Abstract
Mir-17-92 encodes 6 miRNAs inside a single polycistronic transcript, the proper expression of which is critical for early B-cell development and lymphocyte homeostasis. However, during the T-cell antigen response, the physiologic function of endogenous miR-17-92 and the roles of the individual miRNAs remain elusive. In the present study, we functionally dissected the miR-17-92 cluster and revealed that miR-17 and miR-19b are the key players controlling Th1 responses through multiple coordinated biologic processes. These include: promoting proliferation, protecting cells from activation-induced cell death, supporting IFN-γ production, and suppressing inducible regulatory T-cell differentiation. Mechanistically, we identified Pten (phosphatase and tensin homolog) as the functionally important target of miR-19b, whereas the function of miR-17 is mediated by TGFβRII and the novel target CREB1. Because of its vigorous control over the Th1 cell–inducible regulatory T cell balance, the loss of miR-17-92 in CD4 T cells results in tumor evasion. Our results suggest that miR-19b and miR-17 could be harnessed to enhance the efficacy of T cell–based tumor therapy.
Introduction
CD4+ T cells are essential components of the adaptive immune system that regulate immune responses against foreign pathogens and tumors. Upon antigen recognition, naive CD4+ T cells undergo activation and expansion, and then contract via programmed cell death.1 Specific antigen challenges also induce CD4+ T cells to differentiate into distinct Th cell lineages characterized by unique cytokine production profiles.2 Among these lineages, Th1 cells, the differentiation of which is controlled by the master transcription factor T-bet,3 are specialized for the clearance of intracellular infections and are implicated as the major effectors against tumors.4 In addition, the conversion of effector T cells to Foxp3+ inducible regulatory T cells (iTregs) is an important mechanism used to balance immune responses5 that is exploited by tumors as a strategy for immune evasion.6 Whereas the protein-based regulatory machinery that operates during the T-cell response has been vigorously explored, we have recently become aware of a novel and crucial element modulating T-cell function: miRNA.7,8
miRNAs are 18- to 24-nucleotide noncoding RNAs that regulate gene expression by destabilizing target mRNAs, leading to mRNA degradation and/or translational repression.9 Recent studies suggest that miRNA-mediated gene regulation represents a fundamental layer of posttranscriptional regulatory programs in metazoan genomes.10 Global disruption of miRNAs caused by defective biogenesis had profound effects on the development of B cells,11 Th1/Th2 differentiation,12,13 and Treg function.14,15 In addition to these demonstrations of the importance of miRNA biogenesis in general, accumulating evidence shows that many specific miRNAs are differentially regulated in hematopoietic lineages and play important roles in controlling the development and function of immune cells.7,8,16–18 One such regulator is the miR-17-92 cluster. This cluster of miRNAs is encoded by a polycistronic miRNA gene and generates a single transcript that yields 6 individual mature miRNAs. These miRNAs are categorized into 3 families based on sequence homology: the miR-17 family (miR-17, miR-20, and miR-18a), the miR-19 family (miR-19a and miR-19b), and the miR-25 family (miR-92a; supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). miR-17-92 is well recognized as an “onco-miR” because of its genomic amplification in certain tumor tissues and its potent acceleration of c-My–induced B-cell lymphoma.19 Genetic ablation has clearly established the critical roles of miR-17-92 in embryonic development.20 In immune cells, miR-17-92 plays an integral part in the development of myeloid cells and B cells.20,21 Mice with germline deletion of miR-17-92 exhibit a severe defect in adult B-cell development with an augmentation of apoptosis in the pro–B-cell fraction and consequently a blockade at the pro-B to pre-B transition.20 In addition, transgenic mice overexpressing the miR-17-92 cluster in lymphocytes develop lymphoproliferative disease and autoimmunity as early as 18 weeks of age. It was suggested that the overexpression of miR-17-92 unbalances lymphocyte homeostasis via control of the tumor suppressor Pten (phosphatase and tensin homolog) and the proapoptotic protein Bim.22
Because of its extensive roles in promoting malignant transformation in hematologic tumors,23 the miR-17-92 cluster has been suggested as a potential candidate for miRNA-based antitumor therapy. However, the global inhibition of miR-17-92 for cancer therapy is largely limited by the lack of knowledge regarding the physiologic function of endogenous miR-17-92 in normal tissues, especially in the immune system, which may compromise the efficacy of the therapy. Furthermore, even less is known about the differences or similarities in the functions of individual miRNAs within the cluster during antigen responses. In the present study, we combined gain- and loss-of-function approaches to analyze the physiologic roles of individual miRNAs within the miR-17-92 cluster in the regulation of T-cell effector function. Our data establish the miR-17-92 cluster as a multifaceted and indispensible positive regulator of CD4+ T-cell antigen responses, particularly in the context of Th1 T cell–mediated tumor rejection, suggesting that global inhibition of miR-17-92 is likely to subvert the immune response against tumors. Furthermore, we demonstrate profound functional divergence among the individual miRNAs in this cluster, with miR-19b and miR-17 accounting almost entirely for the pro-Th1 influence of mir-17-92, whereas miR-18a acts as an internal antagonist of the cluster's function.
Methods
Mice
Mice homozygous for the floxed mir-17-92 allele20 were purchased from The Jackson Laboratory (Mir17-92tm1.1Tyj/J), lck-cre (B6.Cg-Tg(lck-cre)1Cwi N9) and cd4-cre mice(C57BL/6Tac-Tg(cd4-cre)N9) were from Taconic. T cell–specific, miR-17-92–deficient C57BL/6 mice were generated by crossing floxed mir-17-92 mice with the cd4-cre or lck-cre mice for > 10 generations. 5C.C7 TCR-transgenic mice were from Taconic (B10.ARag2tm1Fwa H2-T18a Tg, Tcra5CC7, Tcrb5CC7lwep). Wild-type (WT) B10.A mice were also from Taconic. All mice experiments were approved by the Institutional Animal Care and Use Committee at Duke University.
miRNA target predictions, luciferase assays, and T-cell transductions
miRNA target candidates were predicted using multiple methods assembled on the miRecords website (http://mirecords.biolead.org/). The 3′UTRs of mouse pten, tgfbr2, and creb1 containing the predicted binding sites of miR-19b and miR-17 were amplified from a 3′-RACE–ready cDNA library generated from mouse T-cell total RNAs. For normalization, firefly luciferase reporter was cotransfected into individual cell lines with a renilla luciferase gene for which expression is not affected by miRNA targeting. Luciferase activity was determined 48 hours after transfection using a dual luciferase assay kit (Promega). For retrovirus transduction, naive T cells were activated for 18 hours and then spin-infected with retrovirus in a 24-well plate at 1258g rpm and 37°C.
qPCR and Western blot
The total RNA of cells was isolated with an miRVana extraction kit (Ambion) according to the manufacturer's instructions. We used the SYBR-based real-time quantitative PCR (qPCR) method to quantify mature miRNA expression. E coli polyA polymerase was used to add adenines at the 3′ end of RNA molecules lacking a polyA tail. After oligodT annealing, a universal tag was attached to the 3′ end of cDNAs during cDNA synthesis using retro-transcriptase Superscript III (Invitrogen). With this universal tag, qPCR was performed with miRNA-specific forward primers and a reverse universal primer mix.
