Abstract
SUMMARY
Ail is an outer membrane protein from Yersinia pestis that is highly expressed in a rodent model of bubonic plague, making it a good candidate for vaccine development. Ail is important for attaching to host cells and evading host immune responses, facilitating rapid progression of a plague infection. Binding to host cells is important for injection of cytotoxic Yersinia outer proteins. To learn more about how Ail mediates adhesion, we solved two high-resolution crystal structures of Ail, with no ligand bound and in complex with a heparin analog called sucrose octasulfate. We identified multiple adhesion targets, including laminin and heparin, and showed that a 40 kDa domain of laminin called LG4-5 specifically binds to Ail. We also evaluated the contribution of laminin to delivery of Yops to HEp-2 cells. This work constitutes a structural description of how a bacterial outer membrane protein uses a multivalent approach to bind host cells.
Keywords: ail, plague, Yersinia pestis, adhesion, invasion, extracellular matrix proteins, outer membrane protein, crystal structure
INTRODUCTION
Yersinia pestis, a Gram-negative bacillus, is delivered by infected fleas to cause bubonic plague or by aerosol to cause pneumonic plague. Y. pestis has been classified by the Centers for Disease Control and Prevention as a Category A microbe because it is readily transmitted by aerosol, is extremely virulent, and carries a high mortality rate. Bubonic plague infects 1000-3000 people per year worldwide. Without antibiotics, the mortality rate for bubonic plague is 40-60%, whereas pneumonic plague is particularly virulent with a > 90% mortality rate if left untreated. Y. pestis can be aerosolized and could be used as an effective bioweapon to spread pneumonic plague. The recent emergence of antibiotic-resistant strains (Galimand et al, 2006) and the lack of an effective vaccine increase the urgency for developing better medical countermeasures against plague.
Once established in a mammalian host, Y. pestis alters expression levels of many outer membrane proteins to ensure survival (Sebbane et al, 2006). One of the most highly up-regulated outer membrane proteins is Ail (adherence and invasion locus) (Pieper et al, 2009), a 17 kDa protein required for virulence in Y. pestis that was originally identified as a virulence factor in Y. enterocolitica (Miller & Falkow, 1988). Y. pestis Ail has been demonstrated to promote adhesion to target cells (Felek & Krukonis, 2009; Kolodziejek et al, 2007) and to confer immune evasion by interfering with the host complement cascade (Bartra et al, 2008; Kirjavainen et al, 2008; Kolodziejek et al, 2007). Although bacterial outer membrane proteins are often used to promote host-microbe interactions, there is currently no structural description of the process due to the difficulty to express, purify, biochemically characterize, and crystallize the outer membrane component(s).
Ail belongs to a family of proteins found in Gram-negative bacteria that have a variety of functions conferring virulence: Y. enterocolitica Ail and Salmonella typhimurium Rck are responsible for both adhesion and serum resistance (Heffernan et al, 1992; Miller et al, 2001), PagC from S. typhimurium is required for survival within macrophages (Pulkkinen & Miller, 1991), and E. coli OmpX, the only structurally characterized member of this family, has unknown function(s), although it does not confer serum resistance or adherence to mammalian cells (Mecsas et al, 1995). OmpX forms an 8-stranded β-barrel with four extracellular loops (Vogt & Schulz, 1999) and other family members are predicted to share the same fold. These proteins display significant sequence similarity within the transmembrane β-barrel, while the extracellular loops show less than 10% sequence identity. Since residues implicated in adhesion and complement resistance are found in the loop regions, the structural basis of virulence for this protein family was not elucidated with the structure of OmpX. In Y. pestis, four ail genes have been found by genome sequencing (Parkhill et al, 2001). Two recent studies showed that identical homologs from different strains (designated Ail in CO92; OmpX in KIM) are responsible for the adhesion, invasion, and complement resistance phenotypes of Y. pestis (Bartra et al, 2008; Kolodziejek et al, 2007).
Proteins from the Ail family have been shown to adhere to target cells by binding to extracellular matrix proteins (ECM) such as laminin and fibronectin (Crago & Koronakis, 1999; Tsang et al, 2010). Heparan sulfate proteoglycan, another ECM component, is also exploited for adhesion by a number of microorganisms (Pettersson et al, 1996). Interactions between ECM components and bacteria can act as bridges to host cells, and in the case of Y. pestis this contact is required for injection of Yersinia outer protein (Yop) pathogenicity factors (Felek et al, 2010; Pettersson et al, 1996; Tsang et al, 2010). However, although several of these interactions have been functionally characterized, a structural description of the host-pathogen interaction is lacking (Crago & Koronakis, 1999).
To better understand how Y. pestis Ail mediates adhesion to host cells, we solved crystal structures of Ail alone and in complex with a heparin analog, sucrose octasulfate (SOS). Like OmpX, Ail forms an 8-stranded β-barrel that spans the outer membrane, with residues important for adhesion located in the extracellular loops (Miller et al, 2001). From our crystal structures and biochemical assays, we obtained the following results: (1) We identified binding partners for Ail using a bacterial binding assay. When expressed in the outer membrane of E. coli, Ail preferentially binds total ECM, followed by laminin and then fibronectin, with no detectable binding to collagen. (2) Through limited proteolysis of laminin (a 900 kDa cruciform-shaped ECM protein), we identified a 40 kDa fragment at the tip of the cruciform called LG4-5 that specifically binds to Ail. (3) The Ail-laminin interaction facilitates delivery of Yop proteins to mammalian cells. (4) Heparin binds at two positively charged sites on the extracellular surface, near the bilayer interface. Our work suggests that Ail uses a multivalent approach to bind ECM components, potentially facilitating adhesion to a variety of cell types.
