Abstract
Protein arginine methyltransferase 10 (PRMT10) is a type-I arginine methyltransferase essential for regulating flowering time in Arabidopsis thaliana (At). We present a 2.6 Å resolution crystal structure of AtPRMT10 in complex with a reaction product, S-adenosylhomocysteine. The structure reveals a dimerization arm 12–20 residues longer than PRMT structures elucidated previously; as a result, the essential AtPRMT10 dimer exhibits a large central cavity and a distinctly accessible active site. We employ molecular dynamics to examine how dimerization facilitates AtPRMT10 motions necessary for activity, and show that these motions are conserved in other PRMT enzymes. Finally, functional data reveal that the N-terminal ten residues of AtPRMT10 influence substrate specificity, and that enzyme activity is dependent on substrate protein sequences distal from the methylation site. Taken together, these data provide insights into the molecular mechanism of Arabidopsis thaliana PRMT10 as well as other members of the PRMT family of enzymes. They highlight differences between AtPRMT10 and other PRMTs, but also indicate that motions are a conserved element of PRMT function.
Keywords: Arabidopsis, epigenetics, flowering time, protein structure, protein motion
Introduction
Protein arginine methyltransferases (PRMTs) catalyze the transfer of methyl groups from S-adenosylmethionine (SAM) to arginine residues of target proteins, and release S-adenosylhomocysteine (SAH) as a product 1. The post-translational methylation of arginines is observed widely in eukaryotes and plays essential roles in many biological processes, such as signal transduction, chromatin remodeling, RNA processing, gene transcription, DNA repair and cellular transport 1–8. PRMTs are generally classified as type I or type II 1. Both types catalyze the production of ω-NG-monomethylarginine, but they generate distinct dimethyl arginine derivatives. Type I enzymes (e.g., PRMT1, 3, 4, 6, 8) specifically produce asymmetric ω-NG,N′G-dimethylarginine, while type II enzymes (e.g., PRMT5, 7 and FBXO11) only produce symmetric dimethylarginine 9. The dysfunctions of mammalian PRMTs have been correlated with the development of cancer as well as autoimmune, cardiovascular, pulmonary and neuro-developmental diseases 10–16.
While PRMTs have a relatively conserved catalytic core, the portions of each enzyme N-terminal to the catalytic core (the “N-terminal additions”) are divergent in sequence and have been demonstrated to be important for the substrate specificity. For example, the zinc-finger domain within the N-terminal addition of PRMT3 is essential for its recognition of RNA-associated targets 17. Previous structural studies have shown that the PRMT catalytic core is composed of three domains: an N-terminal SAM binding domain, a central arm domain, and a C-terminal β-barrel domain 18–23. The main substrate binding site is located in a cleft formed between the SAM binding domain and the β-barrel domain 19,20. Dimerization is a conserved feature in PRMTs and has been established to be essential for the methyltransferase activity of PRMTs by facilitating SAM binding 19,20.
PRMT methyltransferase activity is regulated by several characteristics of the target protein. For example, the local sequence of the methylation site is an important determinant of arginine methylation 24,25. PRMT-catalyzed reactions typically occur within glycine- and arginine-rich motifs, such as “RG”, “RGG” and “RXR” 26, although exceptions have been noted 25. The activity of PRMTs can also be affected by the sequences distal to the methylation site 27 and by protein binding partners 28,29. Circumstantial evidence has suggested that PRMTs often form complexes with other proteins in vivo, and that these proteins impact subcellular location and substrate recognition 30,31.
AtPRMT10 is a plant-specific type I PRMT that plays an essential role in the regulation of flowering time in Arabidopsis 32. Genetic disruption of AtPRMT10 causes delayed flowering due to up-regulated transcription of a major flowering repressor, FLOWERING LOCUS C (FLC) 32. Biochemical studies showed that AtPRMT10 can specifically methylate arginine-3 of both histone H4 and histone H2A in vitro, and preferentially produces asymmetrical dimethylarginines. Besides AtPRMT10, eight other AtPRMTs have been identified in the Arabidopsis genome, including AtPRMT1a, AtPRMT1b, AtPRMT3, AtPRMT4a, AtPRMT4b, AtPRMT5, AtPRMT6 and AtPRMT7. These AtPRMT paralogs likely have diverse properties in cellular location, substrate specificity and protein-protein interaction 1,7.
Here we report the crystal structure of AtPRMT10 in complex with a product of its enzymatic reaction, SAH. This structure provides insights into how AtPRMT10 interacts with peptides, and reveals structural features that may confer unique substrate specificity to AtPRMT10, including the role of the AtPRMT10 N-terminal addition in the enzyme function. Our studies also show that AtPRMT10 exists predominantly in a dimeric state in solution, and disruption of dimerization causes loss of activity. We further examine the impact AtPRMT10 dimerization has on enzyme motion using molecular dynamics (MD) simulations. Our results highlight distinct differences between AtPRMT10 and other structurally-characterized PRMTs, but also indicate that motions are a conserved element of PRMT function.
Results
Crystal Structure of the AtPRMT10-SAH Complex
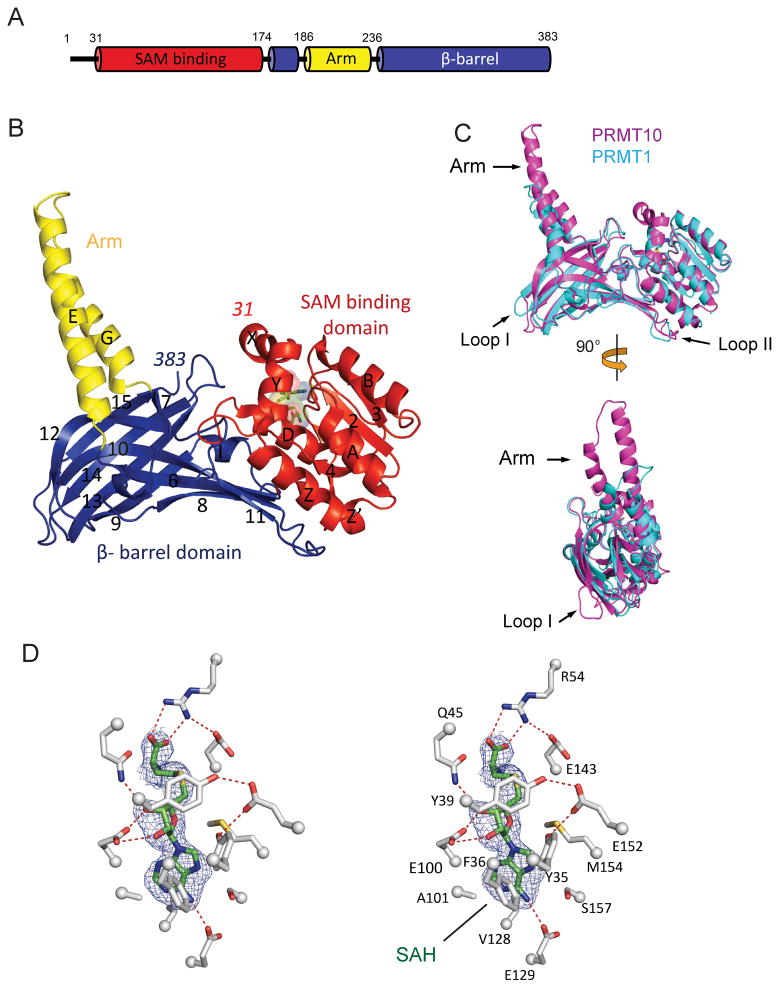
The structure of AtPRMT10 (residues 11–383) in complex with SAH was determined by molecular replacement and refined to 2.6 Å resolution (Table I). The N-terminal twenty residues of the AtPRMT10 construct employed (residues 11–30) lack electron density and were not placed in the final refined model. AtPRMT10 exhibits three sequentially folded domains: an N-terminal SAM binding domain (residue 31–174), a central arm domain (residues 187–236), and a C-terminal β-barrel domain (residues 175–186 and residues 237–383) (Figure 1A & B). The SAM binding domain is composed of two N-terminal helices (αX & αY, residues 31–50) followed by a classical Rossman fold (residues 51–174) consisting of five α helices (αZ, αZ′, αA, αB, αD) and five β strands (β1 to β5). The consensus Rossman fold has been observed in other known SAM-dependent methyltransferases 33,34, while the two N-terminal helices (αX & αY) are unique to PRMTs 20. The β-barrel domain, forming close contacts with the SAM-binding domain at one end of its barrel, harbors ten β-strands (β6 to β15) and two short α-helices (αH and αI). The arm domain, exhibiting a helix-turn-helix fold, is inserted in between β6 and β7 of the β-barrel domain and protrudes from the main body of the protein. Sequence analysis reveals four PRMT signature motifs in AtPRMT10 (Figure 2). Motif I (YFxxY) and Motif II (DVGxGxG) are directly involved in the binding of cofactor SAM. Motif III (SExMGxxLxxExM), harbors two critical catalytic residues E143 and E152. Mutation of either of these two residues completely disrupted the methyltransferase activity of AtPRMT10 (data not shown). Motif IV (or the THW motif) is the most highly conserved sequence among PRMTs and is directly involved in the formation of the active site. As expected, disruption of motif IV is accompanied with complete loss of the methyltransferase activity of AtPRMT10 (data not shown).
