Abstract
Amyloid fibrils are self-assembled filamentous structures associated with protein deposition conditions including Alzheimer's disease and the transmissible spongiform encephalopathies. Despite the immense medical importance of amyloid fibrils, no atomic-resolution structures are available for these materials, because the intact fibrils are insoluble and do not form diffraction-quality 3D crystals. Here we report the high-resolution structure of a peptide fragment of the amyloidogenic protein transthyretin, TTR(105–115), in its fibrillar form, determined by magic angle spinning NMR spectroscopy. The structure resolves not only the backbone fold but also the precise conformation of the side chains. Nearly complete 13C and 15N resonance assignments for TTR(105–115) formed the basis for the extraction of a set of distance and dihedral angle restraints. A total of 76 self-consistent experimental measurements, including 41 restraints on 19 backbone dihedral angles and 35 13C–15N distances between 3 and 6 Å were obtained from 2D and 3D NMR spectra recorded on three fibril samples uniformly 13C, 15N-labeled in consecutive stretches of four amino acids and used to calculate an ensemble of peptide structures. Our results indicate that TTR(105–115) adopts an extended β-strand conformation in the amyloid fibrils such that both the main- and side-chain torsion angles are close to their optimal values. Moreover, the structure of this peptide in the fibrillar form has a degree of long-range order that is generally associated only with crystalline materials. These findings provide an explanation of the unusual stability and characteristic properties of this form of polypeptide assembly.
Amyloid fibrils are highly organized filamentous structures formed by the self-assembly of polypeptide molecules that in their soluble forms can have a wide variety of secondary structures and functions. Approximately twenty peptides and proteins aggregate into amyloid fibrils in vivo and are associated with protein deposition diseases including type II diabetes, Alzheimer's disease, and the transmissible spongiform encephalopathies (1–3). In addition, numerous peptides and proteins without connection to any known disease have been shown to be capable of forming amyloid fibrils in vitro under appropriate conditions (1–5), leading to the suggestion that the ability to form fibrils is an inherent property of polypeptide chains (3, 6). Regardless of the polypeptide precursor, all amyloid fibrils exhibit several common characteristics (1–3): (i) fibrils bind the dye Congo red, resulting in a green birefringence under polarized light; (ii) electron microscopy (EM) and atomic force microscopy reveal that fibrils are typically long, unbranched, and ≈100 Å in diameter; and (iii) x-ray diffraction patterns of oriented fibrils indicate an ordered “cross-β” structure, which consists of β-sheets running parallel to the fibril axis, with the individual peptide strands oriented perpendicular to the fibril axis (1, 7). According to the most recent x-ray and EM models of amyloid fibril structure (3, 8–14) on the supramolecular level, the fibrils appear to consist of a small number of protofilaments (typically two to six) wound around a core that may in some cases be hollow. The protofilaments are thought to be composed of relatively flat β-sheets with an overall long-range twist, which can be accomplished by minimal rotations (of ≈1–3°) between successive peptide strands (8, 13, 14).
Atomic-resolution structural information about the individual peptide strands that make up the protofilaments has proved difficult to obtain because although the fibrils are large multimolecular assemblies, they lack complete 3D order and are thus not amenable to routine characterization by x-ray crystallography and solution-state NMR spectroscopy. Recent advances in magic angle spinning (MAS) solid-state NMR instrumentation and methodology have, however, permitted the de novo determination of the 3D structures of biological molecules in the microcrystalline states, notably of a tripeptide N-formyl-l-Met-l-Leu-l-Phe (15), and a 62-residue α-spectrin Src homology 3 (SH3) domain (16). Such techniques can also be applied to the structural characterization of amyloid fibrils and other incompletely ordered biological systems. Indeed, site-specific structural measurements have been performed for various amyloidogenic peptides (17–26), and a structural model for fibrils formed by the full length Alzheimer's β-amyloid peptide has recently been proposed (24). In this article we report the success of these techniques in permitting the determination of the high-resolution structure of a peptide in an amyloid fibril, elucidating not only the backbone fold but also the precise conformation of the side chains.
The peptide involved is an 11-residue fragment of human transthyretin (TTR) (27). TTR is a 55-kDa homotetramer of 127-residue subunits with extensive β-sheet structure (28) and is involved in the transport of thyroxine and retinol in plasma. Wild-type TTR and many of its fragments and naturally occurring mutants self-assemble into amyloid fibrils in vivo and in vitro and are associated with diseases including familial amyloid polyneuropathy and senile systemic amyloidosis (1, 2, 27). TTR(105–115) (amino acid sequence Tyr-Thr-Ile-Ala-Ala-Leu-Leu-Ser-Pro-Tyr-Ser), which comprises residues 105–115 of TTR, corresponds to a β-strand located at the surface of the thyroxine-binding channel formed by the homotetramer. This peptide can be assembled into homogeneous amyloid fibrils (27) with favorable spectroscopic properties (26) and hence represents an important model system for the investigation of structural details in fibrils at atomic resolution. Recently we have described the nearly complete sequence-specific backbone and side-chain 13C and 15N NMR chemical shift assignments for TTR(105–115) in the fibrillar state (26), an essential step in the determination of the complete 3D structure. In this article we describe a series of NMR experiments that has allowed us to extract a large number of internuclear distances and dihedral angles that relate to this peptide in the fibrils. These restraints were used to calculate an ensemble of low-energy peptide structures using simulated annealing molecular dynamics. The results indicate the presence of remarkably well defined backbone and side-chain conformations for the majority of the residues in the peptide. The main chain of TTR(105–115) in the amyloid fibril is in an extended β-strand conformation. This observation is in agreement with other measurements, including our preliminary experiments (26), which indicate that amyloid fibrils consist primarily of β-sheet secondary structure (1–3, 7, 9, 13, 14, 17–25). In addition, the atomic-resolution structure reveals previously inaccessible details about the precise conformation of the backbone and side chains.