To quantify absolute copy numbers of miRNAs, the mature forms of these molecules were chemically synthesized (by Integrated Device Technology), and 2 dosages, 0.1 or 0.01 pmol, were included as standard inputs to produce corresponsive cDNAs in parallel. qPCR measurements were performed as described in the previous paragraph. Standard curves measuring each miRNA were generated using linear regression with a semilog scale on the miRNA copy numbers. The absolute copy numbers of individual miRNAs in the samples of interest were calculated from the standard curves.
Western blot was performed with the following primary Abs: anti-PTEN, anti-TGFβRII, anti-CREB1, anti-Bim, anti–p-Smad3(S423/425), anti-Smad3 (Cell Signaling Technology), and anti–β-actin (Sigma-Aldrich). Anti–rabbit Alexa Fluor 680 and anti–goat Alexa Fluor 800 (Invitrogen) were used as the secondary Abs, and the fluorescence intensity was measured on an Odyssey system (LI-COR).
Live-cell video microscopy and image analysis
Live-cell imaging was performed on a Zeiss Axiovert-100TV station equipped with a Zeiss 40× EC Plan-Neofluar objective lens (numerical aperture = 1.30), a CoolSnapHQ CCD camera (Roper Scientific) and a high-speed piezo Z-motor for Z-stack recording as described previously.24 The pleckstrin homology green fluorescent protein (PH-GFP) probe was transduced into 5C.C7 T cells in the presence or absence of miR-19b coexpression. CH27 cells were preloaded with 10μM moth cytochrome-c (MCC) peptide for 2 hours at 37°C to serve as APCs. The experiments were performed in the minimal imaging medium composed of 1× HANKS buffer, 2.0mM CaCl2, 1.0mM MgCl2, and 2% FBS. The imaging process was controlled by MetaMorph Version 7.6 software (Universal Imaging) to record the dynamics of probe mobilization every 30 seconds in a 3D manner. Using the MetaMorph software suite, images of the best focus plane were used for data analysis. The activity of PI3K was represented by measuring the ratio of the average GFP florescent intensity in the synaptic region versus the average intensity in the rest cell area. The highest level of GFP synaptic accumulation within the first 10 minutes of T:APC contact was used as the mark for the maximal activity of PI3K activation in the initiation stage (before the formation of mature immunologic synapse) of TCR signaling. A similar measurement was performed 1 hour after the initiation of TCR signaling to assess the maintenance of PI3K signaling.
B16 melanoma tumor model
The B16/F10 melanoma cell line was a gift from Dr Thomas Tedder (Duke University). The ovalbumin (OVA)–secreting B16/F0/OVA cell line was kindly provided by Dr Edith Lord (University of Rochester, Rochester, NY). In the subcutaneous melanoma tumor model, WT, f/+, and knockout (KO) mouse littermates were anesthetized and injected subcutaneously on the shaved right lateral flank with 2 × 105 B16/F10 tumor cells or 3 × 105 B16/F0/OVA cells in 200 μL of sterile PBS. Tumor volumes were monitored and calculated using the equation: V = 4 (L1 × L22)/3, where V is volume (mm3), L1 is the longest radius (mm), and L2 is the shortest radius (mm).
Histology
The footpad was excised, inflated with 4% paraformaldehyde in PBS, fixed overnight at room temperature, placed in 70% ethanol, embedded with paraffin, and sent for H&E staining. Photographs were taken under a 40× lens.
Statistical analysis
Unpaired 2-tailed t tests were used to determine whether the difference between a given set of means was statistically significant. P < .05 was considered statistically significant.
Results
miR-19b and miR-17 facilitate T-cell expansion upon antigen challenge
Upon antigen challenge, we observed a rapid decline in the levels of most miRNA species in CD4+ T cells. Among the few miRNAs showing significant augmentation, the miRNAs comprising the miR-17-92 cluster form a distinct group, although the dynamics of mature miRNA elevation vary individually (supplemental Figure 1B). In addition, the absolute copy numbers of individual miRNAs in T cells are quite diversified (supplemental Figure 1C-D). These phenomena prompted us to explore the intrinsic function of the miR-17-92 cluster during the effector T-cell response and to further analyze the individual functions of the component miRNAs.
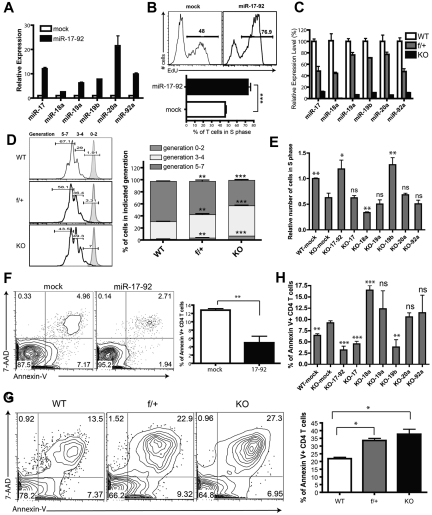
To characterize the role of miR-17-92 in CD4+ T cell proliferation after antigen engagement, we first infected lymph node (LN) T cells from 5C.C7 TCR-transgenic mice with a retrovirus encoding the whole primary miR-17-92 transcript. This resulted in a 3- to 20-fold expression increase of each of the 6 miRNAs in CD4+ T cells (Figure 1A). Interestingly, the intracellular level of individual miRNAs varied greatly (Figure 1A and supplemental Figure D-E), suggesting a potential intrinsic diversity in posttranscriptional processing efficiency within the cluster. T cells were then stimulated with CH27 APCs preloaded with the agonist peptide MCC for 24 hours, and the proliferation of CD4+ T cells was analyzed by 5-ethynyl-2′-deoxyuridine staining. As reported previously,22 we noted a higher percentage of CD4+ T cells in the S phase for those cells overexpressing miR-17-92 (Figure 1B). We then examined the impact of reduced miR-17-92 expression on CD4+ T-cell proliferation by analyzing mir-17-92 conditional KO mice in which the mir-17-92 locus is floxed20 and CD4-Cre mediates T cell–specific deletion (Figure 1C). CD4+CD25− conventional T cells from the LNs and spleen were sorted by FACS and stained with CFSE, stimulated by plate-bound anti–CD3/CD28, and cell proliferation was assessed. Reciprocal to the enhanced proliferation of CD4+ T cells overexpressing miR-17-92, CD4+ T cells heterozygous or null for mir-17-92 showed a gene-dosage-dependent reduction in proliferation (Figure 1D). Furthermore, we also noted a moderate defect in the proliferation of primed, miR-17-92–deficient CD4+ T cells during the recall response (supplemental Figure 2A). These results suggested that miR-17-92 facilitates CD4+ T cell proliferation in vitro.
Figure 1.