Table I.
Crystallographic data and refinement statistics
| Resolution (Å) (highest shell) | 33.0–2.61 (2.67–2.61) |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a= 80.55 b=86.69 c=114.74 β=89.97 |
| Twinning fraction | 0.5 |
| Number of total reflections | 48404 |
| Number of unique reflections | 46744 |
| Rsyma (%) (highest shell) | 12.0 (58.0) |
| Completeness (%) (highest shell) | 96.6 (91.0) |
| Mean I/σ (highest shell) | 13.0 (2.30) |
| Average redundancy | 2.90 (2.60) |
| Rcryst b(%) (highest shell) | 18.2 (35.0) |
| Rfreec (%) (highest shell) | 22.4 (41.2) |
| RMSD | |
| Bond lengths (Å) | 0.007 |
| Bond angles ( ° ) | 1.17 |
| Dihedral angles ( ° ) | 16.2 |
| Number of atoms per AU | |
| Protein | 10751 |
| Ligand | 104 |
| Solvent | 174 |
Rsym = Σ|I − 〈I〉|/ΣI where I is the observed intensity and 〈I〉 is the average intensity of multiple symmetry-related observation of the reflection.
Rcryst = Σ||Fobs| − |Fcalc||/Σ|Fobs| where Fobs and Fcalc are the observed and calculated structure factors, respectively.
Rfree = Σ||Fobs| − |Fcalc||/Σ|Fobs| for 5% of the data not used at any stage of the refinement.
Figure 1. Crystal structure of AtPRMT10.
(A) Domain architecture of AtPRMT10 from Arabidopsis thaliana. The SAM binding domain (residues 31–174) is shown in red, the β-barrel domain (residues 175–186, 237–383) in blue, and the arm domain (or dimerization domain, residues 187–236) in yellow. (B) The crystal structure of the AtPRMT10-SAH complex with key secondary structure elements labeled (helices are labeled X through I and β-strands are numbered 1 through 15). The bound SAH is shown as sticks and spheres. The structure is colored as in Figures 1A–B. The first and last residues of AtPRMT10 are indicated. (C) Two views of the superimposition of AtPRMT10 (residues 31–383, magenta) with rat PRMT1 (residues 41–353, cyan, PDB entry 1ORI). Key structural differences between AtPRMT10 and PRMT1 (located in the dimerization domain and two loops in the β-barrel domain) are indicated by arrows. (D) A stereo-view representation of SAH binding. A 2.6 Å resolution simulated annealing omit map of SAH contoured at 2.5σ (blue mesh) is shown. Hydrogen bonds are indicated by red dashed lines.
Figure 2. Structure-based sequence alignment of AtPRMT10, rat PRMT1 (PDB: 1ORI), rat PRMT3 (PDB: 1F3L), yeast RMT1 (1G6Q) and mouse CARM1 (PDB: 3B3F).
Secondary-structure elements are shown across the top of the aligned sequences. Residue numbers are provided on the right. Invariant and similar residues are highlighted in black and gray, respectively. Protein domains are colored as in Figures 1A–B. The four PRMT signature motifs are labeled. Residues involved in SAM binding and dimerization are highlighted by red and black stars, respectively.
The structure of AtPRMT10 exhibits a similar overall fold relative to other PRMTs of known structure, exhibiting, for example, a 1.8 Å root-mean-square deviation over 245 Cα positions with PRMT1 (residues 41–354). However, a strikingly unique feature of AtPRMT10 is its dimerization arm, consisting of two straight anti-parallel α-helices, which is significantly longer (41 Å) than that of other PRMTs (e.g., PRMT1, 22 Å; PRMT3, 22 Å; CARM1, 34 Å) (Figure 1C). AtPRMT10 also differs from other PRMTs in two loop regions of the β-barrel domain (Figure 1C). Sequence alignment indicates that these loops are relatively conserved among AtPRMT10 orthologs (Figure S1), but highly divergent among PRMT paralogs (Figure 2). Loop I is located adjacent to a conserved substrate binding site of PRMTs (see below). Acidic residues in Loop II have been shown to be important for the interaction of PRMT1 with its substrates 35.
AtPRMT10 Active Site
In the AtPRMT10-SAH complex, SAH binds within a deep pocket formed by the three N-terminal α-helices (αX, αY and αZ) and the carboxyl ends of the parallel β-strands (β1 to β5) (Figure 1D). Most of the residues involved in SAH binding are highly conserved among type I PRMTs (Figure 2), indicating that members of the type I PRMT family likely share similar mechanisms in cofactor binding and catalysis. Hydrogen bonding plays a major role in the interaction of AtPRMT10 with SAH, with six such interactions formed between AtPRMT10 and the three moieties of SAH (adenine, ribose and homocysteine). R54 of the helix αZ forms bifurcated hydrogen bonds with the terminal carboxylate group of the homocysteine moiety. For the ribose moiety, hydrogen bonds are observed between the two main-chain hydroxyl groups and the side chains of E100 of strand β2 and Q45 of helix αY. The adenine group is recognized by the E129 from the loop between β2 and β4. In addition to hydrogen bonding, the main-chain of the glycine rich loop (G78 and G80) and the side-chains of seven other residues (A101, V128, F36, M154, S157, Y35 and Y39) form van der Waals contacts with SAH. Given the small difference between the chemical structure of SAM and SAH, it is expected that SAM binds to the active site in a manner similar to that observed here for SAH.
AtPRMT10 Dimer
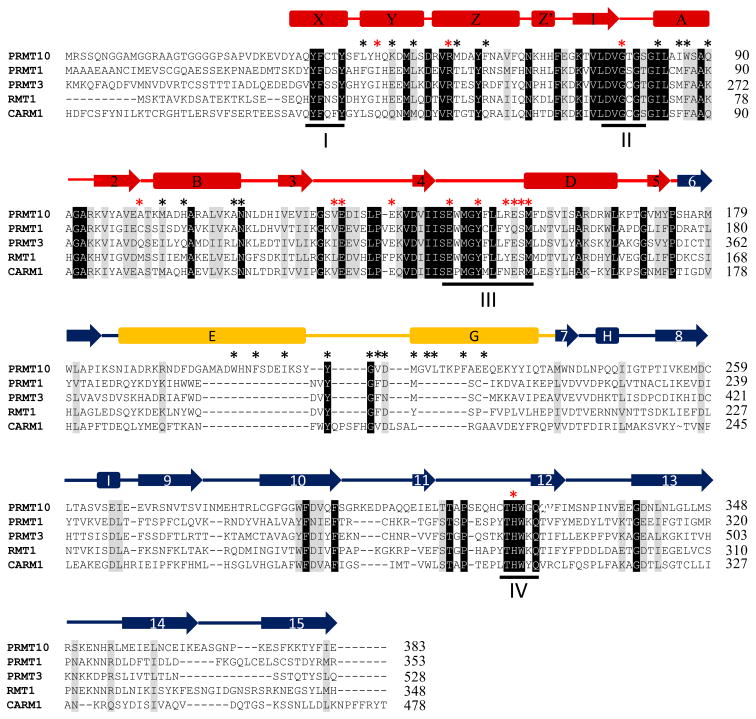
AtPRMT10 forms a ring-like homodimer through the interaction between the dimerization arm (αE-loop-αG) of one monomer and the outer surface (αY, αZ, αA & αD) of the SAM binding domain of the other monomer (Figures 3A & 3B). Both active sites are located at the periphery of the central cavity formed upon dimerization of AtPRMT10. As observed for other PRMTs, hydrophobic interactions are a major force during the formation of the AtPRMT10 dimer. A network of three hydrogen bonds is also observed at the PRMT dimer interface, with the side-chains of Q90 and N115 forming hydrogen bonds with the main-chains of G215 and D217 respectively (Figure 3C). The hydrogen bonds between N115 and D217 are highly conserved among PRMTs (Figure 2). Another conserved residue at the dimer interface is G215, whose small side-chain is apparently favorable for the formation of the sharp turn at the tip of the dimerization arm. Overall, the residues on the surface of the SAM binding domain that produce the AtPRMT10 dimer interface are highly conserved when compared to other PRMTs. In contrast, however, the residues that form the dimerization arm of AtPRMT10 exhibit little or no conservation with homologous enzymes (Figure 2 and Figure S2).