Materials and Methods
TTR(105–115) Amyloid Fibrils. Isotopically labeled TTR(105–115) peptides (amino acid sequence Tyr-Thr-Ile-Ala-Ala-Leu-Leu-Ser-Pro-Tyr-Ser) used in the present work were synthesized by using standard solid-phase methods and purified by HPLC (CS Bio, San Carlos, CA). The details of the preparation and characterization of TTR(105–115) fibrils have been presented (26). Most structural restraints were obtained by using three types of isotopically labeled peptides referred to as TTR(105–115)YTIA, TTR(105–115)AALL, and TTR(105–115)LSPY, which were uniformly 13C, 15N (U-13C, 15N)-labeled in consecutive stretches of four amino acids as indicated by the subscripts [e.g., for TTR(105–115)YTIA, residues Tyr-105 through Ala-108 are (U-13C, 15N)-labeled].
NMR Spectroscopy. NMR experiments were performed on a custom-designed spectrometer (courtesy of D. J. Ruben, Francis Bitter Magnet Laboratory, Massachusetts Institute of Technology) operating at the 1H Larmor frequency of 500 MHz, using a Varian-Chemagnetics (Fort Collins, CO) triple-resonance 1H/13C/15N T3 probe equipped with a 4-mm spinner module. The spinning frequencies used for all experiments were in the range of 9–11 kHz, regulated to ±5 Hz by using a Doty Scientific (Columbia, SC) spinning frequency controller. The sample temperature during the experiments was maintained at 2°C by using a stream of cooled nitrogen gas, delivered to the sample via a Varian-Chemagnetics variable-temperature stack.
The NMR pulse sequences used for the measurement of the structural constraints have been described in detail (29–34) and are not discussed extensively here. Briefly, the majority of 13C–15N distances were determined by using the 3D z-filtered transferred-echo double-resonance (3D ZF TEDOR) technique (30), and the backbone torsion angles were measured by using 3D dipolar–chemical shift correlation methods, 1HNi–15Ni–13Cαi–1Hαi for φ (31, 32) and 1HNi+1–15Ni+1–13Cαi–1Hαi (32) and 15Ni–13Cαi–13C′i–15Ni+1 (33, 34) for ψ.
Structure Calculations. Structures were calculated by using simulated annealing molecular dynamics in Cartesian coordinates as implemented in the program cns (35). The refinement protocol involved high-temperature molecular dynamics at 3,000 K for a total of 1,000 steps with the duration of 2 ps per step, followed by a cooling stage, where the temperature was gradually decreased from 3,000 to 0 K in 24,000 steps. In addition to the experimental distance and dihedral angle restraints and database-derived restraints (discussed below), which were modeled with square well-quadratic potentials, the total energy target function included energy terms representing covalent bonds, three-atom bond angles and dihedral (improper) angles required to maintain correct geometries, and van der Waals interactions (35). All 20 low-energy structures in the calculated ensemble exhibited acceptable covalent bond geometries with 100% of the (φ, ψ) pairs in the most favored regions of the Ramachandran space as evaluated by the program procheck (36), and no distance or dihedral angle violations >0.1 Å or 5°, respectively (Figs. 4 and 5, which are published as supporting information on the PNAS web site).
Results and Discussion
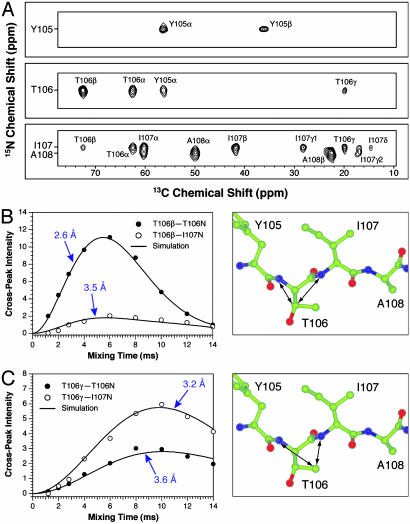
Internuclear Distance Measurements. The majority of C–N distances in the 3–6 Å range in TTR(105–115) fibrils (Table 1 and Table 2, which is published as supporting information on the PNAS web site) were obtained by using the 3D ZF TEDOR technique (30), and representative results are shown in Fig. 1. Fig. 1 A shows a single 2D plane from the 3D ZF TEDOR data set recorded on the TTR(105–115)YTIA sample, consisting of a 15N–13C chemical shift correlation spectrum acquired with a TEDOR mixing time of 6.0 ms. Site-specific distance information is encoded in the intensities of cross-peaks present at characteristic 15N and 13C frequencies (ΩN, ΩC). For example, the cross-peak between I107 13Cδ and I107 15N, which corresponds to a ≈4.5 Å distance, has an intensity significantly lower than the T106 13Cγ–T106 15N and T106 13Cγ–I107 15N crosspeaks at calculated distances of ≈3.5 and 3 Å, respectively. As demonstrated previously (30), precise and accurate distances of up to 4–6 Å can be determined by recording a series of 2D 15N–13C correlation spectra as a function of the TEDOR mixing time (also referred to as cross-peak buildup curves).
