with program DnaSP25 and genetic parameters were analyzed with either maximum composite likelihood (d) or Nei-Gojobori method (dN, dS) implemented in the Molecular Evolutionary Genetics Analysis software package (MEGA, version 4.0).26
3.4. Phylogenetic analysis
Phylogenetic analysis was performed for the combination of datasets 1 and 2. The best-fit nucleotide substitution model was first estimated through a hierarchical likelihood ratio test (hLRT) with Modeltest.27 Under the best-fit model, the unrooted maximum-likelihood (ML) tree was generated in program PHYML (20) and used as the template to evaluate the extent of clock-like evolution between the datasets 1 and 2 through a regression analysis of root-to-tip distance against sampling dates in program Path-O-Gen (http://tree.bio.ed.ac.uk/software/pathogen). Bayesian Markov chain Monte Carlo (MCMC) phylogenetic trees were simulated in BEAST package under the best-fit nucleotide substitution model as well as additional parameter settings, including a relaxed molecular clock (uncorrelated, lognormal), a Bayesian skyline coalescent prior, and a total run of 50 million generations to reach relevant parameter convergence as estimated by Tracer.28 The inferred MCMC trees then served as the input to estimate the strength of HCV strain clustering in terms of disease patterns or sample dates in program BaTS with 1000 replications and the removal of the first 10% trees as burn-in.29 Both the association index (AI)30 and the parsimony score (PS)31 were computed to see whether disease patterns or sampling dates are more strongly associated with the underlying phylogeny than expected by chance alone.
3.5. Statistical analysis
The significance of changes in either nucleotide or amino acid frequency was examined by Chi-square test. Other differences with regard to genetic parameters were assessed for statistical significance using two-tailed Student’s t-test.
4. Results
4.1. Lack of amplification bias by different primer sets
The potential effect of different primer sets on the amplification of HCV Core/E1 domain was tested in five serum samples. Direct sequencing of amplicons with the primer sets from either dataset 2 or 3 showed the complete identity. Over 1935 bp amplicon sequence (387 bp x 5), the primer set from dataset 1 generated one silent mutation (A→T), indicating a 99.95% match in comparison with the primer sets from datasets 2 and 3. Therefore, the use of different primer sets did not result in noticeable bias on the amplification of the targeted domain, allowing valid comparative analysis to be performed based on these three datasets.
4.2. Comparative analysis and Taq DNA polymerase-associated errors
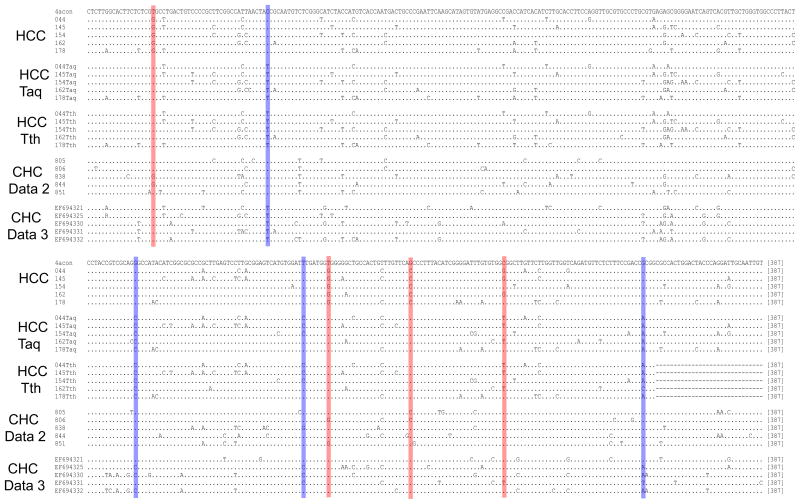
In comparison with dataset 1, the HCC and cirrhosis groups in dataset 2 showed four distinct nucleotide substitutions at positions 891 (T→G, p<0.0001), 1138 (T→G, p<0.0001), 1161 (G→C, p<0.0001) and 1187 (C→G, p<0.0001). Due to the potential significance of such a nearly complete sweep-out in HCV strains associated with the end-stage liver disease, we repeated the experiment in five samples carrying HCV strains with all four substitutions. Surprisingly, none of these samples showed the initially observed substitutions. We then conducted additional experiments. First, an additional 25 samples were processed starting from the step of RNA extraction. Sequence alignment showed the same result, with the lack of nucleotide substitutions seen in the initial analysis. Instead, there were four alternative nucleotide substitutions at positions 923, 1084, 1131 and 1226 (Fig. 1). Second, these 30 samples were re-analyzed using a new RT-PCR protocol in which M-MLV reverse transcriptase and AmpliTaq DNA polymerase were replaced with SuperScript III reverse transcriptase (Invitrogen) and rTth DNA polymerase, XL (Applied Biosystems), which contains Deep Vent DNA polymerase with exonuclease activity. This experiment confirmed the result from the repeated experiment with AmpliTaq DNA polymerase (Fig. 1). Finally, with the use of AmpliTaq DNA polymerase or rTth DNA polymerase, XL, the PCR step, consisting of 70 cycles of two rounds, was used to amplify two independent HCV clones from our previous study.32 Direct amplicon sequencing indicated complete identity to the cloned HCV sequences (data not shown).