Figure 3. Dimer formation of AtPRMT10.
(A) The crystal structure of an AtPRMT10 dimer. The structure is colored and labeled as in Figures 1A–B. The location of each dimer interface is highlighted by an arrow. The dimer is formed by the interaction between the dimerization arm of one monomer and the outer surface of the SAM binding domain of the other monomer. (B) A surface representation of two views of an AtPRMT10 dimer, with two monomers are colored in gray and pink, respectively. (C) An expanded stereo-view of the dimer interface. Two monomers are colored as in Figure 2B. Residues involved in dimer formation are shown as sticks and labeled. Hydrogen bonds are indicated by red dashed lines.
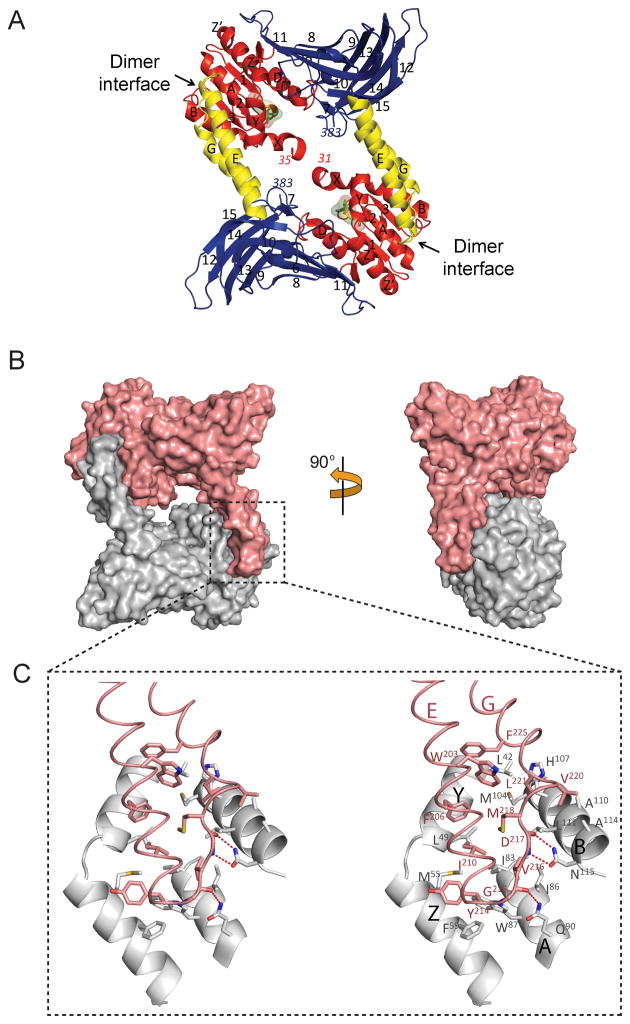
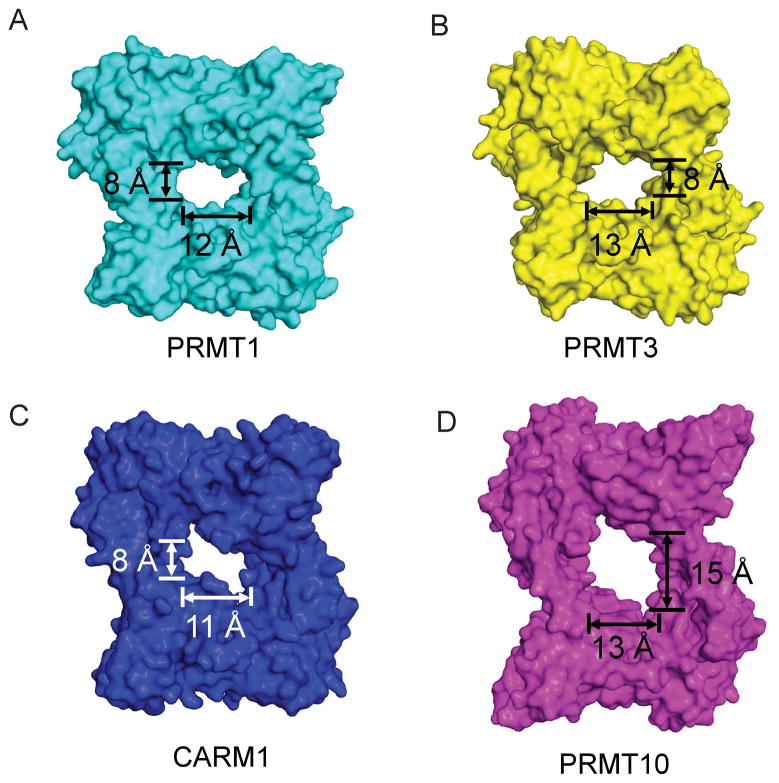
Notably, the central cavity of AtPRMT10 is significantly larger than those of other PRMTs with known structure (Figure 4). AtPRMT10 creates a cavity 15 Å high by 13 Å wide (15 × 13 Å), while those of PRMT1, PRMT3 and CARM1 exhibit cavities that are 8×12, 8×13 and 8×11 Å, respectively (Figure 4). The dimerization arms of PRMTs are composed of a helical segment and a connecting loop at the tip of the dimerization arm (Figure 1B &1C). Although the connecting loop is involved in forming the dimer interface, the size of the central cavity is mainly determined by the length of the helical segment. Relative to other PRMTs, CARM1 has a much longer connecting loop at the tip of the dimerization motif. As a result, although the dimerization arm of CARM1 (34 Å) is longer than that of PRMT1 (22 Å) and PRMT3 (22 Å), its helix segment (18 Å) has similar size with those of PRMT1 (17 Å) and PRMT3 (17 Å). Accordingly, the central cavity of CARM1 is comparable in size to those of PRMT1 and PRMT3. Consistent with the dimer observed in the crystal structure, our results from dynamic light scattering and gel filtration experiments confirmed that AtPRMT10 exists predominately as dimer in solution, and that the oligomeric state of the enzyme is independent of SAH binding (Table II).
Figure 4. Surface representation of various PRMT paralogs, including rat PRMT1 (PDB: 1ORI) (A), rat PRMT3 (PDB: 1F3L) (B), mouse CARM1 (PDB: 3B3F) (C) and AtPRMT10 (D).
For consistency, the sequences N-terminal to helix X (including helix X) are deleted from the structure. The dimensions of the central cavities are indicated.
Table II.
Oligomeric states of AtPRMT10 mutants
| Monomer size (kDa) | GF size (−SAH) (kDa) | GF size/Monomer size | −SAH | +SAH (1 mM) | |||
|---|---|---|---|---|---|---|---|
| DLS size (kDa) | DLS size/Monomer size | DLS size (kDa) | DLS size/Monomer size | ||||
| wt | 43.1 | 80 | 1.8 | 93 | 2.2 | 92 | 2.1 |
| ΔN10 | 42.1 | N.D. | N.D. | 89 | 2.1 | 87 | 2.1 |
| ΔN20 | 41.4 | N.D. | N.D. | 83 | 2.0 | 83 | 2.0 |
| ΔN30 | 40.4 | 68 | 1.7 | 74 | 1.8 | 80 | 2.0 |
| Δ203-225 | 40.8 | 34 | 0.9 | 66 | 1.6 | 61 | 1.5 |
N.D., not determined; DLS, dynamic light scattering; GF, gel filtration
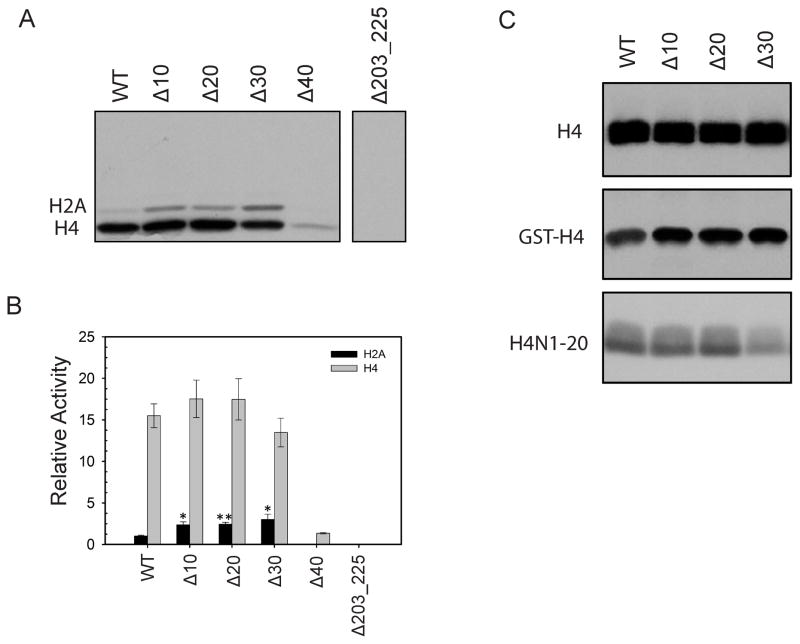
To test the importance of the dimer interface observed in the crystal structure during AtPRMT10 function we designed an arm mutant, Δ203-225, in which the part of the dimerization arm that forms the dimer interface was replaced with a stretch of glycine and serine residues (GGSGGS). AtPRMT10 Δ203-225 was stably over-expressed in E. coli, suggesting that it was well folded. The oligomeric state of AtPRMT10 Δ203-225 was examined using dynamic light scattering and gel filtration experiments (Table II). Our results show that mutation of the dimerization arm disrupted dimer formation. The impact of dimerization on the methyltransferase activity of AtPRMT10 was examined by measuring the activity of the arm mutant Δ203-225. The arm mutant displayed no observable activity toward H2A and H4 (Figure 5A and 5B), indicating that dimerization is essential for the methyltransferase activity of AtPRMT10.