The functionality of miR-19b and miR-17 in antigen-stimulated CD4+ T-cell proliferation and activation-induced cell death. (A,C) Assessment of miR-17-92 relative expression in CD4+ T cells by qPCR. Data were normalized to a reference small RNA U6. Bar graph shows means ± SEM from 3 independent experiments. (A) LN T cells from 5C.C7 TCR-transgenic mice were primed by syngeneic APCs loaded with agonist peptide MCC (10μM) and transduced with retrovirus encoding GFP alone (mock) or the mir-17-92 gene with GFP. Three days later, CD4+GFP+ T cells were FACS sorted, and the expression of miRNA was analyzed by qPCR. Data were normalized to the mock group. (B) mir-17-92 genes were introduced into 5C.C7 T cells as described in panel A. Cells were selected by puromycin for 48 hours, and restimulated with MCC-loaded CH27 APCs (10μM) for 24 hours. 5-ethynyl-2′-deoxyuridine (EdU) was supplied into the culture medium 3 hours before cell fixation. The percentage of CD4+GFP+ T cells in the S phase was measured with the Click-iT EdU flow cytometry assay (Invitrogen). Top: representative FACS plot; bottom: statistical analysis of 5 independent experiments. (C) Expression of miRNAs in CD4+ T cells from LNs and spleens of miR-17-92f/fCD4-Cre− (WT), miR-17-92f/+CD4-Cre+ (f/+), and miR-17-92f/fCD4-Cre+ (KO) littermates. Data were normalized to WT. (D) CD4+CD25− T cells from LNs and spleens of WT, f/+, and KO littermates were labeled for 10 minutes at 37°C with CFSE at a ratio of 3 × 106 cells/4μM chemical, followed by washes with complete culture medium, and were then activated by 1 μg/mL of plate-bound anti-CD3 and anti-D28 Abs (eBioscience) for 72 hours. Left: representative FACS plot showing CFSE dilution. Tinted peaks represent CFSE-stained T cells without stimulation; right: statistical analysis of 3 independent experiments. (E) CD4+CD25− T cells from WT or KO mice were primed and transduced with indicated retroviruses. After 2 days of puromycin selection, cells were restimulated with anti–CD3/CD28 for 24 hours, and the number of T cells in the S phase was determined by staining of pulsed EdU. Bar graphs summarize means ± SEM from 4 independent experiments and data were normalized to WT-mock. The statistical significance was assessed in comparison with the KO-mock group. (F) LN T cells from 5C.C7 TCR-transgenic mice were primed and transduced with retrovirus encoding GFP or miR-17-92 as described in panel A. Three days later, viable cells were enriched by density gradient centrifugation and then restimulated with CH27 loaded with MCC (10μM) for 24 hours, and the status of AICD was assessed by annexin V and 7AAD staining (BioLegend). The bar graph summarizes the means ± SEM from 4 independent experiments. (G) CD4+CD25− T cells from LNs and spleens of WT, f/+, and KO littermates were stimulated with anti–CD3/CD28 Abs for 72 hours, and the percentages of CD4+ T cells undergoing apoptosis were assessed by annexin V and 7AAD staining. The bar graph summarizes means ± SEM from 3 independent experiments. (H) CD4+CD25− T cells from LNs and spleens of KO mice were primed and transduced with retrovirus encoding individual miRNAs of miR-17-92 as described in panel E. Three days after transduction, viable CD4+ T cells were enriched and restimulated with anti-CD3/CD28 for 24 hours. The profiles of restimulation-induced apoptosis were measured by annexin V staining. Bar graphs summarize the means ± SEM from 3-5 independent experiments. The statistical significance was assessed in comparison with the KO-mock group. *P < .05; **P < .01; ***P < .001; and ns, no significance.
To determine the contribution of each individual miRNA of the miR-17-92 cluster in promoting CD4+ T-cell proliferation, we retrovirally transduced the whole cluster or individual miRNAs back into miR-17-92–deficient CD4+ T cells and analyzed CD4+ T-cell proliferation. As expected, reexpressing the whole cluster rescued the proliferation defect of miR-17-92–deficient CD4+ T cells. Moreover, miR-19b alone exhibited functional equivalence to the whole cluster in enhancing CD4+ T-cell proliferation (Figure 1E). Surprisingly, although it only differs from its family member miR-19b by 1 nucleotide at position 11, miR-19a failed to promote the proliferation of CD4+ T cells; the expression of miR-18a exerted an inhibitory effect on proliferation. These data suggested that miR-19b is the functional representative of miR-17-92 in regulating CD4+ T cell proliferation upon antigen stimulation.
During the course of an immune response, antigen-reactive T cells clonally expand and then apoptotically contract to maintain immune homeostasis. To determine whether miR-17-92 affected activation-induced cell death (AICD) for CD4+ T cells, we challenged 5C.C7 T cells overexpressing miR-17-92 with MCC peptide and measured AICD via annexin V/7-amino-actinomycin D (7AAD) staining. miR-17-92 overexpression reduced the proportion of annexin V+CD4+ T cells compared with cells expressing the mock vector (Figure 1F), whereas peripheral CD4+ T cells from miR-17-92–deficient mice showed reduced survival after stimulation (Figure 1G). When AICD was measured for miR-17-92–deficient CD4+ T cells replenished with individual miRNAs, miR-19b and miR-17 enhanced CD4+ T-cell survival after stimulation; again, miR-18a exhibited antagonizing behavior, enhancing the apoptosis of CD4+ T cells (Figure 1H).
miR-19b promotes IFN-γ production
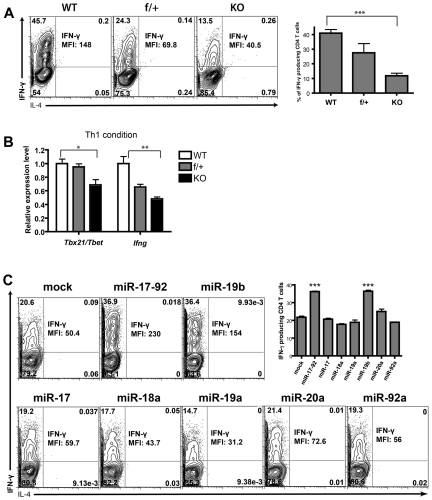
The diversity of pathogens demands discrete cytokine combinations from Th cells, and so an effective adaptive immune response requires appropriate CD4+ T-cell functional differentiation. To determine whether miR-17-92 is involved in this process, we cultured naive CD4+ T cells from miR-17-92–deficient mice under Th1 conditions in vitro. We observed a dramatic decrease in the production of IFN-γ and TNF by miR-17-92–deficient Th1 cells (supplemental Figure 3A). To exclude the possibility that this decrease was caused by a defect in proliferation and survival, we examined cytokine production ability on a single-cell basis by intracellular staining. Whereas approximately half of the WT CD4+ T cells were IFN-γ+, only 10% of miR-17-92–deficient CD4+ T cells produced IFN-γ and at much lower levels (Figure 2A). Generational analysis confirmed that the IFN-γ production defect of miR-17-92–deficient CD4+ T cells is independent of its defect of cell proliferation (supplemental Figure 4). In agreement with these observations, the transcription levels of ifng and the key Th1 lineage–specific factor tbx21 were significantly reduced in miR-17-92–deficient Th1 cells (Figure 2B). We did not detect any percentile diminution of TNF-producing cells or any disadvantage in TNF expression on a per-cell basis (supplemental Figure 3B-C). This suggests that the decrease in the total amount of TNF produced by miR-17-92–deficient cells was a result of proliferation and/or survival defects. We assessed the individual contributions of the miRNAs in supporting IFN-γ production. Only miR-19b was able to rescue the capacity of IFN-γ production in miR-17-92–deficient Th1 cells (Figure 2C and supplemental Figure 5). These results demonstrated an indispensible role for miR-19b in promoting effector T cell IFN-γ production.