Fig. 1.
Alignment of full analytical domain (387 bp) of five representative sequences from each group. The HCV genotype 4a consensus sequence is shown on the top line. HCC, the HCC group from dataset 2; Taq and Tth indicate the same HCC samples repeated with either AmpliTaq DNA polymerase and rTth DNA polymerase, XL, respectively; CHC, chronic HCV infection without end-stage liver disease. The four nucleotide substitutions from either initial comparative analysis or the repeated experiments are marked as red and blue, respectively. Dot indicates nucleotide identity.
4.3. Genetic and phylogenetic analysis
All genetic and phylogenetic analyses showed similar results with either inclusion (the sequence being analyzed: 387 bp in length) or exclusion (the sequence being analyzed: 363 bp length) of the codons containing the eight potential PCR-associated errors as described above. For simplicity, only results generated under 363-bp analytical domain were presented.
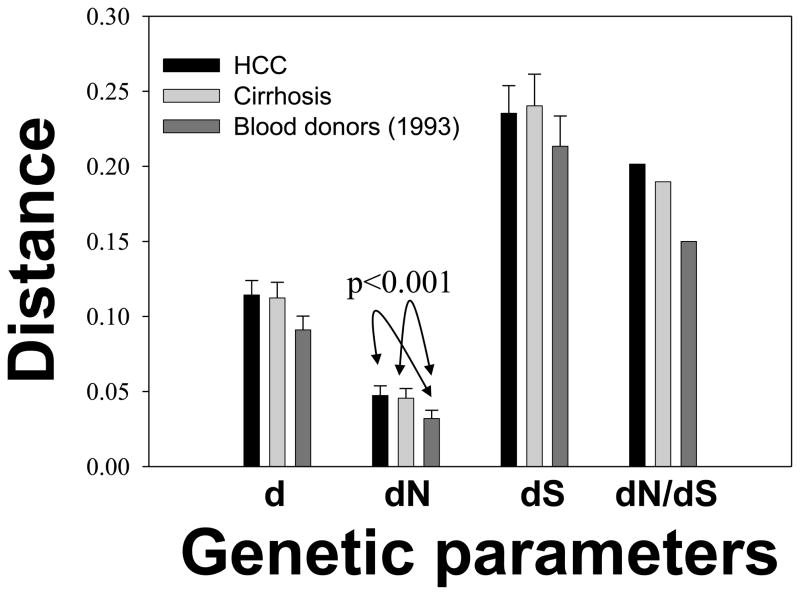
There was no obvious difference between the HCC and cirrhosis groups from dataset 2 in terms of genetic diversity and Tajima’s D test (Fig. 2). In comparison with the dataset 1, both HCC and cirrhosis groups from the dataset 2 had higher genetic diversity, especially with significantly increased dN values (p<0.001) (Fig. 2). Accordingly, the HCC and the cirrhosis group of dataset 2 had increased dN/dS values, corresponding to Tajama’s D test that showed the stronger negative values in groups HCC (−1.46) and cirrhosis (−1.35) than the dataset 1 (−1.25).
Fig. 2.