Figure 5. Methyltransferase activities of different AtPRMT10 constructs in vitro.
(A) Indicated AtPRMT10 proteins (5 μg) were used for in vitro methylation activity assay. The reaction mixture was separated on a 15% SDS-PAGE and the autoradiograph of the gel is recorded. The experiments were performed in triplicate and a typical result is shown here. (B) Quantification of the results from Figure 5A. The relative activities presented here were calculated by considering the activity of wild-type AtPRMT10 over H2A as one. The activities of AtPRMT10 mutants over H2A are significantly higher than that of wild-type enzyme (n=3; error bars represent SEM; Student t test *p < 0.03, **p < 0.002). (C) Methyltransferase activities of different AtPRMT10 constructs on H4, GST-H4 and H4N1-20.
AtPRMT10 Surface Electrostatics
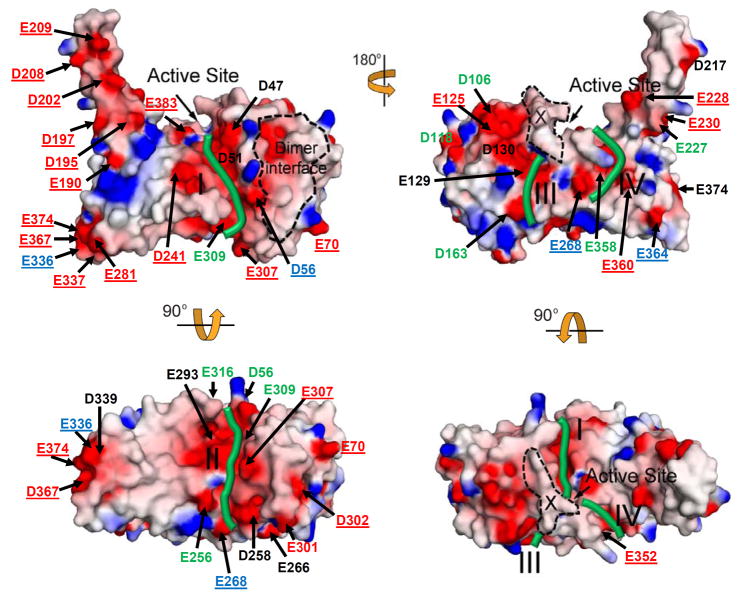
Surface charge distribution appears to impact the function of PRMTs. For example, published data have suggested that surface charges are crucial for the interaction of PRMT with substrates and other proteins 19,20. Figure 6 illustrates the surface charge distribution of AtPRMT10. As seen in other PRMTs, the surface of AtPRMT10 contains numerous acidic patches, especially around the active site. However, there are notable differences in the surface charge distribution of AtPRMT10 compared to other PRMTs of known structure (Figure 6). In particular, the unusually long dimerization arm of AtPRMT10 contains ten acidic residues (E190, D195, D197, D202, D208, E209, D217, E227, E228, E230) (Figure 6) that generate a relatively large acidic surface along this domain relative to other PRMTs. A second difference is observed at one end of the β-barrel domain, where AtPRMT10 has a large acidic patch formed by residues E281, E336, E337, D339, E367 and E374 (Figure 6). Other PRMTs contain fewer acidic residues in this region (Figure 2). Acidic amino acid residues in this location have been shown to be important for the substrate interaction of PRMT135.
Figure 6. Surface electrostatic potential of AtPRMT10.
Acidic surfaces are represented in red; basic surfaces in blue and neutral surfaces in gray. Acidic surface residues are labeled and colored based on their conservation among five PRMT paralogs as indicted by the sequence alignment in Figure 2 (black, conservation ≥80%; green, 40%<conservation<80%; blue, 20%<conservation ≤40%; unique for AtPRMT10, red). The location of putative substrate binding grooves, the active site, the dimer interface and helix αX are highlighted and labeled. The top left view has the same orientation as in Figure 1B.
Structural studies of PRMT1 have indicated the location of the substrate binding groove of this enzyme 19. Based on the location of acidic patches and the shape of the AtPRMT10 surface in light of other PRMTs of known structure, we have identified four putative substrate binding grooves on the surface of AtPRMT10 (Figure 6). Binding grooves I and II are located in the cleft formed between the SAM binding domain and the β-barrel domain and are directly connected to the active site. Binding grooves III and IV lie on the surface of the β-barrel domain. Substrates can also enter the active site through binding groove III. A high degree of conservation is maintained in the residues that form binding grooves I and II (Figure 6 and S2), suggesting the conserved role for these two binding grooves during substrate interaction. In contrast, little conservation is observed for the residues that form binding grooves III and IV (Figure 6 and S2). It is possible that the unique compositions of binding grooves III and IV may confer unique substrate specificities upon AtPRMT10 compared to other PRMTs.
Increased Active Site Accessibility in AtPRMT10
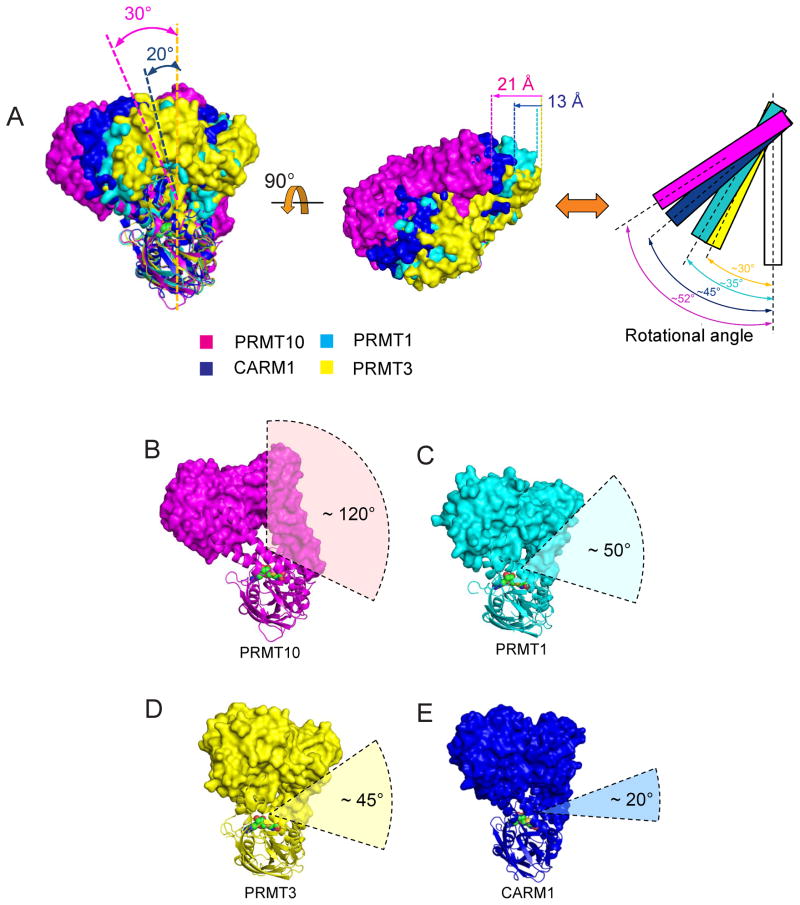
While the PRMT family shares a three-domain architecture and a dimeric oligomerization state, the relative orientation of the two monomers in a functional dimer significantly varies between different PRMTs due to the diversity in dimerization arm length and composition. Consequently, the dimeric forms of different PRMTs do not superimpose well. When we align different PRMTs based on one of their two monomers (the “bottom monomers” in Figure 7A, left panel), the other monomers (the “top monomers”) are translated to distinct locations. In Figure 7A (left panel), the top monomers of PRMT1 (cyan) and PRMT3 (yellow) are located directly above their bottom monomers, while the top monomers of AtPRMT10 (magenta) and CARM1 (blue) are positioned away from the vertical by 30° and 20°, respectively. The top monomers of AtPRMT10 and CARM1 are also observed to be translated leftward 21 Å and 13 Å to the left, respectively, relative to the position of PRMT1 and PRMT3 (Figure 7A, middle panel). Finally, the angles formed by the two monomers of a PRMT dimer vary significantly among enzyme paralogs, ranging from 30° in PRMT3 to 52° in AtPRMT10 (shown schematically in Figure 7A, right panel).