RESULTS
Ail crystallizes as an 8-stranded antiparallel β-barrel
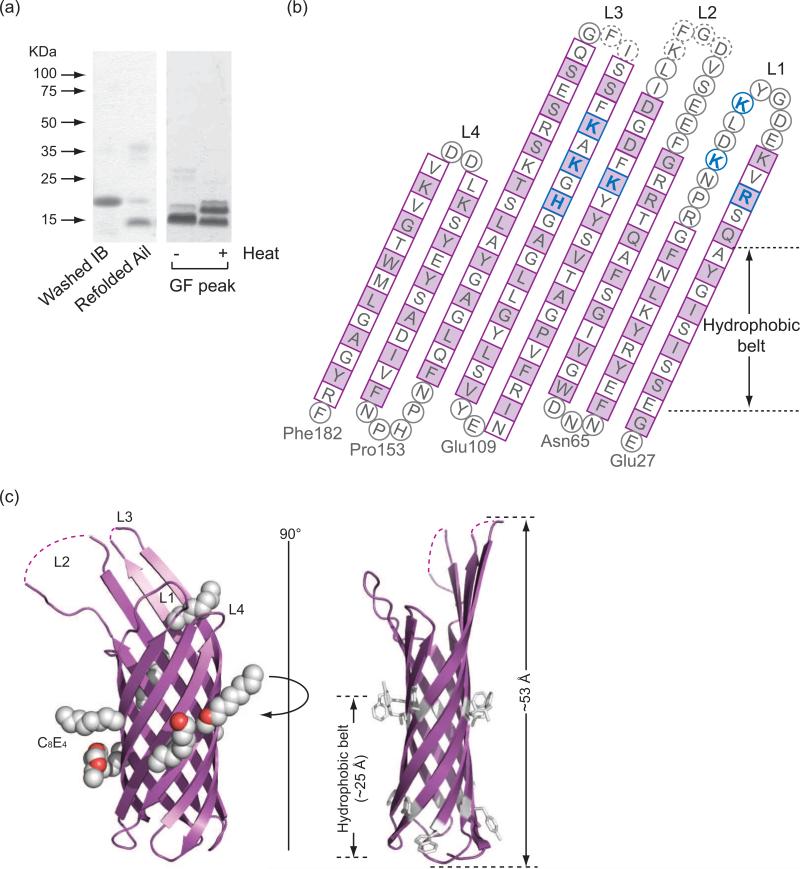
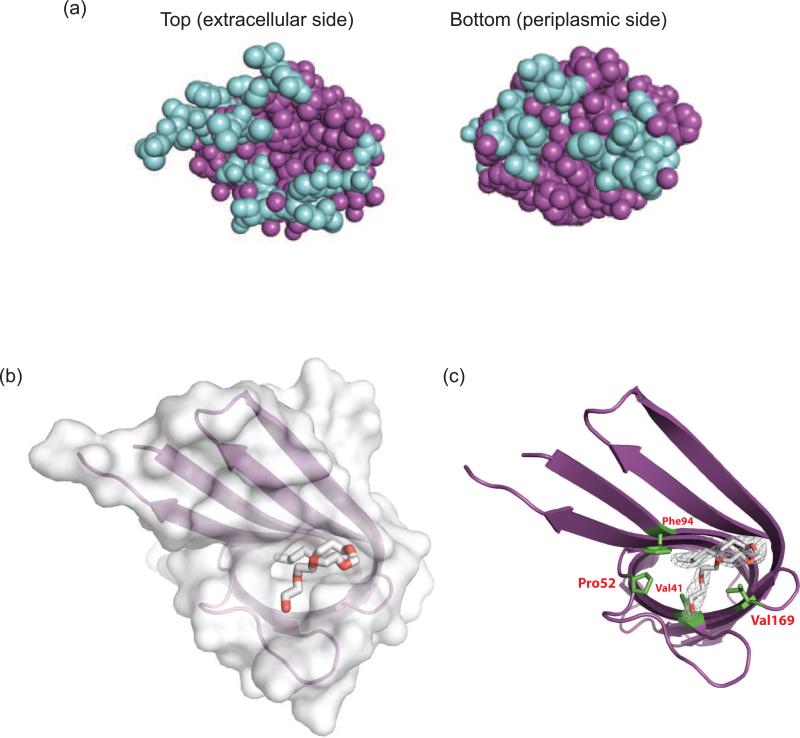
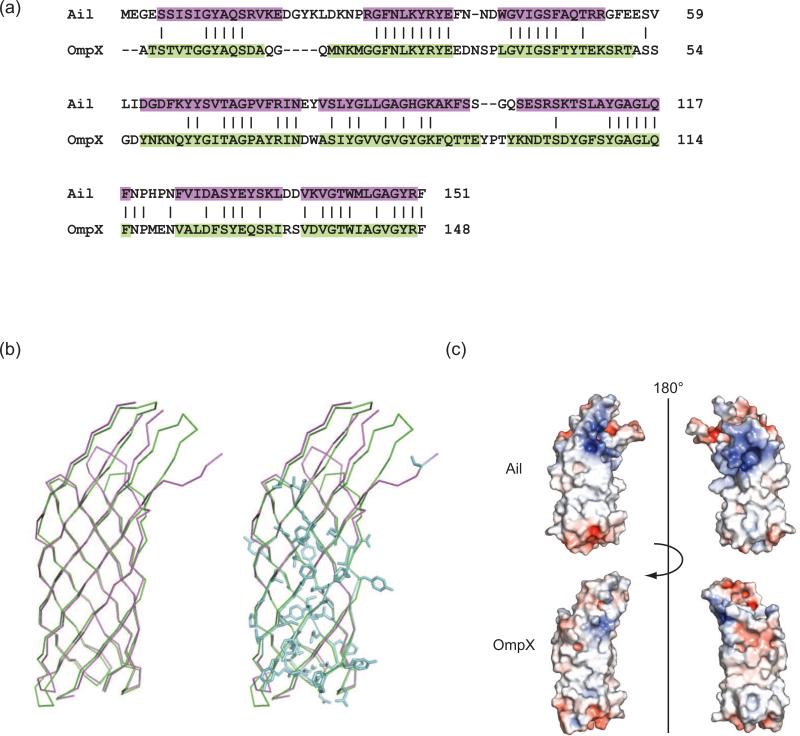
When targeted to E. coli outer membranes, Ail is produced in amounts sufficient for functional experiments but insufficient for structural studies. We therefore expressed Ail as cytoplasmic inclusion bodies and refolded the protein in the presence of the detergent dodecyl maltoside (Fig. 1a). Purified Ail was heat modifiable on SDS-PAGE, which is characteristic of tightly folded β-barrel proteins. Crystals were grown from the detergent tetraethylene glycol monooctyl ether (C8E4) and diffracted to a resolution of 1.80 Å (crystallographic statistics, Table 1; electron density quality, Fig. S1). Apo-Ail crystallized in spacegroup P43212 with one molecule per asymmentric unit. This outer membrane protein forms an 8-stranded antiparallel β-barrel with four extracellular loops. The model is complete with the exception of four residues in extracellular loop 2 and two residues in loop 3, which are disordered (Fig. 1b, c). The β-strands range in length from 10 to 18 residues, forming a barrel that is 53 Å high, with an elliptical cross section measuring 12 Å by 20 Å. When viewed in space filling representation from the extracellular or periplasmic side of the membrane, there is no obvious channel through the barrel (Fig. 2a). As is often observed for membrane proteins, aromatic residues delineate the bilayer interfaces. We located seven C8E4 detergent molecules bound to Ail, six of which are located on the outside surface of the barrel (Fig. 1c). However, one detergent molecule sits in a hydrophobic cleft on the extracellular surface (Fig. 1c, Fig. 2b, 2c), and potentially mimics binding of some target adhesion proteins (discussed below). Ail exhibits significant structural similarity to E. coli OmpX, with an rmsd for Cα positions in the barrel of 1.7 Å. As seen for OmpX, extracellular loops L2 and L3 are significantly longer than L1 and L4 (here we define loops as residues connecting the transmembrane portions of two β-strands, regardless of secondary structure). There is substantial sequence similarity between Ail and OmpX (the two proteins are 41% identical), but conserved residues are found almost exclusively in the barrel (Fig. 3a, b), with little conservation in the extracellular loops. When viewing the electrostatic surface properties of Ail and OmpX, we noticed that Ail displays two areas of positive charge on the extracellular surface that are absent in OmpX (Fig. 3c). If adhesion to ECM proteins is mediated by electrostatic interactions, the variations in surface charge may help to explain why Ail functions as an adhesin, whereas OmpX does not.