Figure 2.
miR-19b is indispensible for IFNγ production from differentiated Th1 cells. (A-B) CD4+CD25− T cells were sorted from LNs and spleens of WT, f/+, and KO littermates, activated by anti–CD3/CD28 Abs under Th1-skewing conditions for 4 days with 50 ng/mL of recombinant mouse IL-12 (Peprotech), 10 μg/mL of purified anti–IL-4 (11B11), and 50 U/mL of recombinant mouse IL-2 (Peprotech). (A) The percentage of viable cells producing IFN-γ and the mean florescence intensity (MFI) of IFN-γ were determined by intracellular staining after 4 hours of stimulation with 0.9nM PMA and 0.5 μg/mL of ionomycin (Sigma-Aldrich) in the presence of 5 μg/mL of brefeldin A (Sigma-Aldrich) and 2μM monensin (eBioscience). The Abs used were anti–IFNγ-APC (BioLegend) and anti–IL-4-PE (BD). The bar graph summarizes the means ± SEM from 3 independent experiments. (B) The mRNA levels of T-bet and IFN-γ from CD4+ T cells differentiated under the Th1-skewing conditions were quantified by qPCR. Data were normalized to a reference gene, SDHA, and are shown as relative to WT. The bar graph shows means ± SEM from 3 independent experiments. (C) As described in Figure 1E, CD4+CD25− conventional T cells of KO mice were primed and transduced with retrovirus encoding individual miRNAs within the miR-17-92 cluster, and then cultured under the Th1-skewing condition for 4 days. The percentages of IFN-γ- or IL-4–producing cells and the MFI of IFN-γ signal were measured by intracellular cytokine staining. Left: representative FACS plot; right: means ± SEM from 3 independent experiments. Statistical analysis was done by comparison with mock. ***P < .001.
miR-17 and miR-19b promote the in vivo Th1 response during DTH
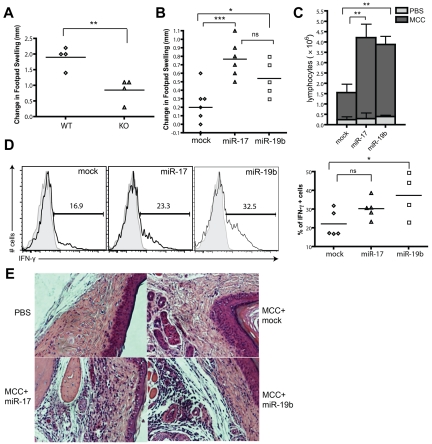
To investigate whether miR-17-92 facilitates Th1 responses in vivo, we assessed the ability of miR-17-92–deficient mice to mount delayed-type hypersensitivity (DTH) responses. WT and miR-17-92f/f CD4-Cre+ mice were immunized with keyhole limpet hemocyanin emulsified in complete Freund adjuvant, and then rechallenged in one footpad with keyhole limpet hemocyanin and with PBS in the contralateral footpad. In agreement with a critical role for miR-17-92 in promoting Th1-mediated effector function, miR-17-92–deficient mice have significantly reduced footpad swelling compared with WT mice (Figure 3A). To determine the function of individual miRNAs in vivo, 5C.C7 T cells were transduced with retrovirus encoding miR-17 or miR-19b and then adoptively transferred into B10A recipients. These mice were then immunized with MCC peptide in complete Freund adjuvant, and DTH was assessed after footpad rechallenge. With transferred T cells expressing miR-17 and miR-19b, we observed intensified footpad swelling, augmented numbers of lymphocytes in the draining LNs (dLNs), as well as enhanced infiltration in the footpad (Figure 3B,C,E). In addition, consistent with its IFN-γ–promoting function in vitro, miR-19b substantially enhanced the capacity of T cells to produce IFN-γ during the DTH response when examined ex vivo (Figure 3D). These data support a general role for miR-17 and miR-19b in facilitating Th1-mediated inflammatory responses in vivo.
Figure 3.
miR-19b and miR-17 enhance DTH responses in vivo. (A) WT and KO mice were immunized subcutaneously with 100μg of KLH in CFA (Sigma-Aldrich) and 8 days later, were injected with 50μg of KLH in 1 footpad and PBS in the contralateral footpad. Increase in footpad thickness was measured for both groups at 48 hours after secondary challenge (n = 4). (B-E) WT B.10A mice were transferred through the tail vein with 0.5 × 106 CD4+ T cells from 5C.C7 Rag2−/− mice infected with GFP-, miR-17-, or miR-19b–expressing retrovirus and immunized subcutaneously with 20 μg of MCC peptide in CFA. Five days after immunization, mice were injected with 20 μg of MCC and PBS in each lateral footpad. Seventy-two hours later, the swelling of footpads (in panel B) and the number of total lymphocytes from the popliteal LN (in panel C) were measured (n ≥ 5). The percentage of IFN-γ–producing cells within the GFP+CD4+ population in the DLN was assayed (in panel D) by intracellular staining (n ≥ 4). Representative images of footpad tissues with H&E staining were shown in panel E. *P < .05; **P < .01; ***P < .001; and ns, no significance. Each experiment was repeated 3 times.
miR-17 and miR-19b inhibit iTreg differentiation
To examine the role of the miR-17-92 cluster in the development and homeostasis of Tregs, we analyzed the Treg profile of miR-17-92f/fCD4-Cre+ mice and miR-17-92f/fLck-Cre+ mice, in which the gene deletion is initiated at the DN4 or late DN2 stages, respectively. Apart from a minor reduction in cellularity, T cells developed normally in the absence of miR-17-92 and we observed no reduction in the percentage of natural Tregs (nTregs) in the thymus, spleen, or LNs in these mice (supplemental Figures 6 and 7).
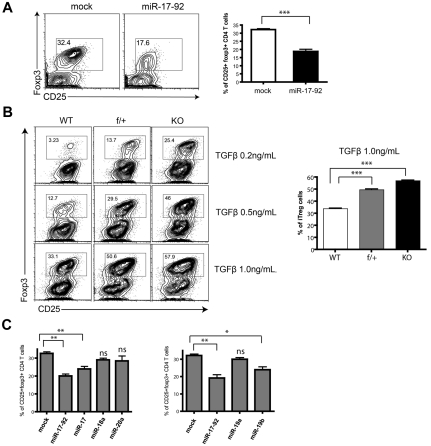
Foxp3+ iTregs are generated through the conversion of CD4+CD25− conventional T cells in the periphery. We used an in vitro differentiation assay25 to examine the role of miR-17-92 in this process. Overexpression of miR-17-92 significantly impaired the induction of Foxp3+CD25+ cells (Figure 4A). We then cultured peripheral CD4+CD25− T cells from miR-17-92–deficient mice in Treg-skewing conditions for 6 days. Consistent with the gain-of-function results, ablation of the miR-17-92 cluster in CD4+ T cells dramatically enhanced Foxp3 expression for various dosages of TGF-β treatment (Figure 4B). When the 6 miRNAs were expressed individually, miR-17 and miR-19b again emerged as the functional representatives inhibiting iTreg differentiation (Figure 4C and supplemental Figure 8).