Figure 7. AtPRMT10 exhibits a uniquely accessible active site.
(A) Superimposition of AtPRMT10 (magenta) with rat PRMT1 (cyan, PDB: 1ORI), rat PRMT3 (yellow, PDB: 1F3L) and mouse CARM1 (blue, PDB: 3B3F). The bottom monomers are used for the alignment. The left view rotates about 90° along the vertical axis with respect to the view shown in Figure 3A. The angles of the rotation of AtPRMT10 and CARM1 relative to PRMT3 are shown. For clarity, the bottom monomers are not shown in the middle panel. The left edges of four monomers are indicated by vertical dashed lines with corresponding colors. The distance of the leftward translation of AtPRMT10 and CARM1 relative to PRMT3 are shown. The right panel is a schematic representation of the middle panel, with PRMT monomers shown as rectangles. The superimposed bottom monomers are shown and colored in white. (B)–(E) Views of the dimeric AtPRMT10, rat PRMT1, rat PRMT3 and mouse CARM1. The top and bottom monomers are shown in surface and ribbon representation, respectively. The cofactor analog SAH is shown in surface representation. The accessibility angle for the active site of each PRMT is labeled.
The differences in the relative orientation of the two monomers in PRMT dimers, together with the differences in the size of the central enzyme cavities, result in significant variations in active site accessibility across the enzymes of known structure. To provide a quantitative measure of active sites accessibility for different PRMTs, we determined an accessibility angle for AtPRMT10, CARM1, PRMT1 and PRMT3. With the bottom monomers in the same orientation, a vertex was placed in the center of dimer cavity, and from that point the largest angle allowed by the molecular surface of the dimer in two dimensions in this view was traced for each structure (Figures 7B–7E). For AtPRMT10, the accessibility angle was ~120° (Figure 7B). However, for PRMT1 and PRMT3 and CARM1, the accessibility angles were ~50°, ~45°, and ~20°, respectively (Figures 7C–7E). Thus, the unique size and orientation of the AtPRMT10 central cavity creates a significantly larger accessibility to this enzyme’s active site relative to the PRMTs of currently known structure.
AtPRMT10 Motion
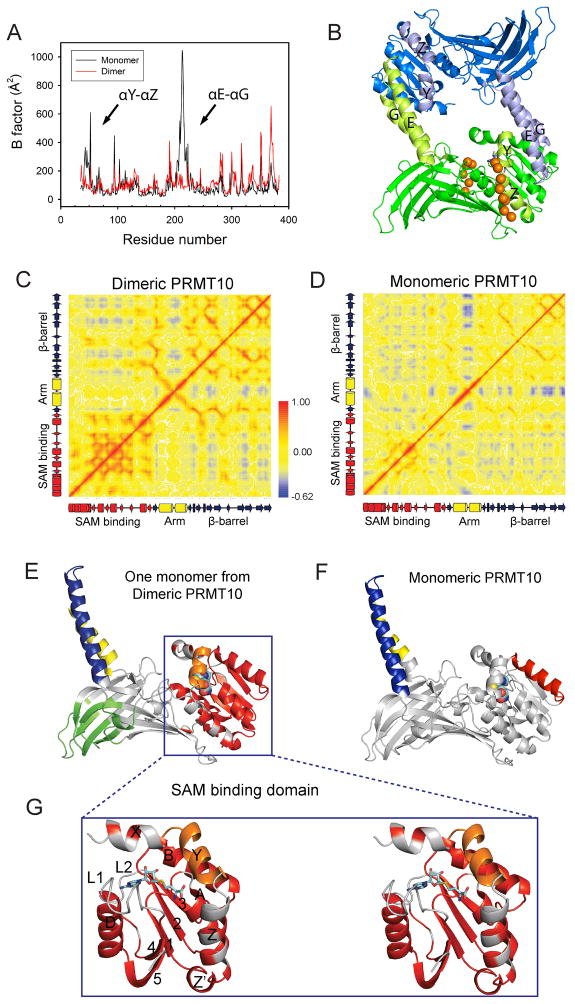
Because dimerization has been shown to be essential for the methyltransferase activity of PRMTs, we examined the impact dimerization has on the motion of AtPRMT10 using 30 ns molecular dynamics (MD) simulations. Monomeric and dimeric forms of AtPRMT10 were examined. The total energy of each system, calculated as the sum of the kinetic and potential energy at each time point, was relatively constant after the first 5 ns, particularly over the last 10 ns (data not shown). Therefore, the averages of the MD trajectories in the last 10 ns were used for the following analysis. The effect of dimerization on the degree of motion of AtPRMT10 was determined by computing the atomic position fluctuations (APFs) of Cα atoms of the monomer and dimer form. Overall, AtPRMT10 exhibits similar APFs in monomeric and dimeric states; however, in the dimeric form, two regions (αY-loop-αZ, residues 40–68; the dimerization arm, residues 187–235) displayed significantly lower APFs than when in the monomeric form (Figures 8A & 8B). The reduced fluctuations within these two regions likely result from their direct involvement in the formation of the dimer interface (Figure 3C). Notably, the region αY-loop-αZ (residues 40–68) is directly involved in the binding of SAH and in the formation of substrate binding groove I. Therefore, stabilization of this region by dimerization likely improves the binding of SAH and substrate proteins.
Figure 8. Effects of dimerization on the motion of AtPRMT10.
(A) Local fluctuation of residues in dimeric and monomeric AtPRMT10. Major differences between monomeric and dimeric AtPRMT10, including the two N-terminal helices αY-αZ and the dimerization arm (αE-αG), are highlighted by arrows and labeled. (B) The structure of a dimeric AtPRMT10. The two monomers are colored in blue and green respectively. The regions that displayed dramatically reduced local fluctuations in dimeric PMT10 are colored in light green in one monomer and in light blue in the other monomer. The putative substrate sites are represented in orange balls. Covariance analysis of dimeric AtPRMT10 (C) and monomeric AtPRMT10 (D). The values of residue-residue correlation coefficients range from blue (anticorrelated, −0.62) to red (correlated, +1.0), with non-correlated residue pairs colored in yellow. Schematic representations of the secondary structure corresponding to the residues on x-axis and y-axis are presented from left to right and bottom to top. (E), (F) Clustering of correlated residues in dimeric AtPRMT10 (E) and monomeric AtPRMT10 (F). Clusters with correlation coefficients higher than 0.7 are shown in different colors (other than gray) and all other regions are colored in gray. SAH is shown as sticks and spheres to highlight the location of the SAM binding pocket, although it is removed from the crystal structures before MD simulations. (G) An expanded stereo-view of the SAM binding domain of Figure 8E, with the secondary structures labeled.
We computed normalized covariance matrices to classify the motions of all residue pairs in the protein (Figures 8C & 8D). Normalized covariance matrices generate the residue-residue correlation coefficients (Cijs), which inform the relative motion between a residual pair. Based on the value of Cijs, the motions of all residue pairs can be classified into three groups: correlated motion (two residues moving toward the same direction) as indicated by Cij approaching 1, anti-correlated motion (two residues moving toward the opposite direction) as indicated by Cij approaching −1, and uncorrelated motion (two residues moving with the lack of a dynamic relationship) with Cij values near zero. The SAM binding domain of dimeric AtPRMT10 exhibits considerably greater residue-residue correlations relative to that of monomeric AtPRMT10 (Figure 8C). Increased residue-residue correlations are also observed in several discrete regions of the β-barrel domain.