Table 1. Summary of restraints and structural refinement statistics for TTR(105–115).
| Restraints | |
| Total experimental restraints* | 76 |
| Total C-N distance restraints | 35 |
| Restraints in the class 2.0 ≤ r < 3.0 Å | 3 |
| Restraints in the class 3.0 ≤ r < 4.0 Å | 22 |
| Restraints in the class 4.0 ≤ r < 6.0 Å | 10 |
| Total backbone dihedral angle restraints† | 41 |
| Chemical shift based (TALOS) restraints | 16 |
| Restraints based on 3D dipolar-chemical shift NMR | 25 |
| Distance restraint violations >0.1 Å | 0 |
| Dihedral angle violations >5° | 0 |
| rmsd from experimental distance restraints, Å | 0.0195 ± 0.0005 |
| rmsd from experimental dihedral angle restraints, Å | 0.85 ± 0.05 |
| Energies | |
| Final energies, kcal·mol-1 | (20)‡ |
| Etotal | 24.8 ± 0.8 |
| Ebond | 1.4 ± 0.1 |
| Eangle | 12.6 ± 0.4 |
| Eimproper | 0.51 ± 0.05 |
| Evan der Waals | 4.6 ± 0.4 |
| Edistance | 3.8 ± 0.2 |
| Edihedral | 1.9 ± 0.2 |
| rmsd from ideal covalent geometry | (20) |
| Bonds, Å | 0.0029 ± 0.0001 |
| Angles, ° | 0.512 ± 0.008 |
| Improper angles, ° | 0.19 ± 0.01 |
| Coordinate rmsd§ | (20) |
| Backbone (residues I107-P113), Å | 0.40 |
| Backbone (residues Y105-S115), Å | 0.69 |
| All heavy atom (residues I107-P113), Å | 0.63 |
| All heavy atom (residues Y105-S115), Å | 1.24 |
| ϕ/ψ in most favored regions of Ramachandran space, %¶ | 100 |
In addition to the 76 restraints used in the structure calculation, 3D ZF TEDOR experiments (Table 2) provided 35 one-bond and two-bond C-N distances (largely independent of peptide conformation), resulting in a total of 111 experimental restraints.
Prior to structure calculation the 41 dihedral restraints were combined into 19 dihedral restraints (9 for ϕ and 10 for ψ) for each backbone dihedral angle except ϕS115 (Table 3). Chemical shift assignments for TTR (105-115) and TALOS (38) predictions for backbone dihedral angles have been presented (26).
(20) represents the average for 20 low-energy structures.
rmsd for the ensemble of 20 low-energy structures were evaluated by using the program MOLMOL (39) for residues Y105-S115 and I107-P113 as indicated in parentheses.
As evaluated by the program PROCHECK (36).
Fig. 1.
Three-dimensional ZF TEDOR 13C–15N distance measurements in TTR(105–115)YTIA fibrils. (A) Strips from a 2D 15N–13C chemical shift correlation spectrum (30) acquired with a TEDOR mixing time of 6.0 ms. Cross-peaks corresponding to distances of ≈4–6 Å are observed for mixing times in the 6- to 12-ms regime. The resonance assignments for TTR(105–115) have been presented (26). (B) Experimental (○ and •) and simulated (—) intensities for the T106 13Cβ–T106 15N(•) and T106 13Cβ–I107 15N(○) cross-peaks as a function of the TEDOR mixing time, and the relevant molecular fragment showing the measured distances. (C) Same as in B but for T106 13Cγ. The distance measurements are summarized in Table 1 and presented in detail in Table 2.
Fig. 1 B and C shows representative buildup curves for cross-peaks corresponding to the T106 13Cβ–T106 15N, T106 13Cβ–I107 15N, T106 13Cγ–T106 15N, and T106 13Cγ–I107 15N distances. For each buildup curve the cross-peak intensity observed in the 2D 15N–13C correlation spectrum is indicated with open or filled circles. In the 3D ZF TEDOR experiment the build up of cross-peak intensity at frequencies (ΩN, ΩC), characteristic of a particular pair of nuclear spins, depends primarily on the active dipolar coupling between the spins and hence the corresponding internuclear distance (a rapid cross-peak buildup translates into a short distance). Therefore, it is immediately apparent that the T106 13Cβ–T106 15N distance is shorter than the T106 13Cβ–I107 15N distance, and that T106 13Cγ is closer to the I107 15N atom than to the T106 15N atom. Quantitative distance measurements are obtained by a least-squares fit of experimental cross-peak buildup curves to theoretical trajectories calculated as a function of the internuclear distance (30) (the best-fit simulations are indicated with solid lines in Fig. 1 B and C). For Thr-106, the distance measurements involving the 13Cβ and 13Cγ nuclei place very strong constraints on the possible side-chain conformations. Note that because of the extended β-strand nature of the peptide, most distance constraints are of the intraresidue and sequential type (i.e., between side-chain 13C sites of residue i and backbone 15N amides of residues i and i + 1). Furthermore, relatively few distance restraints are available for Tyr-105 and Tyr-114 because of spectral overlap in the aromatic region, and no restraints are available for the C-terminal Ser-115 residue, which was not uniformly 13C, 15N (U-13C, 15N)-labeled. Consequently, the conformations of residues T106–P113 in the central region of the peptide are much more accurately defined than those of the N- and C-terminal residues.