Figure 4.
miR-19b and miR-17 suppress iTreg differentiation. (A) 5C.C7 T cells were primed, transduced with retrovirus encoding GFP alone or miR-17-92/GFP, and cultured under iTreg-differentiation conditions for 6 days with 0.2-1.0 ng/mL of recombinant human TGFβ (Peprotech), 50 U/mL of recombinant mouse IL-2 (Peprotech), 10 μg/mL of anti–IL-4 (11B11), 10 μg/mL of anti–IFN-γ (XMG1.2), and 10 μg/mL of anti–IL-6 (BD Biosciences). The percentage of Treg cells within the CD4+GFP+ population was measured by Foxp3 staining (eBioscience). The bar graph summarizes means ± SEM from 4 independent experiments. (B) CD4+CD25− T cells sorted from LNs and spleens of WT, f/+, and KO littermates were cultured under iTreg-differentiation conditions for 6 days with the indicated TGF-β doses, and the percentage of CD25+Foxp3+ Treg cells was assessed. Left: representative FACS plot; right: statistical analysis of 4 independent experiments at the indicated TGF-β dose. (C) 5C.C7 T cells were transduced with individual miRNAs from the miR-17 or miR-19 families, and cultured under iTreg-differentiation conditions for 6 days. The percentage of CD25+Foxp3+ cells was measured by flow cytometry. Bar graphs summarize the means ± SEM of 3 independent experiments. *P < .05; **P < .01; ***P < .00; and ns, no significance.
Pten is the principal miR-19b target regulating CD4+ T-cell function
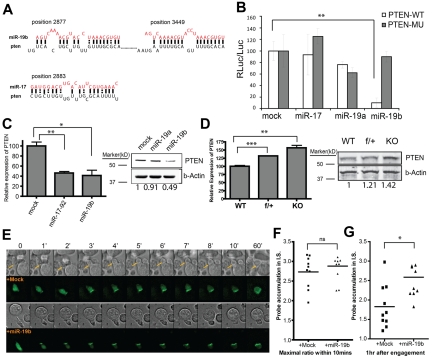
Because miR-19b had a profound impact on CD4+ T cell effector responses, we reasoned that the specific genes suppressed by miR-19b should be dominant inhibitors that negatively regulate multiple complicated processes. Previous reports have identified Pten, a negative regulator of PI3K signaling, as a target of the miR-17-92 cluster.22 Computational prediction revealed that the 3′UTR of pten mRNA contains 2 miR-19b– and 2 miR-17–binding sites highly conserved between mouse and human (Figure 5A). To assess miRNA binding to the 3′UTR, we constructed luciferase reporters with the full-length pten 3′UTR. The reporter was tested in NIH3T3 cell lines stably overexpressing miR-19b, miR-19a, or miR-17. We observed significant suppression of luciferase production by miR-19b, but not miR-17 or miR-19a (Figure 5B). In addition, site-directed mutagenesis of the miR-19–binding motifs within the 3′UTR of pten26 completely abolished this suppression (Figure 5B), suggesting that miR-19b directly binds to these 2 sites. Consistently, overexpression of miR-19b in T cells down-regulated endogenous Pten mRNA and protein levels (Figure 5C). Reciprocally, mRNA and protein levels of Pten were significantly increased in CD4+ T cells lacking the miR-17-92 gene (Figure 5D).
Figure 5.
Pten is the primary target of miR-19b in regulating CD4+ T cell effector functions. (A) Schematic illustration of the predicted targeting sites for miR-19b and miR-17 within the 3′UTR of pten mRNA. (B) The full-length 3′UTR of pten (Pten-WT) or 3′UTR with mutations at the 2 miR-19b target sites (Pten-MU) were cloned downstream of a luciferase reporter and transfected into NIH3T3 cell lines stably expressing the indicated miRNAs. The luciferase activity was measured 72 hours after transfection. Bar graphs show the means ± SD of 3 independent experiments. (C) 5C.C7 T cells transduced with mock, mir-17-92, or mir-19b were sorted by FACS, and total RNA and protein were extracted for qPCR and Western blot. (D) Relative expression of Pten mRNA in CD4+CD25− T cells from LNs and spleens of WT, f/+, and KO mice was measured by qPCR. T cells activated with plate-bound anti–CD3/CD28 Abs for 72 hours were lysed for protein quantification. qPCR data were normalized to SDHA and are shown as relative to mock or WT. Bar graph shows the means ± SEM for 3 independent experiments. (E) miR-19b–mediated regulation on PI3K signaling on antigen engagement was visualized by fluorescence video microscopy. As described previously,27 the PH domain of Akt kinase was fused with GFP as an imaging probe to monitor the production of PIP3 through PI3K activation. miR-19b or mock vectors were expressed simultaneously with this PH-GFP–imaging probe in 5C.C7 T cells, which were challenged by CH27 APCs preloaded with MCC agonist peptide. Live-cell imaging was performed to monitor the initial signaling strength and the duration of PI3K activation. T cells with PH-GFP expression and stimulated by contact with a single APC were monitored for > 1 hour in a 3D manner, and the best focus plane was used for data analysis. The activity of PI3K was represented by measuring the ratio of the average probe fluorescent intensity in the synaptic region versus the average intensity in the rest cell area. Top panels: representative montages from the DIC channel; bottom panels: representative montages from the GFP channel. (F) The highest level of probe synaptic accumulation within the first 10 minutes of T:APC contact was used as the mark for the maximal activity of PI3K activation in the initiation stage (before the formation of mature immunologic synapse) of TCR signaling. (G) A similar measurement was performed at 1 hour after the initiation of TCR signaling. *P < .05; **P < .01; and ***P < .001.
To assess directly the biochemical consequence of miR-19b targeting, we performed live-cell imaging to visualize the perturbation by miR-19b of PI3K signaling initiated by TCR-antigen recognition. The PH domain of Akt kinase was fused with GFP as a probe to monitor the local production of PIP3 through PI3K activation. miR-19b and this PH-GFP probe were simultaneously expressed in 5C.C7 T cells, which were then challenged with MCC peptide loaded on APCs. Within 10 seconds, agonist engagement of the TCR led to PI3K activation, which was sustained for hours inside the gradually assembled immunologic synapse.27 When we expressed miR-19b in 5C.C7 T cells and followed the dynamics of PH-GFP probe recruitment upon stimulation, we did not detect any enhancement of PI3K activation during the initiation stage (ie, within 10 minutes of T:APC engagement, Figure 5E-F); however, when measured 1 hour after T:APC engagement, there was a significant enhancement of sustained PI3K activity (Figure 5E, G). These single-cell assays strongly suggest that the Pten-mediated opposition of PI3K signaling was weakened by miR-19b expression.