To better understand the biological significance of residue-residue correlations, single-linkage clustering analysis was then conducted to identify groups of residues that move together. Clustering of dimeric AtPRMT10 at a correlation coefficient above 0.7 resulted in five clusters, while clustering of monomeric AtPRMT10 under the same criterion only resulted in three clusters (Figures 8E & 8F). One notable difference between monomeric AtPRMT10 and dimeric AtPRMT10 lies in the SAM binding domain. Most of this region, except the two N-terminal helices (αX and αY) and two loop regions (L1 and L2), are clustered in dimeric AtPRMT10 (Figure 8G), while only helix B is self-clustered in monomeric AtPRMT10. In addition, one end of the β-barrel domain is clustered in dimeric AtPRMT10, but not in monomer AtPRMT10. These data establish that the SAM binding domain and one end of the β-barrel domain to move as a cohesive unit in dimeric AtPRMT10, but not in monomeric AtPRMT10.
To extend these investigations into other PRMTs, we examined the motion of monomeric and dimeric PRMT3 using the same MD simulation protocol described above. Similar to AtPRMT10, dimerization significantly lowered the APFs in the N-terminal region (αX-αY-αZ, residues 208–245) and the dimerization arm (residues 370–394) (Figure S3A). In addition, normalized covariance analysis clearly shows that dimerization promotes coherent protein motions in the SAM binding domain and several discrete regions of the β-barrel domain (Figure S3B & S3C). Taken together, our results show that dimerization productively impacts the motion of the PRMTs. In particular, the SAM binding domain in both AtPRMT10 and PRMT3 move as a cohesive unit in the enzyme dimer but not the monomer.
PRMT10 N-terminus in Enzyme Function
Finally, we examined the impact of the N-terminal addition (residues 1–30) on the dimeric state and methyltransferase activity of AtPRMT10. We created three N-terminal deletion mutants, including ΔN10 (residues 11–383), ΔN20 (residues 21–383) and ΔN30 (residues 31–383), and compared their biophysical properties and methyltransferase activities to those of full-length AtPRMT10. The oligomeric states of these mutants were investigated using dynamic light scattering (DLS) and gel filtration experiments (Table II). As observed in the wild-type enzyme, all N-terminal deletion mutants form dimers in solution. Moreover, the oligomeric states of these mutants are SAH-independent. Together, these data suggest that the N-terminal addition does not impact AtPRMT10 dimerization.
The methyltransferase activities of wild-type AtPRMT10 and the three N-terminal deletion mutants were measured as described previously 32; while these initial studies do not provide kinetic values, they are sufficient to highlight relative differences in enzyme function (Figure 5A and 5B). Purified calf thymus core histones, which are a mixture of histones H2A, H2B, H3 and H4, were chosen as the substrate. Of these four histones, H2A and H4 are known to be methylated by AtPRMT10. Upon analysis of the experiment by SDS-PAGE, the methylation state of H2A and H4 can be quantified individually, due to their difference in molecular weight. Interestingly, ΔN10 had approximately 3-fold greater activities toward H2A relative to the wild-type enzyme. Additional deletions of the N-terminus (ΔN20 and ΔN30) did not enhance the methylation of H2A by AtPRMT10. When H4 was used as the substrate, however, all three N-terminus mutants displayed wild-type level activities (Figure 5A and 5B). These results indicate that first ten residues of AtPRMT10 impact enzyme methyltransferase activity in a protein substrate-dependent manner. In the AtPRMT10 crystal structure, the helix αX (residues 32–40) covers the opening of the SAM binding pocket and stabilizes SAH binding with van der Waals interactions (Figure 1). As expected, the deletion of helix αX (Δ40) causes a dramatic drop in the activity for both H2A and H4.
Previous studies of PRMT1 have shown that amino acids distal to the methylation site can affect the methylation of H427. Thus, we examined whether the substrate sequence outside of the methylation site also impacts the activity of AtPRMT10. We examined the purified full-length histone H4 as well as H4N1-20, a peptide covering only the N-terminal twenty residues of histone H4. We found that the activity of AtPRMT10 on the full-length H4 substrate was markedly higher than that on the H4N1-20 substrate (Figure 5C), in spite of a 10-fold higher concentration of H4N1-20 was present in these assays. Thus, it appeared that the sequence downstream of the N-terminal 20 residues of histone H4 enhanced the methyltransferase activity of AtPRMT10. As shown in Figure 4, AtPRMT10 likely has multiple spatially adjacent substrate binding grooves on its surface, which can together accommodate a long substrate. The region of H4 beyond the C-terminal 20 residues can possibly facilitate methylation by forming additional contacts with the substrate binding grooves and thereby increasing the binding affinity between AtPRMT10 and H4.
The methylation site of AtPRMT10 in both histone H2A and histone H4 is located at the far N-terminus of these proteins. To examine whether a bulky protein fused to the N-terminus of H4 would impact AtPRMT10 activity at arginine-3 on H4, we compared the methylation of histone H4 and N-terminally GST-tagged histone H4 (GST-H4) by AtPRMT10 (Figure 5C). Our results show that the presence of a N-terminal GST tag modestly reduced the activity of AtPRMT10 by ~2-fold relative to untagged H4. These data indicate that AtPRMT10 can methylate R3 of H4 even when it is not located at the far N-terminus of this histone protein.
Discussion
We present the first structure of a plant protein arginine methyltransferase, that of AtPRMT10, and highlight unique features of this enzyme, including a long dimerization arm and a distinctly open conformation in the catalytic dimer. We also establish for the first time that the family of PRMTs exhibit conserved domain motions, particularly within the enzyme region that binds the SAM cofactor that donates the methyl group to arginines on target proteins. Together, these data advance our understanding of features shared by the PRMT enzymes, which function as both epigenetic and non-epigenetic factors, as well as unique aspects particular family members may employ to impact substrate preference.
In a functional PRMT dimer, the enzyme active sites are located at the periphery of a central cavity (Figure 3). This configuration likely impacts access of substrate proteins to the PRMT catalytic site. Indeed, most known methylation sites are located in disordered regions of substrates, and the structural flexibility around the methylation site has been shown to be essential for PRMT function 25. Comparing PRMT dimers of known structure demonstrates that PRMT paralogs exhibit a range of accessibility in the active site, which can be roughly summarized as: AtPRMT10>PRMT1>PRMT3>CARM1 (Figure 7). These variations result primarily from differences in the relative orientation of the two monomers in a functional PRMT dimer and differences in the dimerization arm length. Previous studies have suggested that the activity and substrate specificity of PRMTs are directly correlated with active site accessibility 36. Thus, it is possible that the more accessible AtPRMT10 active site may allow this enzyme to methylate arginine residues that do not serve as substrates for other PRMTs.
We show that AtPRMT10, like other PRMTs, functions only as a dimer (Figure 5). MD simulations on both the monomeric and dimeric forms of AtPRMT10 and PRMT3 show that dimer formation produces coherent motions in key catalytic domains (Figure 8 and Figure S3). PRMT dimers exhibit reduced fluctuations in the N-terminal αY-loop-αZ region, which not only forms direct contacts with the SAM methyl donor, but also forms a portion of substrate binding groove I that is conserved among PRMTs. Furthermore, dimerization results in more correlated motions throughout the SAM binding domain. Previous studies have shown that oligomerization can facilitate protein-ligand interaction by increasing the correlation in the motion of the structural elements involved in ligand binding 37. Importantly, our results show that the effects of dimerization on the motion of AtPRMT10 can be generalized into other members of the PRMT family (Figure S3). Dimerization appears to facilitate the methyltransferase activity of PRMTs by producing coherent protein motions in the SAM binding region.
Members of the PRMT family have a relatively conserved catalytic core, but exhibit remarkable diversity in the length and sequence of their N-terminal regions. Multiple lines of evidence suggest that the variations in the N-terminus diversify the functions of the PRMT family by modulating the substrate specificities 17,18,38,39. AtPRMT10 has a 30-residue N-terminal addition, which is one of the shortest among known PRMTs. Secondary structure analysis predicts that the N-terminal addition of AtPRMT10 remains in a disordered state. In support of this prediction, the AtPRMT10 N-terminal addition is prone to proteolysis (data not shown), and is not ordered in our crystal structure. Although PRMT1 also has a short N-terminal region (~30 residues), its length varies more among different PRMT1 isoforms and these variations have been shown to alter the substrate specificity of PRMT140.
The results presented here indicate that residues 1–10 can affect the substrate specificity of AtPRMT10 (Figure 5A & 5B). The deletion of the N-terminal addition enhances the activity of AtPRMT10 toward histone H2A, but does not significantly alter AtPRMT10 activity toward histone H4. This variation may result from the difference in the way that H4 and H2A interact with AtPRMT10. Based on the crystal structure of dimeric AtPRMT10, the 30-residue N-terminal addition is likely located at one side of the ring, adjacent to substrate binding grooves III and IV, but distant from substrate binding grooves I and II. Thus, H2A may employ AtPRMT10 substrate binding groove III or IV, while H4 employs substrate binding groove I. The local sequence of the methylation site in H2A (SGR3GKGG) is identical to that of H4 (SGR3GKGG), indicating that the sequence outside the methylation site is also important for the interaction of PRMT with its substrates. In support of this notion, our results demonstrate that deletion of the sequence C-terminal to residue 20 of H4 dramatically reduced the methylation at arginine 3 by AtPRMT10 (Figure 5C).