Torsion Angle Measurements. Backbone torsion angles in TTR(105–115) fibrils (Table 1 and Table 3, which is published as supporting information on the PNAS web site) were determined by using 3D dipolar–chemical shift correlation experiments, which report on the relative orientations of 1H–15N, 1H–13C, and 13C–15N dipolar tensors (31–34). It is well established that these experiments have regions of highest angular resolution and sensitivity for molecular conformations that correspond to relatively small deviations from the parallel or antiparallel orientation of the two dipole vectors of interest (i.e., projection angles of |Θ| ≈ 0–30°). For the peptide backbone, the experiment which correlates the 1HN–15N and 1Hα–13Cα dipolar coupling tensors within residue i, also referred to as 1HNi–15Ni–13Cαi–1Hαi (31, 32), is most sensitive for peptide conformations characterized by φi ≈ –120 ± 30° (i.e., conformations in the β-sheet region of Ramachandran space). The torsion angle actually probed in the 1HNi–15Ni–13Cαi–1Hαi experiment is not φ but rather θ(HN—N—Cα—Hα), which for l amino acids is related to φ according to φ ≈ θ + 60° (31, 32). Similarly, it can be shown that experiments which correlate the 1HN–15N dipolar coupling of residue i + 1 with the 1Hα–13Cα coupling of residue i (1HNi+1–15Ni+1–13Cαi–1Hαi) (32) and the 15N–13Cα coupling of residue i with the 13C′i–15Ni+1 interaction (15Ni–13Cαi–13C′i–15Ni+1) (33, 34) are particularly strongly dependent on those peptide conformations described by 90° < ψi < 150° and 150° < |ψi| < 180°, respectively (i.e., also in the β-sheet region of Ramachandran space). Thus, these experiments are exquisitely sensitive to the region of conformational space that is most relevant for the polypeptide chains within amyloid fibrils.
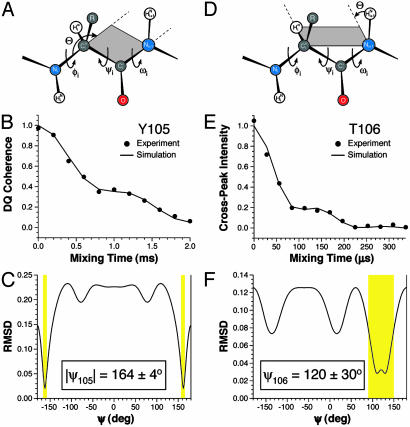
Fig. 2 shows representative measurements of ψ for TTR(105–115)YTIA fibrils determined by using the 3D 1HNi+1–15Ni+1–13Cαi–1Hαi and 15Ni–13Cαi–13C′i–15Ni+1 techniques. All torsion angle experiments used in the present work used two dimensions for frequency labeling with the isotropic chemical shifts and a third dipolar dephasing dimension, which encodes the torsion angle information by evolving correlated nuclear spin states under the relevant dipolar interactions (32). The theoretical background and spectral simulations for these torsion angle experiments have been described in detail (31–34). Experimental and theoretical dipolar dephasing curves for the Tyr-105 and Thr-106 residues are shown in Fig. 2 B and E, respectively. The simulations accurately describe the experimental results, and the dihedral angles are obtained via a least-squares fit of the data to a series of theoretical dephasing trajectories calculated as a function of ψ, displayed as rms deviation (rmsd) plots in Fig. 2 C and F. The dipolar–chemical shift experiments indicate the presence of characteristic β-strand φ and ψ angles for the majority of residues in TTR(105–115) fibrils leading to dihedral restraints of φ ≈ –120 ± 20° and ψ ≈ 125 ± 15° on average for most residues (Table 3). In addition, these experiments reveal an extended conformation for the N-terminal Tyr-105 residue, characterized by ψ = 164°. The φ and ψ angles measured by using the 3D dipolar–chemical shift techniques are in quantitative agreement (in all cases within ±5–10°) with the previously reported restraints based only on chemical shift information (Table 3) (26), as well as the measurements of 13Cβi–15Ni+1 distances (Table 2), which depend primarily on the value of ψi. Note that although the dipolar dephasing trajectories in Fig. 2 are primarily a function of the torsion angle of interest, they also depend on the exact values of the relevant three-atom bond angles. Consequently, the possible 2–3° variations in polypeptide bond angles derived from an extensive statistical survey of the Cambridge Structural Database (37) have been included in the simulations and are reflected in the uncertainties reported in Table 3.
Fig. 2.