miR-17 facilitates effector T-cell responses by targeting TGFβRII and CREB1
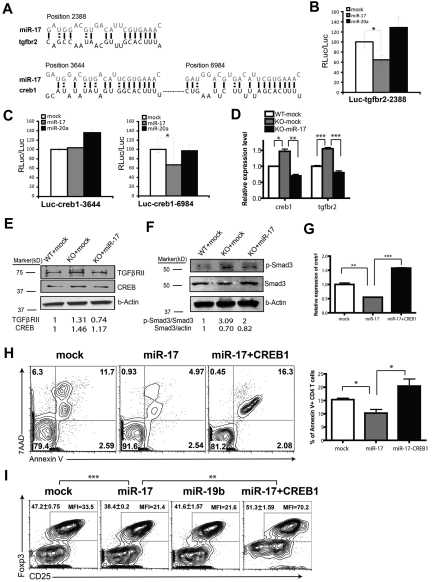
In addition to miR-19b, miR-17 also played a key role in inhibiting AICD of CD4+ T cells and in blocking iTreg differentiation. Although it was reported previously that Bim, a proapoptotic factor, is targeted by the miR-17-92 cluster,20,22 we failed to find any biochemical evidence of such targeting in CD4+ T cells (supplemental Figure 9). To further explore the molecular mechanism of miR-17 regulation, microarray analysis was performed to characterize the gene-expression patterns among WT T cells, T cells with miR-17-92 deletion, and T cells from the KO background with miR-17 added back. All microarray data are available in the Gene Expression Omnibus under accession number GSE32533. From the analysis, 122 genes were up-regulated in the miR-17-92–deficient CD4+ T cells and reciprocally repressed when miR-17 was reintroduced. We next searched these mRNAs for sequences complementary to the seed region of miR-17. Twenty-two genes were selected as candidates that might be controlled directly by miR-17 (supplemental Table 1). Based on the phenotypic impact of miR-17 in T cells, 2 genes in this group attracted our attention: TGF, β receptor II (tgfbr2), and cAMP-responsive element binding protein 1 (creb1), both of which have been implicated previously in the Treg-differentiation pathway.28,29 In addition, C/EBP-β, a CREB1-transcriptional target, has been reported to promote apoptosis of macrophages after IFN-γ stimulation.30 Through bioinformatics approaches, one conserved miR-17–binding site was identified in the 3′UTR of tgfbr2 and 2 sites were found for creb1 (Figure 6A). We verified that miR-17 can bind directly to the 3′UTR of tgfbr2 (position 2388) and creb1 (position 6984) mRNAs (Figure 6B-C) using luciferase reporter assays. In CD4+ T cells lacking mir-17-92, the expression levels of TGFβRII and CREB1 were significantly elevated at both the mRNA and protein levels, and these elevations were completely abolished when the mir-17 gene was reintroduced (Figure 6D-E).
Figure 6.
miR-17 modulates CD4+ T cell effector responses by targeting TGFβRII and CREB1. (A) Schematic representation of the putative miR-17–binding sites in the 3′UTR of tgfbr2 and creb1. (B-C) A portion of the 3′UTR of tgfbr2 (NM009371 position 2171-2552) or creb1 (NM133828 position 3442-3792 containing site 3644 or position 6758-7144 containing site 6984) was cloned downstream of a luciferase reporter and transfected into an NIH3T3 cell line stably expressing miR-17, miR-20a, or mock control. The luciferase activity was measured 72 hours after transfection. Bar graphs show the means ± SD of 3 (B) or 6 (C) independent experiments. (D-E) CD4+CD25− T cells from WT or KO mice were transduced with indicated retrovirus, and CD4+GFP+ T cells were FACS sorted and total RNA and protein were extracted for qPCR (D) and Western blot (E). The bar graph shows means ± SEM from 3 independent experiments. (F) T cells were transduced with indicated virus and cultured under iTreg-differentiation conditions for 5 days. CD4+GFP+ T cells were then sorted, lysed, and analyzed for Smad3 Ser423/425 phosphorylation by Western blot. (G-I) 5C.C7 T cells were primed and transduced with retrovirus containing both the CREB1-IRES-GFP expression cassette and the indicated miR-17 expression cassette, miR-17 alone with GFP marker, or GFP only. (G) Assessment of CREB1 expression in 5C.C7 T cells by qPCR. The graph shows means ± SEM from 3 independent experiments. (H) Death profile of CD4+GFP+ T cells after restimulation with anti–CD3/CD28 Abs. Left: representative FACS plot; right: bar graphs showing means ± SEM from 3 independent experiments. (I) 5C.C7 T cells were transduced as indicated and cultured under iTreg-differentiation conditions for 5 days. The percentage of CD25+Foxp3+ cells inside of the CD4+ T-cell populations and the MFI of the intracellular Foxp3 staining were measured. The numbers on the left corner show the means ± SEM of the percentage of iTreg cells from 3 independent experiments. *P < .05; **P < .01; and ***P < .001.
To validate its functional relevance, we examined whether the moderate reduction of TGFβRII by miR-17 was sufficient to alter TGF-β signaling. TGF-β binds to TGFβRII, which initiates the signaling cascade by recruiting and phosphorylating the type I receptor. During Treg differentiation, this receptor activation leads to Smad3 Ser423/425 phosphorylation and translocation to its binding sites within the foxp3 gene-enhancer region.28,31 We found that Smad3 was hyperphosphorylated on TGFβ treatment in miR-17-92–deficient T cells, and this was diminished by miR-17 ectopic expression (Figure 6F).
The transcription factor CREB1 was shown to bind to the CNS2 region of the foxp3 gene in a DNA-methylation–sensitive manner.29 To demonstrate the functional importance of targeted CREB1 suppression by miR-17, we restored CREB1 expression in the presence of miR-17 expression using a bicistronic expression vector (Figure 6G). In CD4+ T cells, the epichromosomal expression of CREB1 reversed the phenotype of miR-17 expression: there was loss of protection from AICD (Figure 6H) and rescue of iTreg differentiation at both the population and single-cell levels (Figure 6I). We concluded that the moderate inhibition of TGFβRII and CREB1 by miR-17 results in the diminution of iTreg lineage commitment, and that the dampened CREB1 expression by miR-17 is sufficient to aid T-cell survival against excessive contraction.
miR-17-92 is essential for effective CD4+ T-cell antitumor responses
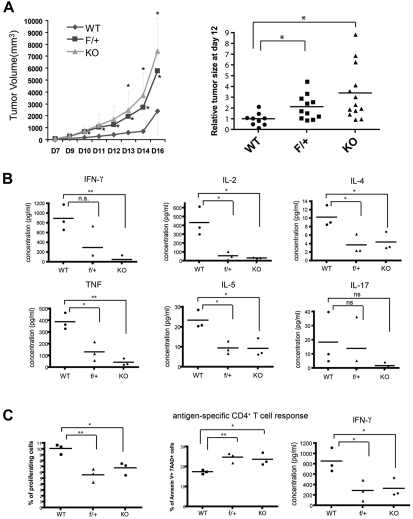
Our phenotypic and mechanistic studies of miR-17-92 indicated that this cluster is a multifaceted promoter of Th1 effector responses. Because Th1-guided effector T-cell function has been well established as a critical defense mechanism for immune surveillance and rejection of tumors, we hypothesized that miR-17-92 might be an important component in the adaptive immune system's attempt to control tumor progression. We subcutaneously transplanted 2 × 105 B16/F10 melanoma tumor cells into miR17-92f/f CD4-Cre+ mice and monitored tumor growth. In mice homozygous or heterozygous for miR-17-92 deletion, the B16 tumors formed faster and had significantly accelerated growth compared with the tumors in WT mice. At day 16, tumor volumes at the site of injection were 2- to 3-fold greater in mice lacking miR-17-92. The loss of a single copy of this gene is sufficient to impair tumor protection (Figure 7A). To detail the functional failure of T cells in the antitumor response, lymphocytes from tumor dLNs were stimulated with plate-bound anti–CD3/CD28 Abs for 24 hours to measure the secretion of the inflammatory cytokines. We found that both Th1 and Th2 cytokines were dramatically reduced in the lymphocyte culture from the dLNs of miR-17-92–deficient mice (Figure 7B). Although there was a declining trend of IL-17 production in the same assay, the change was not statistically significant (Figure 7B). To confirm the essential role of CD4+ T cells in the anti-B16 tumor responses, we collected CD4+ T cells from WT or KO miR-17-92 mice and CD8+ T cells from the tumor antigen–specific Pmel TCR–transgenic mice and cotransferred them into Rag2−/− mice that were then challenged with B16/F10 cells. T-cell responses in the dLNs were analyzed 18 days after tumor transplantation. We found that deficiency of miR-17-92 in CD4+ T cells alone significantly impaired the Th1 response to B16 tumor cells, including a decrease in cell number (supplemental Figure 10A) and IFN-γ production of CD4+ T cells (supplemental Figure 10B) in the dLNs. In addition, the ability of miR-17-92–deficient CD4+ T cells to help the CD8+ T cells was also inhibited (supplemental Figure 10C).