PRMT10 displayed comparable activities toward histone H4 and N-terminally GST-tagged histone H4 (GST-H4) (Figure 5C). In histone H4, the methylation site arginine-3 is located proximal to the N-terminus. Therefore, the standard H4 substrate can bind to the AtPRMT10 binding grooves in a linear fashion, as modeled in the crystal structure of PRMT1-peptide complex 19. Our results with the GST-H4 substrate, however, indicate that the target arginine of AtPRMT10 could be located internally within a larger protein, rather than only within an N-terminal tail. Such an observation expands the scope of potential substrates for AtPRMT10 to include proteins with target arginines located on flexible central loops capable of accessing the AtPRMT10 active site. Such substrate proteins may be identified in Arabidopsis that impact flowering time in an AtPRMT10-dependent manner. In summary, the data presented here indicate that, while the PRMTs share some key traits (e.g., a functional dimer and coherent SAM-binding domain motion), unique features of specific PRMTs, like the larger central cavity of the AtPRMT10 dimer, may lead to unique methylation patterns and target substrate proteins.
Materials and Methods
Cloning, expression and purification of AtPRMT10
The expression plasmids encoding wild-type AtPRMT10 (1–383) and its various mutants and related constructs were created using the standard ligation-independent cloning techniques, as described by Stols et al. 41. All expression plasmids used in this study were sequence verified. AtPRMT10 was overexpressed in Escherichia coli BL21-CondonPlus (DE3) RIPL (Stratagene) and purified as described previously with some modifications 42. The cells were grown at 37° C to an OD600 of 0.6 in Luria-Bertani media containing 50 μg/mL chloramphenicol and 50 μg/mL ampicilin. Protein expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM and the culture was grown for another 16 h at 18 °C. The harvested cells were resuspended in buffer A (50 mM Na phosphate pH 7.4, 500 mM NaCl and 20 mM imidazole) supplemented with 0.5 mM EDTA, 0.1% Triton X-100, 1 mM phenylmethylsulphonyl fluoride (PMSF), one tablet of a protease inhibitor cocktail (Roche), and 1 mg/mL lysozyme. After 45 min of incubation on ice, the resuspended cells were sonicated on ice for 3 min and the lysate was centrifuged at 50,000 × g for 60 min at 4 °C. The supernatant was passed through a 0.2 μm filter (Millipore) and then loaded onto a 5 mL high performance HisTrap™ column (GE Life Sciences), equilibrated with buffer A. The column was washed with 100 mL buffer A to remove nonspecifically bound proteins; the bound protein was then eluted with buffer B (50 mM Na phosphate pH 7.4, 50 mM NaCl and 250 mM imidazole). The elutent was loaded onto a HiPrep™ 26/10 desalting column (GE Healthcare Life Sciences) equilibrated with buffer C (20 mM Tris-HCl pH 8.0 and 150 mM NaCl), and the protein-containing fractions were collected. To remove the His-MBP tag, TEV protease was added into the pooled protein fractions at a ratio of 1:100 (w/w) TEV to AtPRMT10. After 12 hr of incubation at 4 °C, the mixture was reloaded onto the 5 mL high performance HisTrap™ column (GE Life Sciences), equilibrated with buffer A. The flow-through fractions were collected and concentrated in a Centricon YM10 (Amicon) concentrator. Concentrated protein was loaded on a HiLoad™ 16/60 Superdex 200 column (GE, Life Sciences) equilibrated with sizing buffer (20 mM Tris-HCl pH 7.5, 250 mM NaCl, 5% Glycerol). AtPRMT10 containing fractions were concentrated, flash frozen in liquid nitrogen and stored at −80 °C. Purified protein was >95% pure by SDS-PAGE.
Crystallization, data collection, structure determination, and refinement
Diffraction-quality crystals of AtPRMT10 (residues 11–383)-SAH complex were obtained by the hanging-drop vapour-diffusion method at 22 °C, with the mother liquid solution containing 0.1 M Tris-HCl pH 7.6, 2.3 M Na2HPO4 and 0.1 M arginine. Crystals grew to the size of 250 × 200 × 50 μm in approximately 10 days. Since flash-frozen crystals diffracted poorly and could not be used for structural determination, diffraction data were collected from warm-mounted crystals to 2.6 Å resolution using a Rigaku X-ray generator MicroMax-007HF. Data from four different crystals were reduced and merged using the program HKL200043 (Table I). Data quality was examined using the program PHENIX 44. The crystal specimen employed to solve the structure contained nearly 50% pseudomerohedral twinning as indicated by the twinning analysis carried out by the program PHENIX (Table SI)44. Notably, the β angle (89.98°) of the unit cell was very close to 90°. Consequently, the diffraction data could also be reduced into the orthorhombic space group P222 and its derivatives. Serious violations of systematic absences were observed, however, in space groups P212121, P21212, P2221. Consequently, molecular replacement was performed in the space group P222. As predicted by the Matthew’s coefficient, two monomers were identified in each asymmetric unit. All solutions in space group P222, however, were finally rejected owing to the presence of significant main-chain clashes during crystal packing. Taken together, these results indicated that P21 was the correct space group and pesudomerohedral twinning was present. As such, refinement was conducted with the twining refinement function of the program PHENIX 44. No violations in systematic absences for P21 and no clashes between protein monomers with four AtPRMT10-SAH complexes per asymmetric unit were observed; the structure was determined and refined with good geometry and statistics in this space group (Table I). The structure was determined in space group P21 by molecular replacement using the program PHENIX 44. The crystal structure of rat PRMT3 (PDB entry, 1F3L), processed using the program chainsaw of the CCP4 package 45, was used as the template for molecular replacement. Due to the salient difference between AtPRMT10 and PRMT3 in the sequence of the dimerization arm, the dimerization arm of PRMT3 (residues 370–399) was not incorporated into the template. Since psudomerohedral twinning (approximately 50%) was detected with the crystals used for the structure determination, least square twin refinement was performed using the program PHENIX. The structural model was further built manually using the program Coot 46, and refined using the program PHENIX.
Methyltransferase activity assay
In vitro methyltransferase assays were performed as described previously 32. In brief, the reaction mixture contained 20 mM Tris-HCl pH 8.0, 4 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 4 μM S-[Methyl-3H]-Adenosyl-L-methionine (Perkin Elmer [NET155]) and indicated concentrations of AtPRMT10 and protein substrates. The reaction mixtures were separated on a 15% SDS-PAGE and stained with commassie blue. The gel was then treated with Amplifier (GE Healthcare Life Sciences), dried and exposed to Kodak Biomax MS film at −80 °C.
Dynamic light scattering (DLS)
The hydrodynamic radii of various AtPRMT10 constructs were measured by a DynoPro DLS system (Wyatt Technology Corporation). All samples and buffers (20 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.5 mM EDTA) were filtered through 0.2 μM filters (Millipore) or centrifuged at 17,000 × g at 4 °C for 30 min before measurement. Three replicates were performed for each sample. The hydrodynamic radii and molecular weights of AtPRMT10 samples were estimated using the assumption of globular protein shape.
Molecular dynamics (MD) simulations
MD simulations of AtPRMT10 were performed using the AMBER 2003 force field 47 as described previously 37. All production runs were generated using the PMEMD module of Amber 9.048 with a 2 fs time step. The topology and parameter files were created using the LEaP program within AMBER 48. To maintain charge neutrality, the protein molecule was surrounded by a truncated octahedron of water and sodium ions in the simulation system. Electrostatic interactions were calculated using the particle-mesh Ewald algorithm 49 with a cutoff of 10 Å applied to Lennard-Jones interactions. Energy minimization was conducted using the SANDER package within AMBER 48. Equilibration consisted of 20 ps of constant volume conditions with heating from 100 to 300 K and subsequent 100 ps of constant temperature conditions.
Simulation results were analyzed by the PTRAJ package in AMBER 48. The pair-wise correlation coefficient, Cij, was calculated between the α-carbons of two residues as described by Sharma et al. 50. When the two residues i and j move in a correlated fashion (the angle between the motion of i and j is less than 90°), 0<Cij≤1; when they move in an anti-correlated way (the angle between the motion of i and j is more than 90° but less than 180°), −1≤Cij<0; finally, when they move in a non-correlated manner (randomly), Cij=0. The more positive the value of Cij is, the smaller the angle between the motion of the two residues is. Single-linkage clustering analysis was performed to identify groups of residues that move in a correlated or anti-correlated fashion, as described by Leese et al.51.