Figure 1. Refolding, purification and crystal structure of Ail.
(a) Left: Ail inclusion bodies run at a molecular weight of about 17 kDa, whereas purified, refolded Ail runs at about 14 kDa. Right: Heat modifiability of purified Ail. The unheated Ail sample runs primarily as a 14 kDa band, while an identical sample after heating for 10 minutes at 100°C runs as a mixture of folded and unfolded species, demonstrating the unusual stability of this protein. (b) Topology diagram of Ail. Residues in β-strands are shown as squares with residues in turns or loops depicted as circles. Dotted circles represent residues disordered in the crystal structure. Side-chains of residues colored purple point to the outside of the barrel. Residues that bind the heparin analog SOS are colored blue. (c) Two orthogonal views of the refined Ail structure. Left: Ail (purple) binds seven C8E4 detergent molecules (white and red space filling representation). Right: Aromatic residues (white) mark the boundaries of the hydrophobic portion of the lipid bilayer. See also Figure S1.
Table 1.
Data collection and refinement statistics
| Apo-Ail | Ail-SOS | |
|---|---|---|
| Data collection | ||
| Space group | P43212 | P32 |
| Cell dimensions | ||
| a, b, c (Å) | 50.2, 50.2, 170.6 | 91.4, 91.4, 47.5 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å)* | 50-1.80 (1.86-1.80) | 50-1.85 (1.92-1.85) |
| Rsym or Rmerge | 0.058 (0.346) | 0.103 (0.713) |
| I / σI | 39.9 (3.9) | 15.7 (2.11) |
| Completeness (%) | 93.1 (68.0) | 100 (99.9) |
| Redundancy | 10.4 (4.7) | 5.2 (4.5) |
| Refinement | ||
| Resolution (Å) | 35.0 - 1.8 | 33.0 - 1.85 |
| No. reflections | 19652 | 37763 |
| Rwork / Rfree | 0.20 / 0.23 | 0.20 / 0.22 |
| No. atoms | ||
| Protein | 1177 | 2378 |
| Ligand/ion | 69 | 202 |
| Water | 70 | 121 |
| Average B-factors | ||
| Protein | 30.5 | 37.8 |
| Ligand/ion | 53.7 | 49.4 |
| Water | 43.7 | 51.3 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.014 | 0.015 |
| Bond angles (°) | 1.259 | 1.381 |
A single crystal was used for each structure and values in parentheses are for the highest-resolution shell.
Figure 2. The barrel is closed on both sides of the membrane and a detergent molecule occupies a hydrophobic cleft on the extracellular surface.
(a) Space filling representation for extracellular and periplasmic surfaces of Ail. Barrel residues are shown in purple, and loops are shown in blue. (b) View from the extracellular surface showing the C8E4 molecule bound in the hydrophobic cleft on the extracellular surface. Ail is shown in surface representation and transparent ribbon, with the detergent molecule shown as a stick figure. (c) Residues forming the binding site for C8E4 are shown in green.
Figure 3. Structure-based sequence alignment of Ail and OmpX and electrostatic surface properties.
(a) Dalilite (Baker et al, 2001) structure-based sequence alignment of Ail (purple) with E. coli OmpX (green) (PDB ID = 1QJ8). (b) Superposition of Ail (purple) with OmpX (green) using the alignment in (a) identified 65 identical residues (blue) shared between the two proteins for an overall sequence identity of 41%. Nearly all of these conserved residues are located in the transmembrane β-strands of the barrel. Since Ail residues involved in adhesion and complement resistance are located primarily in the extracellular loops, this may explain why OmpX does not have similar functions. (c) Electrostatic surface representations for Ail and OmpX. Regions with potentials above +5kT (blue) and below -5kT (red) were calculated by the program APBS (Holm & Park, 2000). Ail displays two regions of positive charge (blue) on the extracellular surface, approximately separated by a 180° rotation. In contrast, the surface of OmpX displays relatively little positive or negative charge.
Ail binds the extracellular matrix protein laminin
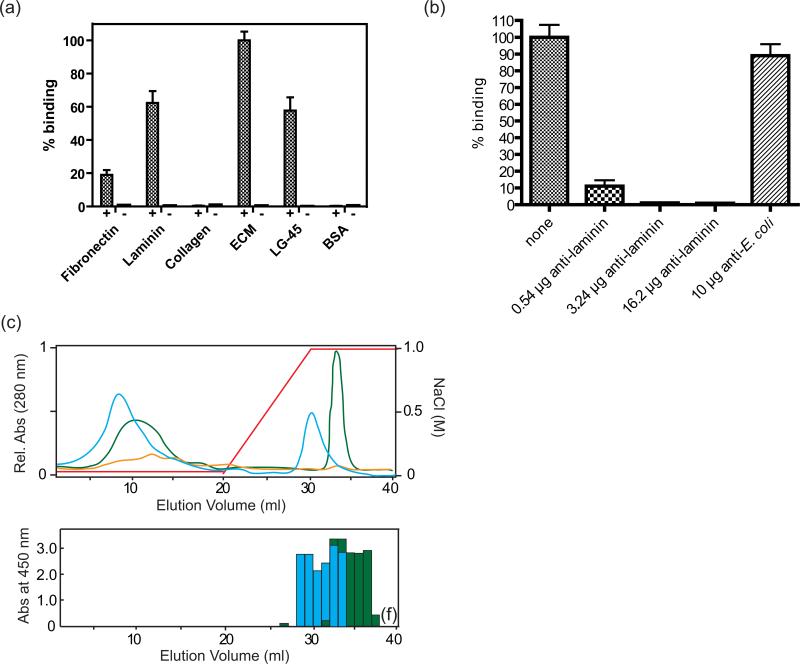
Ail has homologs in Salmonella called Rck and PagC, which are associated with virulence in that organism. When expressed in the outer membrane of E. coli, Rck and PagC induce bacterial binding to the extracellular matrix proteins laminin and fibronectin, with higher specificity for laminin (Crago & Koronakis, 1999). Recently it was also shown that both purified Ail from Y. pestis and bacteria expressing this protein bind to fibronectin, however laminin binding was not investigated (Tsang et al, 2010). Since two Salmonella homologs of Ail share an affinity for laminin, we asked whether Y. pestis Ail might bind to laminin (or other ECM proteins) as an adhesion target. To detect target adhesion proteins, glass slides were coated with human ECM proteins and incubated with E. coli expressing Ail in the outer membrane. The slides were washed and bound cells were stained and counted. We observed that bacteria expressing Ail specifically bound ECM proteins as compared to the control, with a clear preference for total ECM, followed by laminin and then by fibronectin (Fig. 4a). In contrast, cells expressing Ail did not bind to collagen (type IV). Because the laminin component of ECM bound the most Ail-expressing bacteria, we tested the ability of an anti-laminin antibody to block binding to ECM. Figure 4b shows that the anti-laminin antibody completely blocked binding of Ail-expressing cells to ECM coated on glass slides.