Figure 7.
In vivo, the miR-17-92 cluster is essential for the T cell–mediated antitumor response. (A) WT (n = 5), f/+ (n = 8), and KO (n = 7) littermates were injected subcutaneously with 2 × 105 B16/F10 melanoma cells. The tumor volume was measured each day from 7 days after injection up to 16 days and plotted against time. Left panel: the representative tumor growth curve; right panel: compiled data from 3 independent experiments. Individual dot represents the relative tumor volume normalized to that of the WT at 12 days after melanoma cell injection. (B) Lymphocytes from the dLNs of tumor-carrying mice were isolated 16 days after B16/F10 melanoma inoculation and stimulated with 1 μg/mL of anti–CD3/CD28 Abs for 24 hours. Supernatants from the cultures were assayed for the concentration of indicated Th1, Th2, and Th17 cytokines using a cytokine bead array (BD Biosciences and Bender). Each dot represents data obtained from an individual mouse (n = 3). (C) WT, f/+, and KO littermates were injected with 3 × 105 OVA-secreting B16/F0 cells. Sixteen days after injection, T cells were enriched from the dLNs, labeled with CFSE, and stimulated with LB27.4 APCs loaded with 10μM OVA peptide (323-339) for 48 hours. Antigen-specific responses (proliferation, AICD, and IFN-γ production) were measured as described above. Each data point represents the sample from an individual mouse (n = 3). Each experiment was repeated at least 3 times. *P < .05; **P < .01; and ns, no significance.
To further demonstrate that this deficiency of immune protection is because of the weakness of antigen-specific T-cell responses against the tumor, we inoculated mice subcutaneously with 3 × 105 B16/F0/OVA cells that ectopically secrete OVA protein. Sixteen days after transplantation, T cells were enriched from the dLNs, labeled with CFSE, and restimulated with OVAII peptide (aa 323-339)–loaded APCs for 48 hours. Upon antigen-specific recall, OVA-specific CD4+ T cells exhibited a significant defect in proliferation and IFN-γ secretion and enhanced apoptosis, all of which correspond to the function of miR-17-92 as revealed in vitro (Figure 7C). These results suggest that miR-17-92 profoundly regulates T cell antitumor responses through strict enforcement on their Th1-lineage–specific functions.
Discussion
Using gain- and loss-of-function analysis, the results of the present study demonstrate that the miR-17-92 cluster is a multicomponent potentiator that governs T cell responses to antigen challenge. Specifically, this cluster manages the efficacy of Th1 responses by protecting T cells from AICD, enhancing T cell proliferation, facilitating IFN-γ production, and obstructing iTreg differentiation. The extensiveness of the regulation of miR-17-92 was especially evident during T cell–dependent tumor rejection, because local Th1-guided cytotoxicity is crucial for direct tumor elimination and indirect galvanization of macrophage activation.4,32 Mice with a T cell–specific single-allele ablation of miR-17-92 were extremely vulnerable to B16 melanoma transplantation, and further analysis revealed that CD4+ T cells from these recipients' dLNs were defective in all of the aforementioned aspects of Th1 responses upon tumor antigen challenge.
The miR-17-92 cluster is produced from a single transcript designated C13orf25.33 Taking this cluster as a single entity, miR-17-92 provides cohesive guidance for T cell antigen responses. However, our data indicate that the individual miRNAs within the cluster are quite diversified in terms of their functions. Previously, we and others independently identified miR-19 as the key component of this cluster in promoting Myc-induced B-cell lymphomas.26,34 In T cells, miR-19b is also critical, because it comprehensively drives the antigen response in every tested aspect. Furthermore, the regulation provided by miR-19b is greatly facilitated by miR-17 in protecting cells from AICD and especially in suppressing iTreg differentiation. Surprisingly, we noted that miR-18a opposes the cluster's pro-Th1 function, primarily through elevation of AICD and inhibition of proliferation. Given that miR-17-92 has been well recognized as an onco-miR, we suspect that miR-18a might act as a “brake” on the pro-proliferation and antiapoptosis functions of the cluster, which is a common phenomenon for proteinaceous oncogenes.35–37
The miRNAs comprising the miR-17-92 cluster can be grouped based on the similarity of their seed regions (nucleotides 2-8), which are thought to be especially critical for the specificity of mRNA targeting. Our current, limited knowledge predicts that the majority of targets should be shared between family members. However, inside primary CD4+ T cells, and despite the very high degree of homology within the miR-17 family, miR-20a is not capable of performing any of pro-Th1 functions of miR-17, and miR-18 clearly exerts an antagonistic effect. Similar distinction was also observed between miR-19a and miR-19b. One explanation is that the diversity of functionality might simply reflect differences in their expression levels. Expression levels between miR-19a and miR-19b were significantly different in T cells, which therefore prevents us from drawing any solid conclusions about the functional diversity inside this family. Nevertheless, our data do suggest a potential difference in the endogenous processing or maintenance of these 2 miRNAs in effector T cells, with miR-19a being disadvantaged. Conversely, inside the miR-17 family, the overexpression level and the absolute copy number of miR-20a was equal to or even higher than that of miR-17 (supplemental Figure 11 and supplemental Figure 1F). Although our quantification determined that miR-18a is expressed at a lower level, the antagonist effect is so evident that distinctions of mRNA targeting become a more conceivable explanation. In mature miRNAs, only few nucleotides differ between miR-17 and miR-20a/miR-18a, and these differences reside outside the seed region. We suspect that these subtle differences are sufficient to result in a significant affinity difference between the miRNAs and their targets to produce the observed differential targeting. Alternatively, as reported previously,38 the loop sequence of the pre-miRNA may participate in the process of target recognition. In that case, the distinct mRNA-targeting activities of miR-181a and miR-181c were largely determined by their divergent pre-miRNA loop sequences, but not by the one-nucleotide alteration in the mature miRNAs. Because the pre-miRNA loops also differ between miR-17 and miR-20a/miR-18a, it is possible that the targeting preference between them is caused by differences in their pre-miRNA loops. Regardless, our results do argue that the sequences outside the seed region can also be an indispensible component of the miRNA-targeting machinery.