Supplementary Material
Acknowledgments
We thank Dr. Michael Miley at the UNC Biomolecular X-ray Crystallography Facility for assistance in sample preparation and data collection. Also, we thank Dr. Feng Ding for assistance in simulation analysis. This work was supported by NIH grant AI78924, the National Basic Research Program of China (grant no.2009CB941500), and the National Natural Science Foundation of China (grant no. 30921061).
Abbreviations
- PRMT
Protein Arginine Methyltransferases
- SAM
S-adenosylmethionine
- SAH
S-adenosylhomocysteine
- FLC
FLOWERING LOCUS C
- MD
Molecular Dynamics
- APFs
Atomic Position Fluctuations
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- PMSF
Phenylmethylsulphonyl Fluoride
- GF
Gel Filtration
Footnotes
Accession numbers
The coordinates and structure factors for the AtPRMT10-SAH crystal structure have been submitted to the Protein Data Bank and assigned the identifier 3R0Q.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, Brown JW, Cerdan PD, Borevitz JO, Mas P, Ceriani MF, Kornblihtt AR, Yanovsky MJ. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 3.Stallcup MR. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20:3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 4.Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 5.Lake AN, Bedford MT. Protein methylation and DNA repair. Mutat Res. 2007;618:91–101. doi: 10.1016/j.mrfmmm.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Smith BC, Denu JM. Chemical mechanisms of histone lysine and arginine modifications. Biochim Biophys Acta. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 8.Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z, Cui P, Pei Y, Wang B, Hu S, Cao X. Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci U S A. 2010;107:19114–19119. doi: 10.1073/pnas.1009669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf SS. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell Mol Life Sci. 2009;66:2109–2121. doi: 10.1007/s00018-009-0010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teyssier C, Le Romancer M, Sentis S, Jalaguier S, Corbo L, Cavailles V. Protein arginine methylation in estrogen signaling and estrogen-related cancers. Trends Endocrinol Metab. 2009;21:181–189. doi: 10.1016/j.tem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Aletta JM, Hu JC. Protein arginine methylation in health and disease. Biotechnol Annu Rev. 2008;14:203–224. doi: 10.1016/S1387-2656(08)00008-2. [DOI] [PubMed] [Google Scholar]
- 12.Boger RH, Cooke JP, Vallance P. ADMA: an emerging cardiovascular risk factor. Vasc Med. 2005;10:S1–2. doi: 10.1177/1358836X0501000101. [DOI] [PubMed] [Google Scholar]
- 13.Ueda S, Yamagishi S, Matsumoto Y, Fukami K, Okuda S. Asymmetric dimethylarginine (ADMA) is a novel emerging risk factor for cardiovascular disease and the development of renal injury in chronic kidney disease. Clin Exp Nephrol. 2007;11:115–121. doi: 10.1007/s10157-007-0471-x. [DOI] [PubMed] [Google Scholar]
- 14.Pullamsetti S, Kiss L, Ghofrani HA, Voswinckel R, Haredza P, Klepetko W, Aigner C, Fink L, Muyal JP, Weissmann N, Grimminger F, Seeger W, Schermuly RT. Increased levels and reduced catabolism of asymmetric and symmetric dimethylarginines in pulmonary hypertension. FASEB J. 2005;19:1175–1177. doi: 10.1096/fj.04-3223fje. [DOI] [PubMed] [Google Scholar]
- 15.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Lin CH, Hsieh M, Li YC, Li SY, Pearson DL, Pollard KM, Li C. Protein N-arginine methylation in subcellular fractions of lymphoblastoid cells. J Biochem. 2000;128:493–498. doi: 10.1093/oxfordjournals.jbchem.a022779. [DOI] [PubMed] [Google Scholar]
- 17.Frankel A, Clarke S. PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Conferral of substrate specificity by a zinc-finger domain. J Biol Chem. 2000;275:32974–32982. doi: 10.1074/jbc.M006445200. [DOI] [PubMed] [Google Scholar]
- 18.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat Struct Biol. 2000;7:1165–1171. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 2000;19:3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO J. 2007;26:4391–4401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J. 2007;26:4402–4412. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Merrill BM, Rajpurohit R, Kumar A, Stone KL, Papov VV, Schneiders JM, Szer W, Wilson SH, Paik WK, Williams KR. Identification of N(G)-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry. 1997;36:5185–5192. doi: 10.1021/bi9625509. [DOI] [PubMed] [Google Scholar]
- 25.Wooderchak WL, Zang T, Zhou ZS, Acuna M, Tahara SM, Hevel JM. Substrate profiling of PRMT1 reveals amino acid sequences that extend beyond the “RGG” paradigm. Biochemistry. 2008;47:9456–9466. doi: 10.1021/bi800984s. [DOI] [PubMed] [Google Scholar]
- 26.Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- 27.Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry. 2007;46:13370–13381. doi: 10.1021/bi701558t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 29.Singh V, Miranda TB, Jiang W, Frankel A, Roemer ME, Robb VA, Gutmann DH, Herschman HR, Clarke S, Newsham IF. DAL-1/4.1B tumor suppressor interacts with protein arginine N-methyltransferase 3 (PRMT3) and inhibits its ability to methylate substrates in vitro and in vivo. Oncogene. 2004;23:7761–7771. doi: 10.1038/sj.onc.1208057. [DOI] [PubMed] [Google Scholar]
- 30.Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu W, Cho H, Kadam S, Banayo EM, Anderson S, Yates JR, 3rd, Emerson BM, Evans RM. A methylation-mediator complex in hormone signaling. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu L, Lu F, Pei Y, Liu C, Cao X. Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 2007;8:1190–1195. doi: 10.1038/sj.embor.7401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fauman EB, Blumenthal RM, Cheng X. Structure and evolution of Adomet-dependent methyltransferase. S-Adenosylmethionine-Depedent Methyltransferase: Structures and Functions 1999 [Google Scholar]
- 34.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee DY, Ianculescu I, Purcell D, Zhang X, Cheng X, Stallcup MR. Surface-scanning mutational analysis of protein arginine methyltransferase 1: roles of specific amino acids in methyltransferase substrate specificity, oligomerization, and coactivator function. Mol Endocrinol. 2007;21:1381–1393. doi: 10.1210/me.2006-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith JJ, Rucknagel KP, Schierhorn A, Tang J, Nemeth A, Linder M, Herschman HR, Wahle E. Unusual sites of arginine methylation in Poly(A)-binding protein II and in vitro methylation by protein arginine methyltransferases PRMT1 and PRMT3. J Biol Chem. 1999;274:13229–13234. doi: 10.1074/jbc.274.19.13229. [DOI] [PubMed] [Google Scholar]
- 37.Teotico DG, Frazier ML, Ding F, Dokholyan NV, Temple BR, Redinbo MR. Active nuclear receptors exhibit highly correlated AF-2 domain motions. PLoS Comput Biol. 2008;4:e1000111. doi: 10.1371/journal.pcbi.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goulet I, Gauvin G, Boisvenue S, Cote J. Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J Biol Chem. 2007;282:33009–33021. doi: 10.1074/jbc.M704349200. [DOI] [PubMed] [Google Scholar]
- 39.Tang J, Gary JD, Clarke S, Herschman HR. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J Biol Chem. 1998;273:16935–16945. doi: 10.1074/jbc.273.27.16935. [DOI] [PubMed] [Google Scholar]
- 40.Pawlak MR, Banik-Maiti S, Pietenpol JA, Ruley HE. Protein arginine methyltransferase I: substrate specificity and role in hnRNP assembly. J Cell Biochem. 2002;87:394–407. doi: 10.1002/jcb.10307. [DOI] [PubMed] [Google Scholar]
- 41.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y, McNamara DE, Miley MJ, Nash RP, Redinbo MR. Functional characterization of the multi-domain F plasmid TraI relaxase-helicase. The Journal of biological chemistry. 2011;286:12670–12682. doi: 10.1074/jbc.M110.207563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Cater CW Jr, Sweet RM, editors. Methods in Enzymology. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collabroative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 48.Case DA, Darden TA, Cheatham TEI, Simmerling CL, Wang J. AMBER. Vol. 9. University of California; San Francisco: 2006. [Google Scholar]
- 49.Essman U, Perera L, Berkowita ML, Darden TA, Lee H. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 50.Sharma S, Ding F, Dokholyan NV. Multiscale modeling of nucleosome dynamics. Biophys J. 2007;92:1457–1470. doi: 10.1529/biophysj.106.094805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Everitt BS, Landau S, Leese M. Cluster analysis. Oxford University Press; Oxford: 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.