Comparisons of intra-group genetic parameters among HCC (n=97), cirrhosis (n=43) and blood donors of the dataset1 (n=49). The group of chronic HCV infection from the dataset 2 (n=6) and the dataset 3 were not included due to small sample size and restrictive sampling pattern, respectively. All genetic parameters, except for dN/dS, are expressed as mean values and standard errors. The dN values from either the HCC group or the cirrhosis group of dataset 2 were significantly higher than that from dataset 1 (blood donors) (p<0.001).
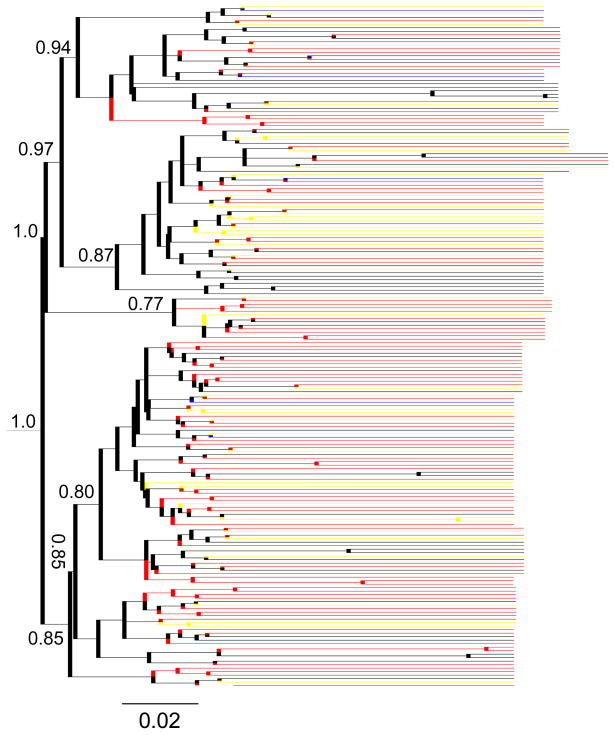
The regression analysis of root-to-tip distance against sampling dates did not support a clock-like evolution in the ML tree constructed with the datasets 1 and 2 (R2=0.026). In subsequent MCMC simulation, a relaxed molecular clock (uncorrelated, lognormal) was then applied. By giving each HCV strain a defined trait, either disease status or sampling time, BaTS analysis was run in two type of data combinations, HCC/cirrhosis and dataset 1/dataset 2. The former did not show obvious branch clustering in terms of disease status (HCC or cirrhosis) in MCMC trees (AI=8, p=0.35; PS=40, p=0.025). When including all HCV strains from the datasets 1 and 2, tree topologies were significantly associated with the distribution of qualitative traits, either disease status (AI=12.5, p<0.001; PS=73, p<0.001) or sampling dates (AI=5.68, p<0.001; PS=35, p<0.001) (Fig. 3).
Fig. 3.
Maximum clade credibility tree of 195 HCV Core/E1 sequences from the HCC group (red), cirrhosis group (yellow), chronic HCV infection (blue) of the dataset 1 as well as the blood donors (black) of the dataset 2. Posterior probability values are shown on major branches.
5. Discussion
Identification of HCC-specific mutations is a challenging endeavor. HCV’s great diversity makes it difficult to perform a comparative analysis among different HCV genotypes or subtypes. The existence of ethnically or geographically specific mutations is also a concern.8 More importantly, even if putative HCC-associated mutations are observed, it is not known if these mutations are responsible for the HCC incidence or a simple result of evolutionary adaptation. The current study was designed to focus on a single HCV genotype (4a) in a single geographical region (Egypt). All three datasets have explicit sampling dates, patterns, and adequate numbers to provide a unique opportunity to explore the possibility of an epidemiological relationship between HCV mutations and HCC incidence.