Figure 4. Laminin binding activity of Ail.
(a) Binding assay for Ail-expressing bacteria to human ECM proteins and LG4-5, a fragment of laminin. Numbers of bound bacteria to laminin, fibronectin, collagen, ECM, LG4-5, and BSA are shown as bars. Standard deviations are shown by error bars. Controls contain cells expressing empty vector. Lanes labels with + contain Ail, while lanes labled with – are controls. (b) Anti-laminin can block binding to ECM by Ail-expressing cells. ECM-coated glass slides were pre-incubated with various amounts of anti-laminin antibody before adding Ail-expressing bacteria. The bar labeled ‘none’ represents no antibody added, whereas an E. coli antibody was used as a positive control. (c) Heparin column purification of laminin digested with elastase and recognition of fragments by Ail. Top: UV absorbance spectra are shown (green: human laminin, blue: mouse laminin, yellow: elastase control). The concentration of NaCl is indicated by a red line. Bottom: ELISA assay using a plate coated with each fraction from the heparin column purification, probed with purified Ail. Binding of Ail was measured as absorbance at 450 nm. Ail binds to laminin fractions (either mouse or human) eluted from a heparin column with high NaCl concentrations.
Since laminin is a very large molecule (~ 900 kDa), the full-length protein was digested by elastase and the digested fragments were separated using a heparin column (Ott et al, 1982). Laminin eluted as two main peaks from the heparin column, with one peak in the unbound fractions and a second peak eluting with 1M NaCl that corresponds to a C-terminal ~40 kDa fragment known as LG4-5 (Laminin G-like domains 4 & 5). Fractions eluted from the column were individually bound to the wells of an ELISA plate and probed for binding to purified Ail. Ail bound specifically to the peak eluting at 1M NaCl (Fig. 4c). To verify the interaction between Ail and LG4-5, we bound LG4-5 to glass slides and evaluated binding to E. coli expressing Ail as described above. LG4-5 bound Ail-expressing cells essentially as well as full-length laminin (Fig. 4a). This work shows that laminin is an adhesion target for Ail, and a major site of interaction is located in LG4-5, although additional domains of laminin may also contribute to binding.
Laminin and fibronectin play a role in Yop delivery to host cells
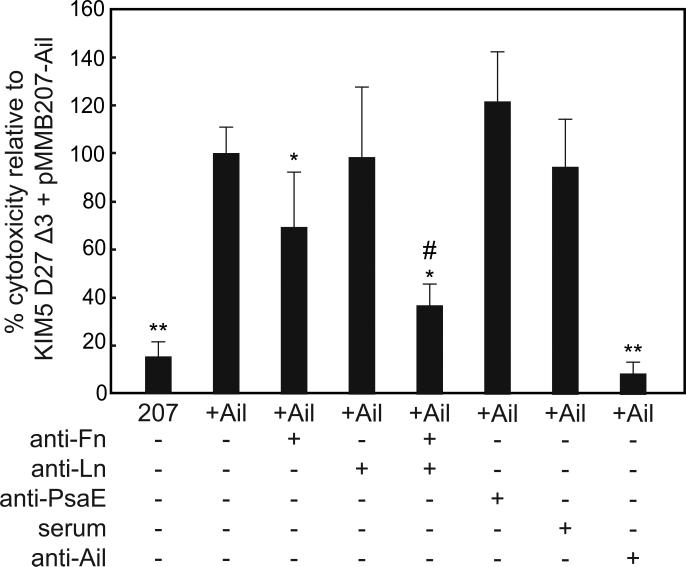
To assess whether Ail interaction with laminin and/or fibronectin plays a role in adhesion-dependent Yop delivery by Y. pestis, we tested the ability of anti-laminin or anti-fibronectin antibodies to inhibit Yop-dependent cytotoxicity. Since several adhesins can contribute to Y. pestis Yop delivery, we used strain KIM5 D27Δ3 (Felek et al, 2010) which lacks Ail, Pla (plasminogen activator), and Psa (pH 6 antigen) to do these experiments. Either empty vector or vector expressing Ail was added back to this strain. Upon treatment with anti-fibronectin antibodies, we observed a reduction in Yop delivery to HEp-2 cells (Fig. 5) as was reported previously (Tsang et al, 2010). Addition of anti-laminin antibodies alone did not inhibit Yop delivery, but a combination of anti- fibronectin and anti-laminin antibodies had a synergistic effect, reducing Yop delivery significantly more than anti-fibronectin antibodies alone. Addition of a negative control antibody against PsaE did not inhibit Yop delivery. Therefore, while fibronectin contributes more strongly to Yop delivery to HEp-2 cells, laminin also contributes to this function of Ail.
Figure 5. Antibodies to fibronectin and laminin can inhibit Y. pestis Yop delivery to host cells.
Y. pestis KIM5 D27Δ3 harboring the empty vector pMMB207 (207) or pMMB207-ail (+Ail) were added to HEp-2 cells at an MOI of 10. A 1:25 dilution of the indicated antibodies was added to HEp-2 cells just prior to the addition of bacteria. At 2.5 - 3.5 hours post-inoculation, cell were fixed and stained with Giemsa. Cytotoxicity (cell rounding) was enumerated for 500-800 cells per treatment in three separate fields and normalized such that Ail-mediated Yop delivery without antibody treatment was 100% (absolute levels of cytotoxicity were 65 - 75%). * p<0.05 relative to KIM5 D27Δ3 +Ail; ** p<0.00002 relative to KIM5 D27Δ3 +Ail; # p<0.01 relative to KIM5 D27Δ3 +Ail + anti-Fn or anti-Ln. Significance was assessed using the Student's t-test.
We also tested the ability of mouse polyclonal antibodies raised against purified Ail to block Ail-mediated Yop delivery. Addition of anti-Ail mouse serum blocked Yop delivery to background levels (>90% inhibition, Fig. 5). Control mouse serum from non-immunized mice had no effect on Ail-mediated Yop delivery. This data shows that antibodies against Ail can efficiently block Yop delivery and emphasizes the role of Ail as an important adhesin in Y. pestis.