Representative torsion angle measurements in TTR(105–115)YTIA fibrils. (A–C) Measurement of ψY105 using a 3D 15Ni–13Cαi–13C′i–15Ni+1 experiment (33, 34). (A) Schematic of the peptide backbone. The projection angle Θ between the 13Cαi–15Ni and 13C′i–15Ni+1 dipole vectors is indicated. (B) Experimental (•) and simulated (—) intensities of the cross-peak corresponding to 13Cα–13C′ double-quantum coherence for residue Y105 in a 13C double quantum–single quantum correlation spectrum (not shown) as a function of the 13C–15N dipolar dephasing time. The time evolution of the double-quantum coherence reports on the projection angle Θ between the 13Cαi–15Ni and 13C′i–15Ni+1 dipole vectors. (C) Plot of the rmsd between the experimental dephasing curve in B and simulated dephasing curves obtained for different values of ψ. Allowed solutions are indicated by yellow rectangles. (D–F) Measurement of ψT106 performed by using a 3D 1HNi+1–15Ni+1–13Cαi–1Hαi experiment (32). (D) Schematic of the peptide backbone. The projection angle Θ between the 15Ni+1–1HNi+1 and 13Cαi–1Hαi dipole vectors is indicated. (E) Experimental (•) and simulated (—) intensities for the I107 15N–T106 13Cα cross-peak in a 15N–13C correlation spectrum as a function of the X–1H dipolar dephasing time (X = 13C, 15N). The time evolution of the cross-peak intensity reports on the projection angle Θ between the 15Ni+1–1HNi+1 and 13Cαi–1Hαi dipole vectors. (F) Plot of the rmsd between the experimental dephasing curve in E and simulated dephasing curves obtained for different values of ψ. Allowed solutions are indicated by the yellow rectangle. The torsion angle measurements are summarized in Table 1 and presented in detail in Table 3.
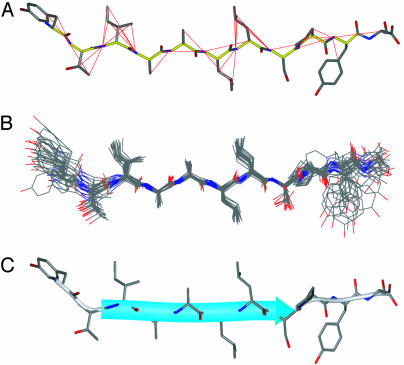
Structure of TTR(105–115) in the Amyloid Fibril. In combination with the backbone restraints determined previously (26) the NMR measurements have yielded a total of 111 self-consistent restraints on the peptide structure. Of the 111 restraints, a total of 76 (i.e., on average 7 restraints per residue) contained information about the peptide conformation (the remaining restraints correspond to one- and two-bond 13C–15N distances that are largely independent of peptide conformation). The data, which include 41 restraints on 19 backbone torsion angles and 35 long-range C–N distances between 3 and 6 Å, are summarized in Table 1 and presented in Tables 2 and 3. An ensemble of twenty low-energy structures of TTR(105–115) in the amyloid fibril calculated by using the NMR restraints and simulated annealing molecular dynamics implemented in the program cns (35) is shown in Fig. 3. Note that the lower density of experimental restraints available for the N- and C-terminal residues (Fig. 3A) is reflected in the ensemble of NMR structures (Fig. 3B) and the atomic RMS deviations for different parts of the peptide (Table 1). Although the majority of the dihedral angles were unambiguously constrained by the NMR measurements, the conformations of the Leu and Tyr side chains could not be defined uniquely by using NMR data alone. Thus, the set of restraints used to generate the final ensemble of structures included database-derived restraints (40) for Leu and Tyr χ1 and χ2 torsion angles. These restraints are based on the observation that for some types of residues certain regions of torsion angle space are highly unlikely to be populated in peptides and proteins (40). For example, and independent of the backbone conformation, the probability that the χ1 angle, θ(N—Cα—Cβ—Cγ), for Leu and Tyr residues is in the 240° interval between χ1 ≈ 120–360° (i.e., χ1 ≈ 180 ± 60° or –60 ± 60°) is ≈98% and 90%, respectively (40), whereas conformations characterized by χ1 ≈ 60 ± 60° are highly unlikely. Furthermore the values of χ1 and χ2 for Leu and Tyr are highly correlated, placing additional restraints on the conformational space accessible to these residues. In a test calculation, where the database-derived restraints were excluded from the calculation protocol, the resulting ensemble consisted of distinct clusters of structures having approximately bimodal distributions for Leu and Tyr χ1 and χ2 angles (note that the Thr and Ile side-chain conformations were uniquely defined in the absence of database-derived restraints for those residues). For example, the resulting structures had χ1 values of about ±60°, 180° and 60°, and ±60°, for Tyr-105, Leu-110, and Leu-111, respectively. Thus, the addition of database-derived restraints is essentially equivalent to filtering the ensemble, to remove those structures where Leu and Tyr have the highly unlikely χ1 ≈ 60° torsion angles. The use of this type of “conformational filter” for the Leu and Tyr side chains neither alters significantly the values of the remaining backbone and side-chain torsion angles in the resulting ensemble of structures nor changes the ensuing discussion of the structural features of TTR(105–115) in the amyloid fibril.