Initial comparative analysis identified four statistically significant nucleotide substitutions in the HCC and cirrhosis groups. However, in repeated experiments, these four mutations were completely lost with the consistent appearance of alternative four mutations (Fig. 1). In sequence chromatograms, almost all eight mutations showed single peaks, suggesting that these mutations are not located in highly variable sites. Experimental contamination is not supported because all other sites from the same HCV isolates appear the same (Fig. 1). Under 70 PCR cycles, four putative false mutations over 387-bp domain give an error rate at 1.5 × 10−4 substitutions per base pair, which is well within the range of Taq DNA polymerase’s misincorporation rate of 2.1 × 10−4 to 2.0 × 10−5 errors per base pair.33–38 Thus the eight nucleotide substitutions observed are most likely not authentic. Under the same experimental procedure, the appearance on different positions in a non-random pattern from repeated experiments may be attributable to the batch to batch difference of AmpliTaq DNA polymerase. Another factor is the subtle alteration of template heterogeneity due to additional 1-year storage. The role of template heterogeneity contributing to the error rate of DNA polymerase has been ignored largely.39–41 Because of the complete sequence identity after 70-cycle PCR upon the use of plasmid DNA as the template, template heterogeneity may be a more possible factor to explain our observation. Finally, the four nucleotide mutations from the initial and repeated experiments are also present on the healthy volunteers from datasets 2 and 3, respectively (Fig. 1). Thus, even assuming a real nature, these mutations may just be a result of adaptive evolution without having any relationship with end-stage liver disease, either HCC or cirrhosis.
At the phylogenetic level, BaTS analysis revealed no apparent clustering in terms of their disease traits in HCC and cirrhosis. However, the inclusion of the dataset 1 (blood donors) resulted in strong association between disease traits (HCC/Cirrhosis or blood donors) (Fig. 3). Since the dataset 1 were sampled in 1993, such an observation may be largely due to different sampling dates rather than disease traits. Because of a small number (n=6) of HCV sequences from blood donors in the dataset 2, a univocal answer may require the analysis with the inclusion of more contemporaneously collected HCV sequences from patients without HCC/cirrhosis.
The HCC group, cirrhosis group and the dataset 1 all have significantly negative Tajima’s D values, indicating an excess of low-frequency mutations and therefore a positive selection pressure. Among datasets the HCC group has the strongest negative Tajima’s D value, corresponding with its highest dN/dS ratio (Fig. 2). Actually, while having similar dS values, the HCC and cirrhosis have significantly higher dN values than the dataset 1 (p<0.001) (Fig. 2). Taken together, these data suggest a strong evolutionary pressure of HCV in patients with end-stage liver diseases, which is consistent with previous reports in HCC patients infected with HCV genotype 1b.8,13 An important implication from this observation is a theoretically enhanced chance for the detection of putative HCC or cirrhosis-specific mutations, which requires caution in data interpretation since the mutations identified may simply be the consequence of adaptation.
It should be noted that our analysis was based on a short HCV domain, the 387-bp partial Core/E1 region. Comprehensive understanding of HCC-specific mutations and/or strains may require a full-length HCV genome scanning as well as the availability of adequate number of samples collected in both simultaneous and longitudinal patterns. In this setting, the current study represents a proof-of-concept investigation in terms of experimental approaches and phylogenetic techniques to address this elusive but clinically important issue.
Acknowledgments
This work was supported by NIH grants R01 DK80711 (XF), R21 AI076834 (AMD) and USA and Egypt Science and Technology Joint Fund BIO6-002-004 (AMD).
Footnotes
Conflict of interest statement