Ail can also use heparan sulfate proteoglycans to bind host cells
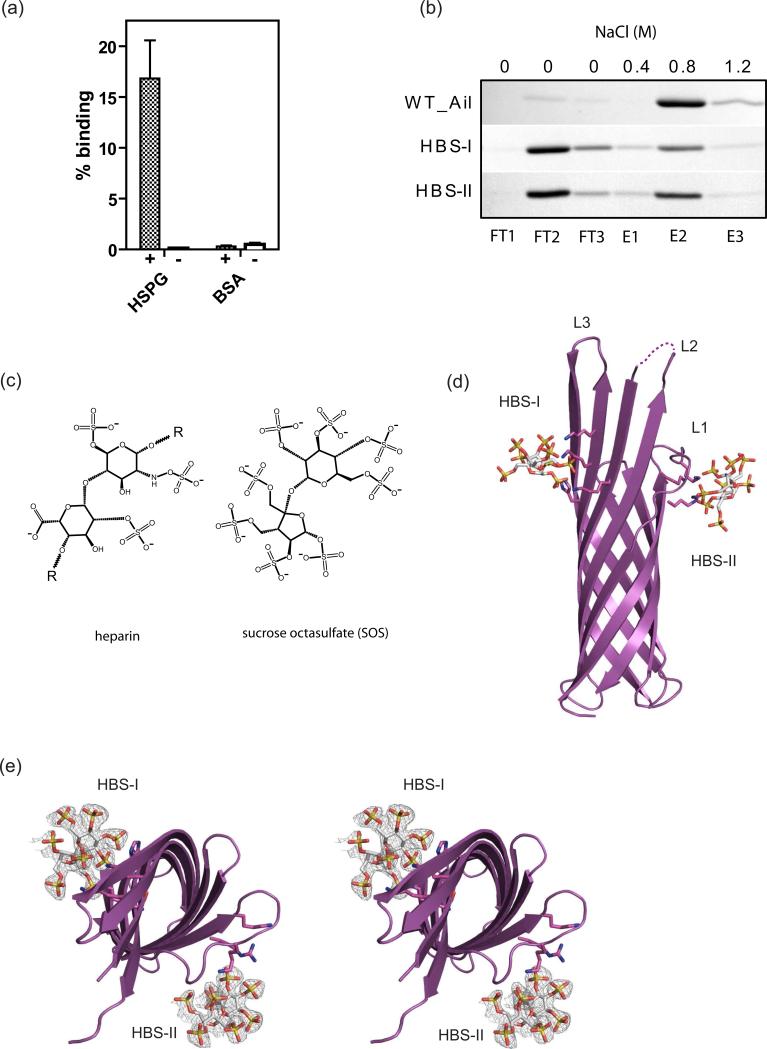
In addition to using Ail to bind to laminin (this work) and fibronectin (Tsang et al, 2010), Y. pestis has been suggested to attach to epithelial cells by binding to heparan sulfate proteoglycans (HSPG) (Zhang et al, 2008). From our crystal structure we speculated that the positively charged regions located on the extracellular surface (Fig. 3c) might interact with negatively charged ligands such as HSPG. We coated glass slides with HSPG and incubated them with E. coli cells expressing Ail in the outer membrane. The slides were washed and bound cells were stained and counted. We observed that cells expressing Ail specifically bound to HSPG but not to control slides (Fig. 6a). We then analyzed binding of purified Ail in detergent to a heparin affinity column (heparin is a highly sulfated glycosaminoglycan closely related to heparan sulfate). Ail bound tightly to the column and was eluted at a NaCl concentration of 0.8 M using a step gradient (Fig. 6b).
Figure 6. Heparin binding activity of Ail.
(a) Bacterial binding assay for Ail-expressing bacteria to heparan sulfate proteoglycan (HSPG). Numbers of bound bacteria to HSPG and BSA are shown in the bar graph. The control represents cells containing empty vector. Lanes labels with + contain Ail, while lanes labled with – are controls. The data were normalized to ECM binding (refer to figure 4a). (b) Binding of WT Ail and HBS mutants to a heparin column. Lanes 1-3 (FT1-FT3) show unbound Ail after sample application and wash steps. Lanes 4-6 (E1-E3) show stepwise elution with increasing NaCl concentration. WT Ail binds quantitatively to the column and is eluted by 0.8 M NaCl, whereas HBS-I and HBS-II bind much more weakly to the heparin column, eluting primarily in the wash (FT2-FT3) fractions. (c) The 2D chemical structures of a heparin subunit (left) and of the heparin analogue, sucrose octasulfate (SOS) (right). Heparin is a polymer of repeating subunits, indicated by ‘R’. Both structures have a disaccharide core and are decorated with negatively charged sulfate functional groups, heparin having three per subunit (and also a carboxyl group) and SOS having eight. (d) Ail binds the heparin analog at two sites. HBS-I is formed by K95, H121, K123 and K125, while HBS-II is formed by R40, K42 and K47. (e) Orthogonal stereo view of heparin (SOS) binding sites. Omit maps of the SOS molecules are shown as mesh models, with SOS molecules colored by atom type. See also Figures S2 and S3.
Ail binds a heparin analog at two extracellular sites near the membrane surface
To better understand the nature of heparin binding to Ail, we attempted co-crystallization with heparin (average molecular weight 10 kDa) and a smaller heparin analog, sucrose octasulfate (Fig. 6c). While we were unable to grow well-ordered Ail-heparin crystals, we obtained crystals of Ail-SOS that diffracted to 1.85 Å resolution (Table 1). Ail-SOS crystallized in spacegroup P32 with two molecules per asymmetric unit and a complex stoichiometry of 1:1. The primary SOS binding site (heparin binding site 1, designated HBS-I) is located on the extracellular surface on the outside of the β-barrel. Extracellular loops 2 and 3 contribute three lysines and one histidine to form HBS-I. In addition, a second SOS molecule is coordinated by two lysines and one arginine in extracellular loop 1 (HBS-II) by a symmetry-related molecule. The two binding sites correspond to the regions of positive charge we identified in the apo-Ail structure (Fig. 3c). Residues located within 3 Å of sulfate groups for both SOS molecules include R40, K47, K50, K95, H121, K123 and K125 (Fig. 1b, 6d, 6e, Fig. S2).
To further characterize binding to heparin, we made a series of mutants in HBS-I and HBS-II to reduce the positive charge. We created a quadruple mutant for HBS-I consisting of K95Q-H121Y-K123Q-K125Q and a triple mutant for HBS-II consisting of R40D-K42-Q-K47Q (substitutions were based on E. coli OmpX). We also created and attempted to characterize an HBS-I + II mutant, but this protein showed low expression levels and ran aberrantly on chromatography columns, suggesting that it did not fold properly. We purified the HBS-I and HBS-II mutant proteins and analyzed binding on a heparin column: both mutants showed significantly weaker binding, eluting primarily in the unbound fractions (Fig 6b). This is further evidence that the sites we identified through crystallography correspond to the functional heparin binding sites on Ail.
DISCUSSION
Y. pestis Ail has been identified as a potential drug and vaccine target because it is highly expressed in a rodent model of bubonic plague, accounting for 20 – 30% of the total outer membrane proteome at 37°C (Pieper et al, 2009; Sebbane et al, 2006). The crystal structures presented here represent the first structures of an Ail protein family member that is required for virulence. As anticipated, Ail is structurally related to E. coli OmpX, having 8 transmembrane β-strands with four extracellular loops that contribute to adhesion properties. Because the loops show very little sequence conservation and OmpX does not exhibit the same functions, determining the structure of Ail was essential for understanding how these activities are accomplished.