It was reported previously that the miR-17-92 cluster influences lymphocyte proliferation and survival by targeting the tumor-suppressor protein Pten.22 In the present study, we showed that Pten is the dominant functionally relevant target of miR-19b in CD4+ T cells. The ability of Pten to suppress T-cell proliferation and survival through its antagonizing effect on the PI3K-Akt pathway is well documented.39 In addition, the PI3K-Akt pathway has also been shown to be critical for facilitating Th1 differentiation and supporting IFN-γ production,40,41 and the PI3K-AKT-mTOR axis was shown to inhibit iTreg differentiation upon TGF-β induction.42,43 As shown by our live-cell imaging experiments, CD4+ T cells with ectopic miR-19b expression have dramatically prolonged PI3K activity at a higher level inside the immunologic synapse. Therefore, although we do not preclude the involvement of other targets, all of the observed phenotypic aspects of miR-19b in effector T cells can be explained by the diminution of Pten expression.
The other novel finding in this study was that miR-17 functions through targeting CREB1. By restoring CREB1 levels in miR-17–coexpressing T cells, we demonstrated that the subtle increase of this protein diminished the protection from AICD afforded by miR-17. This functionally confirmed CREB1 as a target of miR-17 and, more importantly, provided the first evidence that CREB1 is a proapoptotic factor during AICD in CD4+ T cells. The ability to restrain iTreg differentiation was an unexpected role for miR-17. In accordance with previous findings,44,45 we found that miR-17 inhibits TGF-β signaling through targeting TGFβRII. Biochemically, the reduction of either CREB1 or TGFβRII is not dramatic, but the impacts from these moderate adjustments on T-cell function are not negligible. However, despite its clear role during iTreg differentiation, the loss of miR-17-92 did not influence the development of nTregs in our mouse models. One possible explanation for this is that the moderate protein-level changes of TGF-βRII, CREB1, and Pten caused by the loss of miR-17/miR-19b, while critical for Foxp3 induction in peripheral conventional T cells, are not sufficient to affect thymic nTreg development. Alternatively, miR-17 and miR-19b do affect the thymic selection of the nTreg population, but in a more subtle way (eg, via TCR repertoire changes within the mature nTreg pool).
Acknowledgments
The authors thank Drs Thomas Tedder and Edith Lord for their generous gifts of the B16 and B16/OVA cell lines, Drs Wei Jia and Pengyuan Yang for technical support on the adoptive transfer and histology, and Drs Youwen He and Peter J. R. Ebert for critical suggestions and reading of the manuscript.
L.H. is a Searle Scholar and is supported by a pathway to independence grant, an RO1 grant from the National Cancer Institute, and a new faculty award from the California Institute for Regenerative Medicine. Q.-J.L. is a Whitehead Family Foundation Scholar and is supported by grants from the American Cancer Society (RSG-10-157-01-LIB), the American Diabetes Association (1-10-JF-28), and the National Institute of Allergy and Infectious Diseases (R56AI091878).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.J. and C.L. carried out most of the experiments and analyzed the data; V.O. and Y.W. contributed critical reagents and technical support; E.L. and F.F. performed bioinformatic analysis of the microarray data for miR-17 target identification; J.S. provided technical support for the animal studies; L.H. and Q.-J.L. conceptualized the project, designed the experiments, supervised the work, and interpreted the results; and C.L., S.J., E.L., L.H., and Q.-J.L. prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Qi-Jing Li, Department of Immunology, Duke University Medical Center, 207 Research Dr, Jones Bldg, Rm 303, Durham, NC 27710; e-mail: qi-jing.li@duke.edu.
References
- 1.Krammer PH, Arnold R, Lavrik IN. Life and death in peripheral T cells. Nat Rev Immunol. 2007;7(7):532–542. doi: 10.1038/nri2115. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda H, Chamoto K, Tsuji T, et al. The critical role of type-1 innate and acquired immunity in tumor immunotherapy. Cancer Sci. 2004;95(9):697–703. doi: 10.1111/j.1349-7006.2004.tb03248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30(5):636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Gallimore A, Sakaguchi S. Regulation of tumour immunity by CD25+ T cells. Immunology. 2002;107(1):5–9. doi: 10.1046/j.1365-2567.2002.01471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li QJ, Chau J, Ebert PJ, et al. miR-181a Is an Intrinsic Modulator of T Cell Sensitivity and Selection. Cell. 2007;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9(8):839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 9.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koralov SB, Muljo SA, Galler GR, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Cobb BS, Nesterova TB, Thompson E, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201(9):1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202(2):261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205(9):1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X, Jeker LT, Fife BT, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205(9):1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu LF, Thai TH, Calado DP, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30(1):80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu LF, Boldin MP, Chaudhry A, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142(6):914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Connell RM, Kahn D, Gibson WS, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33(4):607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontana L, Pelosi E, Greco P, et al. MicroRNAs 17-5p–20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9(7):775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 22.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42(8):1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li QJ, Dinner AR, Qi S, et al. CD4 enhances T cell sensitivity to antigen by coordinating Lck accumulation at the immunological synapse. Nat Immunol. 2004;5(8):791–799. doi: 10.1038/ni1095. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Jin W, Hardegen N, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198(12):1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huppa JB, Gleimer M, Sumen C, Davis MM. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat Immunol. 2003;4(8):749–755. doi: 10.1038/ni951. [DOI] [PubMed] [Google Scholar]
- 28.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 29.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204(7):1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gade P, Roy SK, Li H, Nallar SC, Kalvakolanu DV. Critical role for transcription factor C/EBP-beta in regulating the expression of death-associated protein kinase 1. Mol Cell Biol. 2008;28(8):2528–2548. doi: 10.1128/MCB.00784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9(2):194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 32.Muranski P, Restifo NP. Adoptive immunotherapy of cancer using CD4(+) T cells. Curr Opin Immunol. 2009;21(2):200–208. doi: 10.1016/j.coi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339(2):327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 34.Mu P, Han YC, Betel D, et al. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23(24):2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frisch SM, Mymryk JS. Adenovirus-5 E1A: paradox and paradigm. Nat Rev Mol Cell Biol. 2002;3(6):441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- 36.La Thangue NB. The yin and yang of E2F-1: balancing life and death. Nat Cell Biol. 2003;5(7):587–589. doi: 10.1038/ncb0703-587. [DOI] [PubMed] [Google Scholar]
- 37.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 38.Liu G, Min H, Yue S, Chen CZ. Pre-miRNA loop nucleotides control the distinct activities of mir-181a-1 and mir-181c in early T cell development. PLoS One. 2008;3(10):e3592. doi: 10.1371/journal.pone.0003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckler JL, Liu X, Turka LA. Regulation of T-cell responses by PTEN. Immunol Rev. 2008;224:239–248. doi: 10.1111/j.1600-065X.2008.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okkenhaug K, Patton DT, Bilancio A, Garcon F, Rowan WC, Vanhaesebroeck B. The p110delta isoform of phosphoinositide 3-kinase controls clonal expansion and differentiation of Th cells. J Immunol. 2006;177(8):5122–5128. doi: 10.4049/jimmunol.177.8.5122. [DOI] [PubMed] [Google Scholar]
- 41.Soond DR, Bjorgo E, Moltu K, et al. PI3K p110delta regulates T-cell cytokine production during primary and secondary immune responses in mice and humans. Blood. 2010;115(11):2203–2213. doi: 10.1182/blood-2009-07-232330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205(3):565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauer S, Bruno L, Hertweck A, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008;105(22):7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mestdagh P, Bostrom AK, Impens F, et al. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol Cell. 2010;40(5):762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dews M, Fox JL, Hultine S, et al. The myc-miR-17∼92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010;70(20):8233–8246. doi: 10.1158/0008-5472.CAN-10-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]