Fig. 3.
Three-dimensional structure of TTR(105–115) in the amyloid fibril determined by using MAS solid-state NMR. (A) Structure of TTR(105–115) (see C below for details) with the experimental NMR restraints (Tables 2 and 3) superimposed. Measured 13C–15N distances are indicated by the red lines, and measured torsion angles between atoms A-B-C-D are indicated by the B—C bond colored yellow. (B) Ensemble of 20 low-energy structures generated by using simulated annealing molecular dynamics implemented in cns (35), based on experimental NMR distance and dihedral angle restraints (Table 1) and database-derived restraints for the Tyr and Leu side chains (40). The peptide structures are superimposed over the backbone atoms of residues I107–P113. The coordinate rmsd for residues I107–P113 was 0.40 Å (backbone) and 0.63 Å (all heavy atom), and for residues Y105–S115 the rmsd values were 0.69 Å (backbone) and 1.24 Å (all heavy atom). (C) Ribbon representation of the structure of TTR(105–115) in the amyloid fibril with side chains shown as stick models. A typical conformer from the ensemble closest to the average structure is shown. The figure was prepared by using the program molmol (39). C, N, and O atoms are indicated in black, blue, and red, respectively. The average torsion angles in TTR(105–115) are listed in Table 4.
The NMR measurements provide the precise backbone and side-chain conformations for most residues of TTR(105–115) in the fibrillar state. The overall conformation of TTR(105–115) in the amyloid fibril is an extended β-strand as depicted in Fig. 3 (Table 4, which is published as supporting information on the PNAS web site). This conclusion is in agreement with our initial experiments (26) and other biophysical studies of a wide range of other systems, which include solid-state NMR measurements, indicating that amyloid fibrils consist primarily of β-sheet secondary structure (1–3, 7, 9, 13, 14, 17–25). The direct measurement of internuclear distances and dihedral angles performed on native fibril samples provides, however, previously inaccessible atomic-resolution structural features, notably the precise sidechain conformations. Of particular significance is that both the main- and side-chain torsion angles are in all cases located within the regions of conformational space known to be preferred by polypeptide chains in natural proteins. It is therefore apparent that the peptide molecules are not only positioned in the fibrils with astonishing regularity (we estimate that the spread of torsion angles within the population of molecules in the fibrillar sample is less that ≈10–20°) but that they achieve this regularity while being accommodated in a structure that is close to that of minimum energy as far as the intramolecular interactions are concerned.
The NMR experiments indicate further that the side chains of the hydrophobic residues in the central region of the peptide, Ile-107 through Leu-111, are situated roughly perpendicular to the plane defined by the C O and N—H bond vectors. These side chains determine the intermolecular interactions between the β-sheets making up the protofilaments, indicating that the interactions between hydrophobic groups are likely to be of particular importance, at least for the peptide considered here. The measurements also reveal that the N terminus is in an extended conformation characterized by ψY105 ≈ 165°, perhaps enabling the polar hydroxyl group to make favorable contacts with water molecules. In addition, we note that the Pro-113 residue is incorporated into the β-strand in the fibril (26) and clearly has a conformation different from that observed in the context of native TTR (28). The pattern required for the most efficient formation of hydrogen bonds between neighboring β-strands within the β-sheet would be a highly regular arrangement of C
O and N—H bond vectors. These side chains determine the intermolecular interactions between the β-sheets making up the protofilaments, indicating that the interactions between hydrophobic groups are likely to be of particular importance, at least for the peptide considered here. The measurements also reveal that the N terminus is in an extended conformation characterized by ψY105 ≈ 165°, perhaps enabling the polar hydroxyl group to make favorable contacts with water molecules. In addition, we note that the Pro-113 residue is incorporated into the β-strand in the fibril (26) and clearly has a conformation different from that observed in the context of native TTR (28). The pattern required for the most efficient formation of hydrogen bonds between neighboring β-strands within the β-sheet would be a highly regular arrangement of C O and N—H group orientations roughly perpendicular to the β-strand ribbon (facing in and out of the page in Fig. 3C). Although the presence of the proline residue does appear to perturb slightly this pattern of C
O and N—H group orientations roughly perpendicular to the β-strand ribbon (facing in and out of the page in Fig. 3C). Although the presence of the proline residue does appear to perturb slightly this pattern of C O and N—H orientations observed for residues T106–S112, the characteristic β-strand torsion angles for the Pro-113 and Tyr-114 residues (Table 4) indicate that no substantial deviation from β-structure arises from the presence of this residue in fibrillar TTR(105–115).
O and N—H orientations observed for residues T106–S112, the characteristic β-strand torsion angles for the Pro-113 and Tyr-114 residues (Table 4) indicate that no substantial deviation from β-structure arises from the presence of this residue in fibrillar TTR(105–115).