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen SL, Morgan TR. The natural history of hepatitis C virus (HCV) infection. Int J Med Sci. 2006;3:47–52. doi: 10.7150/ijms.3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36 (5 Suppl 1 ):S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB. Hepatocellular carcinoma and hepatitis C in the United States. Hepatology. 2002;36:S74–S83. doi: 10.1053/jhep.2002.36807. [DOI] [PubMed] [Google Scholar]
- 4.Castello G, Scala S, Palmieri G, Curley SA, Izzo F. HCV-related hepatocellular carcinoma: from chronic inflammation to cancer. Clin Immunol. 2010;134:237–50. doi: 10.1016/j.clim.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Kuiken C, Simmonds P. Nomenclature and numbering of the hepatitis C virus. Method Mol Biol. 2009;510:33–53. doi: 10.1007/978-1-59745-394-3_4. [DOI] [PubMed] [Google Scholar]
- 6.Alam SS, Nakamura T, Naganuma A, Nozaki A, Nouso K, Shimomura H, et al. Hepatitis C virus quasispecies in cancerous and noncancerous hepatic lesions: the core protein-encoding region. Acta Med Okayama. 2002;56:141–7. doi: 10.18926/AMO/31716. [DOI] [PubMed] [Google Scholar]
- 7.De Mitri MS, Mele L, Chen CH, Piccinini A, Chianese R, D’Errico A, et al. Comparison of serum and liver hepatitis C virus quasispecies in HCV related hepatocellular carcinoma. J Hepatol. 1998;29:887–92. doi: 10.1016/s0168-8278(98)80115-3. [DOI] [PubMed] [Google Scholar]
- 8.Fishman SL, Factor SH, Balestrieri C, Fan X, Dibisceglie AM, Desai SM, et al. Mutations in the hepatitis C virus core gene are associated with advanced liver disease and hepatocellular carcinoma. Clin Cancer Res. 2009;15:3205–13. doi: 10.1158/1078-0432.CCR-08-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuhara T, Takeishi K, Toshima T, Morita K, Ueda S, Iguchi T, et al. Impact of amino acid substitutions in the core region of HCV on multistep hepatocarcinogenesis. Hepatol Res. 2010;40:171–8. doi: 10.1111/j.1872-034X.2009.00575.x. [DOI] [PubMed] [Google Scholar]
- 10.Gimnez-Barcons M, Franco S, Surez Y, Forns X, Ampurdans S, Puig-Basagoiti F, et al. High amino acid variability within the NS5A of hepatitis C virus (HCV) is associated with hepatocellular carcinoma in patients with HCV-1b-related cirrhosis. Hepatology. 2001;34:158–67. doi: 10.1053/jhep.2001.25512. [DOI] [PubMed] [Google Scholar]
- 11.Horie C, Iwahana H, Horie T, Shimizu I, Yoshimoto K, Yogita S, Tashiro S, Ito S, Itakura M. Detection of different quasispecies of hepatitis C virus core region in cancerous and noncancerous lesions. Biochem Biophys Res Commun. 1996;218:674–81. doi: 10.1006/bbrc.1996.0121. [DOI] [PubMed] [Google Scholar]
- 12.Nagayama K, Kurosaki M, Enomoto N, Miyasaka Y, Marumo F, Sato C. Characteristics of hepatitis C viral genome associated with disease progression. Hepatology. 2000;31:745–50. doi: 10.1002/hep.510310327. [DOI] [PubMed] [Google Scholar]
- 13.Ogata S, Nagano-Fujii M, Ku Y, Yoon S, Hotta H. Comparative sequence analysis of the core protein and its frameshift product, the F protein, of hepatitis C virus subtype 1b strains obtained from patients with and without hepatocellular carcinoma. J Clin Microbiol. 2002;40:3625–30. doi: 10.1128/JCM.40.10.3625-3630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rster B, Zeuzem S, Krump-Konvalinkova V, Berg T, Jonas S, Severin K, et al. Comparative sequence analysis of the core-and NS5-region of hepatitis C virus from tumor and adjacent non-tumor tissue. J Med Virol. 2001;63:128–34. [PubMed] [Google Scholar]
- 15.Takahashi K, Iwata K, Matsumoto M, Matsumoto H, Nakao K, Hatahara T, et al. Hepatitis C virus (HCV) genotype 1b sequences from fifteen patients with hepatocellular carcinoma: the ‘progression score’ revisited. Hepatol Res. 2001;20:161–71. doi: 10.1016/s1386-6346(00)00141-8. [DOI] [PubMed] [Google Scholar]
- 16.Nakamoto S, Imazeki F, Fukai K, Fujiwara K, Arai M, Kanda T, Yonemitsu Y, Yokosuka O. 2010 Association between mutations in the core region of hepatitis C virus genotype 1 and hepatocellular carcinoma development. J Hepatol. 2010;52:72–8. doi: 10.1016/j.jhep.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H. Amino acid substitutions in hepatitis C virus core region predict hepatocarcinogenesis following eradication of HCV RNA by antiviral therapy. J Med Virol. 2011;83:1016–22. doi: 10.1002/jmv.22094. [DOI] [PubMed] [Google Scholar]