While individual residues contributing to adhesion in Y. pestis Ail have not been previously characterized, a mutagenesis study was done on Y. enterocolitica Ail to identify residues involved in adhesion, invasion, and serum resistance (Miller et al, 2001). The two Ail proteins have limited sequence similarity, however, and many of the residues identified in the Y. enterocolitia study are not conserved in Y. pestis Ail. For residues affecting adhesion, no point mutants conferred measurable decreases in binding. However a double mutant (D67G V68G, corresponding to D93 and F94 in Y. pestis) severely affected adhesion to CHO cells. The Y. pestis equivalent two residues are located on the 4th β-strand just above the bilayer interface. In our structures, the F94 side-chain extends toward the barrel interior and forms part of the binding site for the detergent molecule we find there. D93 points out from the barrel to the extracellular space. Since this region is hydrogen bonded to neighboring strands 3 and 5, two adjacent glycine mutations could conceivably disrupt the secondary structure and destabilize the fold, leading to the observed decrease in adhesion. The authors made a similar double mutant in Y. enterocolitica Ail, D67N V68G, which displayed a more subtle reduction in adhesion. Still, residues involved in adhesion and invasion to ECM proteins in Y. enterocolitica cluster around a hydrophobic cleft in the middle of the extracellular surface of Y. pestis Ail, where we find a molecule of bound detergent in both structures. The detergent molecule could potentially mimic binding of natural substrates such as laminin and fibronectin, although the idea remains to be tested. This may be a general feature of outer membrane protein adhesins: Neisserial surface protein A is an outer membrane adhesin with 10 β-strands, and it also displays a hydrophobic cleft on the extracellular surface with a bound detergent molecule in the crystal structure (Vandeputte-Rutten et al, 2003). In support of this hypothesis, a number of the residues required for heparin binding, including K123 and K125, do not strongly affect fibronectin binding when mutated to alanine, suggesting that ECM proteins may bind at site(s) distinct from heparin (Tsang and Krukonis, unpublished data).
By expressing Ail in the outer membrane of E. coli, we showed that it binds to the ECM proteins laminin and fibronectin, with a preference for laminin in our assay. Although we obtained lower binding to fibronectin using glass slides and counting stained bacteria, the results are somewhat method dependent (ESK, unpublished results) (Tsang et al, 2010). The Ail-fibronectin interaction appears to be more important for Yop delivery in HEp-2 cells, but a combination of anti-fibronectin and anti-laminin antibodies reduced Yop delivery significantly more than anti-fibronectin antibodies alone, so laminin plays some role in this process. The relative abundances of laminin and fibronectin in HEp-2 cells are unknown. We note that other cell lines may display different ratios of laminin to fibronectin on the surface, so in some cases laminin binding could assume greater importance. Certainly, the ability of Ail to bind both ECM proteins would be of advantage. The suggestion that ECM proteins such as laminin (this work) and fibronectin (Tsang et al, 2010) are natural targets for Ail is further supported by work on Salmonella Ail homologs, which demonstrated binding to the same proteins (Crago & Koronakis, 1999). Because we detected stronger binding to laminin, we went on to characterize this interaction at the protein level. Laminin is a 900 kDa glycoprotein composed of three disulfide-linked protein chains (Beck et al, 1990). To localize the interaction, we digested laminin with elastase to obtain a 40 kDa C-terminal fragment, LG4-5 (laminin G-like domains 4 and 5), and showed that specifically this fragment binds to Ail with an affinity similar to full-length laminin as detected by bacterial binding.
An additional component of the extracellular matrix is heparan sulfate proteoglycan, which has also been suggested to bind to Y. pestis (Zhang et al, 2008). Many bacterial virulence factors adhere to heparan sulfate proteoglycans on the surface of the host cell, (Rostand & Esko, 1997). Another Y. pestis adhesin, Pla, has been shown to bind heparan sulfate proteoglycan when expressed in E. coli (like Ail, it binds total ECM and laminin too) (Lahteenmaki et al, 1998). The Ail-SOS structure shows that Ail binds heparin at two extracellular sites very near the outer membrane, which should be at least partially masked by LPS in cells. Nevertheless, our data show that Ail binds heparin as a purified protein in detergent-containing buffers and also when expressed in the outer membranes of E. coli K12 or B strains, both of which lack O-antigen (Fig. 6a, b) (Boman & Monner, 1975; Prehm et al, 1976). Likewise, Y. pestis lacks O-antigen, so it should expose similar regions of Ail on the cell surface (Prior et al, 2001). The lack of O-antigen in the Y. pestis outer membrane may therefore facilitate Ail-heparin interactions. Thomas et al. reported that attachment of Y. pestis to a human respiratory cell line (a model of pneumonic plague) is inhibited by certain oligosaccharides including heparin (Thomas & Brooks, 2006). This report underscores the physiological relevance of heparin-targeted adhesion for Y. pestis and our work suggests that Ail may contribute to these interactions. Whether other Ail family members can bind heparin is unclear: a multiple seuqence alignment for Ail, OmpX, Rck and PagC indicates that HBS-II is not conserved, but HBS-I is partially conserved for PagC (Fig. S3). Although further investigations are needed to decipher the importance of heparin binding for Ail family members, our Ail-SOS crystal structure provides information that may be useful for anti-adhesion therapy using oligosaccharides.
Ail has at least two functions in Y. pestis – it acts an adhesin to facilitate contact with host cells, and it confers resistance to complement-mediated killing by host serum (Kolodziejek et al, 2007). Bartra and colleagues demonstrated that Ail is required for resistance to complement at 26°C and 37°C by probing a Y. pestis Ail deletion mutant and an E. coli strain that heterologously expressed Ail with serum from various animals (Bartra et al, 2008; Kolodziejek et al, 2007). This function allows Y. pestis to survive in blood, which is critical for establishing infection and for host-to-host transmission. Also necessary for successful infection is the delivery of Yops to host cells, and this function is brought about by the adhesion properties of Ail. Felek and Krukonis showed that a KIM5Δail mutant was severely inhibited in Yop delivery and was greatly attenuated for virulence in mice (Felek & Krukonis, 2009). More recently, Kolodziejek et al. deleted ail from the fully virulent CO92 Y. pestis strain and looked at its ability to induce pneumonic plague in both mice and rats (Kolodziejek et al, 2010). They found that this strain showed a delayed time to death in mice, and completely attenuated virulence in the rat. Since Ail does not display complement resistance against murine serum, the delayed infection was attributed to defects in adhesion, suggesting that adhesion is important but not critical to infection. By contrast, Ail provides resistance to killing by rat serum (as it does for human serum) so the total attenuation in the deletion mutant demonstrated the absolute requirement of complement resistance in virulence. We conclude that both functions of Ail are important for disease progression. Future efforts will include structural characterization of Ail-ECM and Ail-complement interactions to better understand precisely how Ail performs these distinct roles.