Implications for Supramolecular Structure of TTR(105–115) Amyloid Fibrils. In addition to revealing the local details of the peptide structure, our results allow us to speculate on the organization of peptides within the amyloid fibrils self-assembled from TTR(105–115). Amyloid fibrils are thought to be assembled in a hierarchical fashion, with individual polypeptides assembling into protofilaments that twist around each other to form the mature fibrils (3). TTR(105–115) protofilaments have an average diameter of 4.3 ± 1.6 nm, as assessed by transmission electron microscopy (TEM) measurements after partial chemical denaturation of the fibrils (41). The NMR data indicate a peptide length of 3.4 ± 0.2 nm (as defined by the distance between the Tyr-105 N and Ser-115 C′ atoms), increasing to ≈3.8 ± 0.2 nm if the side chains are included, suggesting that each β-sheet within a protofilament is assembled from a hydrogen-bonded array of single peptides in an extended conformation. X-ray fiber diffraction data indicate that the distance between sheets is of the order of 1 nm (42), suggesting that four β-sheets (i.e., ≈4 nm) in a “cross-β” configuration make up each protofilament. Because TEM analysis reveals an average diameter of 10.8 ± 1.2 nm for mature TTR(105–115) fibrils, it appears likely that up to four protofilaments are wound around each other in a close-packed arrangement to form the final structure.
Conclusions
In summary, we have designed and executed a set of experiments that have allowed us to determine the complete atomicresolution structure of a peptide fragment of transthyretin in an amyloid fibril. The current work is the culmination of a series of substantial advances in MAS NMR methodology spanning more than a decade, and in the techniques used to prepare samples of highly ordered fibrils for these studies. The determination of a large number of NMR distance and dihedral restraints recorded on uniformly 13C, 15N (U-13C, 15N)-labeled fibrils reveals a series of important structural features. Of particular interest is the fact that the peptide chain is able to adopt a very low energy molecular conformation, despite the constraints of packing in the cross-β structure. The ability to adopt such a stable structure may explain the very high degree of organization within the fibrillar samples. Such a level of organization in a molecular assembly has previously been associated only with crystalline materials. It appears therefore that the amyloid core structure should be considered as a 2D molecular crystalline form of a polypeptide chain, a result consistent with the well established commonality in morphological and other characteristic features of amyloid fibrils. Extension of the present MAS NMR strategies to permit intermolecular interactions to be defined therefore promises to be of considerable interest. Overall, therefore, the success of the experiments used in the present work for the determination of the 3D structure of TTR(105–115) in the fibrillar state underlines the tremendous opportunities for MAS NMR methodology in the de novo structure determination of amyloid fibrils and other large biological complexes that lack complete 3D order.
Supplementary Material
Acknowledgments
We thank Dr. J. D. Gross for stimulating discussions. C.P.J. was supported by a National Science Foundation Graduate Research Fellowship; C.E.M. was supported by a Royal Society University Research Fellowship; V.S.B. was supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship; and M.T.M. was supported by National Institutes of Health National Research Service Award GM-20818. The research of C.M.D. was supported in part by the Wellcome Trust, and the research of R.G.G. was supported by National Institutes of Health Grants GM-23403 and EB002026.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MAS, magic angle spinning; rmsd, rms deviation; ZF TEDOR, z-filtered transferred-echo double-resonance; TTR, transthyretin.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1RVS).
References
- 1.Sunde, M. & Blake, C. C. F. (1998) Q. Rev. Biophys. 31, 1–39. [DOI] [PubMed] [Google Scholar]
- 2.Kelly, J. W. (1998) Curr. Opin. Struct. Biol. 8, 101–106. [DOI] [PubMed] [Google Scholar]
- 3.Dobson, C. M. (2001) Philos. Trans. R. Soc. London B 356, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guijarro, J. I., Sunde, M., Jones, J. A., Campbell, I. D. & Dobson, C. M. (1998) Proc. Natl. Acad. Sci. USA 95, 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fändrich, M., Fletcher, M. A. & Dobson, C. M. (2001) Nature 410, 165–166. [DOI] [PubMed] [Google Scholar]
- 6.MacPhee, C. E. & Dobson, C. M. (2000) J. Am. Chem. Soc. 122, 12707–12713. [Google Scholar]
- 7.Sunde, M., Serpell, L. C., Bartlam, M., Pepys, M. B., Fraser, P. E. & Blake, C. C. F. (1997) J. Mol. Biol. 273, 729–739. [DOI] [PubMed] [Google Scholar]
- 8.Inouye, H. & Kirschner, D. A. (1996) Ciba Found. Symp. 199, 22–35. [DOI] [PubMed] [Google Scholar]