- 18.Arthur RR, Hassan NF, Abdallah MY, El-Sharkawy, Saad MD, Hackbart BG, et al. Hepatitis C antibody prevalence in blood donors in different governorates in Egypt. Trans R Soc Trop Med Hyg. 1997;91:271–4. doi: 10.1016/s0035-9203(97)90070-5. [DOI] [PubMed] [Google Scholar]
- 19.Ray SC, Arthur RR, Carella A, Bukh J, Thomas DL. Genetic epidemiology of hepatitis C virus throughout Egypt. J Infect Dis. 2000;182:698–707. doi: 10.1086/315786. [DOI] [PubMed] [Google Scholar]
- 20.Ryu SH, Fan X, Xu Y, Elbaz T, Zekri AR, Abdelaziz AO, et al. Lack of association between genotypes and subtypes of HCV and occurrence of hepatocellular carcinoma in Egypt. J Med Virol. 2009;81:844–7. doi: 10.1002/jmv.21451. [DOI] [PubMed] [Google Scholar]
- 21.Plancoulaine S, Mohamed MK, Arafa N, Bakr I, Rekacewicz C, Trgout DA, et al. Dissection of familial correlations in hepatitis C virus (HCV) seroprevalence suggests intrafamilial viral transmission and genetic predisposition to infection. Gut. 2008;57:1268–74. doi: 10.1136/gut.2007.140681. [DOI] [PubMed] [Google Scholar]
- 22.Fan X, Lyra AC, Tan D, Xu Y, Di Bisceglie AM. Differential amplification of hypervariable region 1 of hepatitis C virus by partially mismatched primers. Biochem Biophys Res Commun. 2001;284:694–7. doi: 10.1006/bbrc.2001.5019. [DOI] [PubMed] [Google Scholar]
- 23.Kuiken C, Yusim K, Boykin L, Richardson R. The Los Alamos HCV Sequence Database. Bioinformatics. 2005;21:379–84. doi: 10.1093/bioinformatics/bth485. [DOI] [PubMed] [Google Scholar]
- 24.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4. 0. Mol Biol Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 27.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 28.Drummond AJ, Rambaut A. 2007 BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker J, Rambaut A, Pybus OG. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol. 2008;8:239–46. doi: 10.1016/j.meegid.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Wang TH, Donaldson YK, Brettle RP, Bell JE, Simmonds P. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol. 2001;75:11686–99. doi: 10.1128/JVI.75.23.11686-11699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitch WN. Toward defining the course of evolution: minimal change for a specific tree topology. Syst Zool. 1971;20:406–16. [Google Scholar]
- 32.Chambers TJ, Fan X, Droll DA, Hembrador E, Slater T, Nickells MW, et al. Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection. J Virol. 2005;79:3071–83. doi: 10.1128/JVI.79.5.3071-3083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnes WM. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992;112:29–35. doi: 10.1016/0378-1119(92)90299-5. [DOI] [PubMed] [Google Scholar]
- 34.Cariello NF, Swenberg JA, Skopek TR. Fidelity of Thermococcus litoralis DNA polymerase (Vent) in PCR determined by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1991;19:4193–8. doi: 10.1093/nar/19.15.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keohavong P, Thilly WG. Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci USA. 1989;86:9253–7. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling LL, Keohavong P, Dias C, Thilly WG. Optimization of the polymerase chain reaction with regard to fidelity: modified T7, Taq, and vent DNA polymerases. PCR Methods Appl. 1991;1:63–9. doi: 10.1101/gr.1.1.63. [DOI] [PubMed] [Google Scholar]
- 37.Lundberg KS, Shoemaker DD, Adams MW, Short JM, Sorge JA, Mathur EJ. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991;108:1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 38.Tindall KR, Kunkel TA. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 39.Akbari M, Hansen MD, Halgunset J, Skorpen F, Krokan HE. Low copy number DNA template can render polymerase chain reaction error prone in a sequence-dependent manner. J Mol Diagn. 2005;7:36–9. doi: 10.1016/s1525-1578(10)60006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loewen PC, Switala J. Template secondary structure can increase the error frequency of the DNA polymerase from Thermus aquaticus. Gene. 1995;164:59–63. doi: 10.1016/0378-1119(95)00383-h. [DOI] [PubMed] [Google Scholar]
- 41.Vandenbroucke I, Eygen VV, Rondelez E, Vermeiren H, Baelen KV, Stuyver LJ. Minor Variant Detection at Different Template Concentrations in HIV-1 Phenotypic and Genotypic Tropism Testing. Open Virol J. 2008;2:8–14. doi: 10.2174/1874357900802010008. [DOI] [PMC free article] [PubMed] [Google Scholar]