EXPERIMENTAL PROCEDURES
Plasmids
Plasmids for Ail, Ail mutants, and OmpX are described in the Supplemental Methods online.
Expression, refolding and purification of Ail
Detailed expression and purification protocols are described in the Supplemental Methods online. Briefly, Ail expressed as inclusion bodies was solubilized in guanidine-HCl, diluted into buffer containing dodecyl maltoside, and purified by anion exchange and gel filtration chromatography steps. During gel filtration, the detergent was exchanged for C8E4 (Anatrace) for crystallization. Natively folded Ail was purified from outer membranes by solubilizing with Elugent (Calbiochem), followed by purification in dodecyl maltoside (Anatrace) using Ni affinity chromatography, and detergent exchange into octyl glucoside (Anatrace) on a gel filtration column.
Crystallization, data collection and structure determination
Ail and Ail-SOS crystals were grown by vapor diffusion from PEG solutions and structures were determined by molecular replacement as described in the Supplementary Methods online.
ECM binding assays
BL21(DE3) cells containing pET20b alone or pET20b expressing Ail were grown in LB medium supplemented with 100 μg/ml carbenicillin. The cells were incubated with shaking O/N at 20°C and the temperature was shifted to 37°C the next day (Ail is expressed at low levels in this system without induction). At mid-log phase, the cells were harvested, washed with PBS, an adjusted to an OD600 of 0.6 with PBS. These cells (0.3 ml) were added to coated glass slides (coating procedure described below). For the experiments that included heparin (Sigma, H3393), the cells were pre-incubated with various concentrations of heparin, 10 μg anti-E.coli antibody (ABCAM, ab48416), or a 1:50 dilution of anti-Ail (serum from BALB/c mice injected with purified Ail) for 1 h at 4°C before being added to the slides. The slides were incubated for 30 min at room temperature with gentle shaking and then washed with phosphate buffered saline (PBS). Cells were fixed with formaldehyde and stained with crystal violet. Grids (3X3) were attached to the center of the back of each slide. One image near the center of each grid was collected at 1600X magnification. The bacteria in each image were counted. The results were normalized such that the average number of cells bound to ECM equaled 100% binding. For the experiments that included heparin, the average number of cells bound to laminin without preincubation with heparin was set to 100%.
To prepare the coated glass slides (BD Falcon, 354108), 0.3 ml of a 20 μg/ml solution of the coat protein was added per well. The coat proteins included: (laminin, Sigma L6274; fibronectin, Sigma F2006; collagen, Sigma C7521; ECM, Sigma E1270; heparan sulfate proteoglycan, Sigma H4777; LG4-5, elastase fragment purified as described in this work; BSA, MP Biomedical 160069). The slides were incubated O/N at 4°C, washed 2X with PBS, blocked for 2 h with 5% (wt/vol) BSA in PBS, and washed again with PBS prior to adding the cells. For experiments using the anti-laminin antibody (ABCAM, ab11575), various concentrations of the antibody diluted in 2.5% (wt/vol) BSA were added to the protein coated wells that had been blocked with 5% BSA as described above. An anti E. coli antibody (10 μg) (ABCAM, ab48416) was used as a control. The antibodies were allowed to bind at room temperature for 5 h. The slides were then washed with PBS prior to adding cells.
Purification of Ail on a heparin column
To detect the binding of isolated Ail protein to heparin, Ail purified from OM was applied to a 1 ml HiTrap Heparin column (GE Healthcare) equilibrated with binding buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA and 1% OG. The column was washed with 4 column volumes of binding buffer and eluted with a NaCl step gradient of 0.4, 0.8, and 1.2 M NaCl.
Digestion, purification, and detection of laminin binding
Laminin (0.5 mg) was digested with 0.1 mg of porcine elastase for 16 hrs. Digested laminin was applied to a Hi-trap heparin column (5 ml, GE Healthcare) and eluted using a 1 M NaCl gradient. Unbound and eluted fractions were used to coat a 96-well polystyrene plate at 4°C overnight. Subsequently, the supernatant was removed and wells were blocked with PBST containing 3% BSA for 1 hr. Wells were incubated with 1 μg/ml Ail (50 μl) in blocking buffer supplemented with 0.1% DDM for 1 hr and washed 3 times with PBST/DDM. An anti-His antibody (HRP conjugate) was added to each well and incubated for 1 hr, followed by 3 washes with PBST. To detect antibody binding, 50 μl TMB was added as a substrate for HRP and incubated for 15 min, followed by addition of 50 μl 1 M HCl to each well to stop the reaction. The absorbance was monitored at 450 nm.
HEp-2 cell cytotoxicity
Yop delivery assays by Y. pestis were performed as previously described (Tsang et al, 2010). Briefly, HEp-2 cells were cultivated until they reached about 50% confluence in 24-well tissue culture plates. Y. pestis KIM5 D27 derivatives were cultured in HIB overnight at 28°C. Overnight cultures were diluted 1:50 in fresh HIB and incubated for 3hours at 28°C + 100 μM IPTG. Tissue culture plates were washed twice with PBS and 0.22 - 0.24 ml serum-free MEM tissue culture medium was added to each well (final volume with bacteria and antibodies was 0.25 ml). As appropriate, cells were incubated with a 1:25 dilution (10 μl of ~0.5mg/ml) dilution of anti-fibronectin antibody (Sigma, F3648), anti-laminin antibody (Abcam, ab11575), antiPsaE antibody (Tsang et al, 2010), anti-Ail mouse serum or matched mouse serum from BALB/c mice. 10 μl bacteria resuspended at an OD620=0.6 in were added to each well (an MOI of 10). Plates were incubated at 37°C in 5% CO2 for 2.5-3.5 hours. Cells were fixed with 0.3 ml methanol and stained with 0.3 ml 0.76 mg/ml Geimsa stain. Rounding was observed and pictures were taken with a phase-contrast microscope. Cytotoxicity was enumerated in a blinded fashion by counting total cells and the number of rounded dark-staining cells experiencing cytotoxicity in three microscopic fields (~150 - 300 cells/field). Percent cytotoxicity was calculated by dividing rounded cells by total cells and normalizing to 100% for strain KIM5 D27Δ3 + pMMB207-Ail. The absolute level of cytotoxicity over multiple experiments ranged from 65-75% cytotoxicity for this strain. Data presented are from triplicate measurements over 2 experiments (n = 6). Statistical significance was assessed using the Student's t-test.
Supplementary Material
HIGHLIGHTS.