- 9.Serpell, L. C., Sunde, M., Fraser, P. E., Luther, P. K., Morris, E., Sandgren, O., Lundgren, E. & Blake, C. C. F. (1995) J. Mol. Biol. 254, 113–118. [DOI] [PubMed] [Google Scholar]
- 10.Serpell, L. C., Sunde, M., Benson, M. D., Tennent, G. A., Pepys, M. B. & Fraser, P. E. (2000) J. Mol. Biol. 300, 1033–1039. [DOI] [PubMed] [Google Scholar]
- 11.Perutz, M. F., Finch, J. T., Berriman, J. & Lesk, A. (2002) Proc. Natl. Acad. Sci. USA 99, 5591–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wille, H., Michelitsch, M. D., Guenebaut, V., Supattapone, S., Serban, A., Cohen, F. E., Agard, D. A. & Prusiner, S. B. (2002) Proc. Natl. Acad. Sci. USA 99, 3563–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jimenez, J. L., Guijarro, J. I., Orlova, E., Zurdo, J., Dobson, C. M., Sunde, M. & Saibil, H. R. (1999) EMBO J. 18, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez, J. L., Nettleton, E. J., Bouchard, M., Robinson, C. V., Dobson, C. M. & Saibil, H. R. (2002) Proc. Natl. Acad. Sci. USA 99, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rienstra, C. M., Tucker-Kellogg, L., Jaroniec, C. P., Hohwy, M., Reif, B., Lozano-Perez, T., Tidor, B. & Griffin, R. G. (2002) Proc. Natl. Acad. Sci. USA 99, 10260–10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castellani, F., van Rossum, B., Diehl, A., Schubert, M., Rehbein, K. & Oschkinat, H. (2002) Nature 420, 98–102. [DOI] [PubMed] [Google Scholar]
- 17.Lansbury, P. T., Costa, P. R., Griffiths, J. M., Simon, E. J., Auger, M., Halverson, K. J., Kocisko, D. A., Hendsch, Z. S., Ashburn, T. T., Spencer, R. G. S., et al. (1995) Nat. Struct. Biol. 2, 990–998. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths, J. M., Ashburn, T. T., Auger, M., Costa, P. R., Griffin, R. G. & Lansbury, P. T. (1995) J. Am. Chem. Soc. 117, 3539–3546. [Google Scholar]
- 19.Heller, J., Kolbert, A. C., Larsen, R., Ernst, M., Bekker, T., Baldwin, M., Prusiner, S. B., Pines, A. & Wemmer, D. E. (1996) Protein Sci. 5, 1655–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laws, D. D., Bitter, H. L., Liu, K., Ball, H. L., Kaneko, K., Wille, H., Cohen, F. E., Prusiner, S. B., Pines, A. & Wemmer, D. E. (2001) Proc. Natl. Acad. Sci. USA 98, 11686–11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benzinger, T. L. S., Gregory, D. M., Burkoth, T. S., Miller-Auer, H., Lynn, D. G., Botto, R. E. & Meredith, S. C. (1998) Proc. Natl. Acad. Sci. USA 95, 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antzutkin, O. N., Balbach, J. J., Leapman, R. D., Rizzo, N. W., Reed, J. & Tycko, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tycko, R. (2001) Annu. Rev. Phys. Chem. 52, 575–606. [DOI] [PubMed] [Google Scholar]
- 24.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tycko, R. & Ishii, Y. (2003) J. Am. Chem. Soc. 125, 6606–6607. [DOI] [PubMed] [Google Scholar]
- 26.Jaroniec, C. P., MacPhee, C. E., Astrof, N. S., Dobson, C. M. & Griffin, R. G. (2002) Proc. Natl. Acad. Sci. USA 99, 16748–16753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustavsson, A., Engstrom, U. & Westermark, P. (1991) Biochem. Biophys. Res. Commun. 175, 1159–1164. [DOI] [PubMed] [Google Scholar]
- 28.Blake, C. C. F., Geisow, M. J., Oatley, S. J., Rerat, B. & Rerat, C. (1978) J. Mol. Biol. 121, 339–356. [DOI] [PubMed] [Google Scholar]
- 29.Jaroniec, C. P., Tounge, B. A., Herzfeld, J. & Griffin, R. G. (2001) J. Am. Chem. Soc. 123, 3507–3519. [DOI] [PubMed] [Google Scholar]
- 30.Jaroniec, C. P., Filip, C. & Griffin, R. G. (2002) J. Am. Chem. Soc. 124, 10728–10742. [DOI] [PubMed] [Google Scholar]
- 31.Hong, M., Gross, J. D. & Griffin, R. G. (1997) J. Phys. Chem. B 101, 5869–5874. [Google Scholar]
- 32.Rienstra, C. M., Hohwy, M., Mueller, L. J., Jaroniec, C. P., Reif, B. & Griffin, R. G. (2002) J. Am. Chem. Soc. 124, 11908–11922. [DOI] [PubMed] [Google Scholar]
- 33.Costa, P. R., Gross, J. D., Hong, M. & Griffin, R. G. (1997) Chem. Phys. Lett. 280, 95–103. [Google Scholar]
- 34.Feng, X., Eden, M., Brinkmann, A., Luthman, H., Eriksson, L., Gräslund, A., Antzutkin, O. N. & Levitt, M. H. (1997) J. Am. Chem. Soc. 119, 12006–12007. [Google Scholar]
- 35.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. W. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 37.Engh, R. A. & Huber, R. (1991) Acta Crystallogr. A 47, 392–400. [Google Scholar]
- 38.Cornilescu, G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13, 289–302. [DOI] [PubMed] [Google Scholar]
- 39.Koradi, R., Billeter, M. & Wuthrich, K. (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
- 40.Dunbrack, R. L. & Karplus, M. (1993) J. Mol. Biol. 230, 543–574. [DOI] [PubMed] [Google Scholar]
- 41.MacPhee, C. E. & Dobson, C. M. (2000) J. Mol. Biol. 297, 1203–1215. [DOI] [PubMed] [Google Scholar]
- 42.Jarvis, J. A., Craik, D. J. & Wilce, M. C. J. (1993) Biochem. Biophys. Res. Commun. 192, 991–998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.