Ail is an 8-stranded β-barrel having 4 extracellular loops that function in adhesion
Ail-laminin and Ail-fibronectin interactions mediate delivery of Yops to host cells
Ail has two binding sites for heparin that also confer adhesion to host cells
ACKNOWLEDGMENTS
We thank N. Suzuki and for providing samples, technical support and discussions, and H. Bernstein and J. Fairman for reading the manuscript. S.Y., P.L., T.J.B., N.N., and S.K.B. are supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases. B.J.H. is supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases. S.F., T.M.T., and E.S.K. are supported by NIH grant R21AI090194 to E.S.K. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) beamline 22-ID at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Coordinates and structure factors for Ail and Ail-SOS have been deposited in the Protein Data Bank with accession codes 3QRA and 3QRC.
SUPPLEMENTAL INFORMATION
Supplemental information includes three figures, supplemental methods and references.
References
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartra SS, Styer KL, O'Bryant DM, Nilles ML, Hinnebusch BJ, Aballay A, Plano GV. Resistance of Yersinia pestis to complement-dependent killing is mediated by the Ail outer membrane protein. Infect Immun. 2008;76:612–622. doi: 10.1128/IAI.01125-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K, Hunter I, Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J. 1990;4:148–160. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- Boman HG, Monner DA. Characterization of lipopolysaccharides from Escherichia coli K-12 mutants. J Bacteriol. 1975;121:455–464. doi: 10.1128/jb.121.2.455-464.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crago AM, Koronakis V. Binding of extracellular matrix laminin to Escherichia coli expressing the Salmonella outer membrane proteins Rck and PagC. FEMS Microbiol Lett. 1999;176:495–501. doi: 10.1111/j.1574-6968.1999.tb13703.x. [DOI] [PubMed] [Google Scholar]
- Felek S, Krukonis ES. The Yersinia pestis Ail protein mediates binding and Yop delivery to host cells required for plague virulence. Infect Immun. 2009;77:825–836. doi: 10.1128/IAI.00913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felek S, Tsang TM, Krukonis ES. Three Yersinia pestis adhesins facilitate Yop delivery to eukaryotic cells and contribute to plague virulence. Infect Immun. 2010;78:4134–4150. doi: 10.1128/IAI.00167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M, Carniel E, Courvalin P. Resistance of Yersinia pestis to antimicrobial agents. Antimicrob Agents Chemother. 2006;50:3233–3236. doi: 10.1128/AAC.00306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffernan EJ, Reed S, Hackett J, Fierer J, Roudier C, Guiney D. Mechanism of resistance to complement-mediated killing of bacteria encoded by the Salmonella typhimurium virulence plasmid gene rck. J Clin Invest. 1992;90:953–964. doi: 10.1172/JCI115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Kirjavainen V, Jarva H, Biedzka-Sarek M, Blom AM, Skurnik M, Meri S. Yersinia enterocolitica serum resistance proteins YadA and ail bind the complement regulator C4b-binding protein. PLoS Pathog. 2008;4:e1000140. doi: 10.1371/journal.ppat.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejek AM, Schnider DR, Rohde HN, Wojtowicz AJ, Bohach GA, Minnich SA, Hovde CJ. Outer membrane protein X (Ail) contributes to Yersinia pestis virulence in pneumonic plague and its activity is dependent on the lipopolysaccharide core length. Infection and immunity. 2010;78:5233–5243. doi: 10.1128/IAI.00783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejek AM, Sinclair DJ, Seo KS, Schnider DR, Deobald CF, Rohde HN, Viall AK, Minnich SS, Hovde CJ, Minnich SA, Bohach GA. Phenotypic characterization of OmpX, an Ail homologue of Yersinia pestis KIM. Microbiology. 2007;153:2941–2951. doi: 10.1099/mic.0.2006/005694-0. [DOI] [PubMed] [Google Scholar]
- Lahteenmaki K, Virkola R, Saren A, Emody L, Korhonen TK. Expression of plasminogen activator pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infection and immunity. 1998;66:5755–5762. doi: 10.1128/iai.66.12.5755-5762.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecsas J, Welch R, Erickson JW, Gross CA. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J Bacteriol. 1995;177:799–804. doi: 10.1128/jb.177.3.799-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VL, Beer KB, Heusipp G, Young BM, Wachtel MR. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol Microbiol. 2001;41:1053–1062. doi: 10.1046/j.1365-2958.2001.02575.x. [DOI] [PubMed] [Google Scholar]
- Miller VL, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott U, Odermatt E, Engel J, Furthmayr H, Timpl R. Protease resistance and conformation of laminin. Eur J Biochem. 1982;123:63–72. doi: 10.1111/j.1432-1033.1982.tb06499.x. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeno-Tarraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson KE, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, Perry RD, Fleischmann RD, Peterson SN. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology. 2009;155:498–512. doi: 10.1099/mic.0.022160-0. [DOI] [PubMed] [Google Scholar]
- Prehm P, Schmidt G, Stirm S. On the mutations responsible for the rough phenotype of Escherichia coli B. J Gen Microbiol. 1976;97:121–124. doi: 10.1099/00221287-97-1-121. [DOI] [PubMed] [Google Scholar]
- Prior JL, Hitchen PG, Williamson DE, Reason AJ, Morris HR, Dell A, Wren BW, Titball RW. Characterization of the lipopolysaccharide of Yersinia pestis. Microb Pathog. 2001;30:49–57. doi: 10.1006/mpat.2000.0411. [DOI] [PubMed] [Google Scholar]
- Pulkkinen WS, Miller SI. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J Bacteriol. 1991;173:86–93. doi: 10.1128/jb.173.1.86-93.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostand KS, Esko JD. Microbial adherence to and invasion through proteoglycans. Infect Immun. 1997;65:1–8. doi: 10.1128/iai.65.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbane F, Lemaitre N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc Natl Acad Sci U S A. 2006;103:11766–11771. doi: 10.1073/pnas.0601182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Brooks T. Attachment of Yersinia pestis to human respiratory cell lines is inhibited by certain oligosaccharides. J Med Microbiol. 2006;55:309–315. doi: 10.1099/jmm.0.46102-0. [DOI] [PubMed] [Google Scholar]
- Tsang TM, Felek S, Krukonis ES. Ail binding to fibronectin facilitates Yersinia pestis binding to host cells and Yop delivery. Infect Immun. 2010;78:3358–3368. doi: 10.1128/IAI.00238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte-Rutten L, Bos MP, Tommassen J, Gros P. Crystal structure of Neisserial surface protein A (NspA), a conserved outer membrane protein with vaccine potential. J Biol Chem. 2003;278:24825–24830. doi: 10.1074/jbc.M302803200. [DOI] [PubMed] [Google Scholar]
- Vogt J, Schulz GE. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure. 1999;7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- Zhang P, Skurnik M, Zhang SS, Schwartz O, Kalyanasundaram R, Bulgheresi S, He JJ, Klena JD, Hinnebusch BJ, Chen T. Human dendritic cell-specific intercellular adhesion molecule-grabbing nonintegrin (CD209) is a receptor for Yersinia pestis that promotes phagocytosis by dendritic cells. Infect Immun. 2008;76:2070–2079. doi: 10.1128/IAI.01246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.