Abstract
Prostate cancer, a leading cause of cancer death, displays a broad range of clinical behavior from relatively indolent to aggressive metastatic disease. To explore potential molecular variation underlying this clinical heterogeneity, we profiled gene expression in 62 primary prostate tumors, as well as 41 normal prostate specimens and nine lymph node metastases, using cDNA microarrays containing ≈26,000 genes. Unsupervised hierarchical clustering readily distinguished tumors from normal samples, and further identified three subclasses of prostate tumors based on distinct patterns of gene expression. High-grade and advanced stage tumors, as well as tumors associated with recurrence, were disproportionately represented among two of the three subtypes, one of which also included most lymph node metastases. To further characterize the clinical relevance of tumor subtypes, we evaluated as surrogate markers two genes differentially expressed among tumor subgroups by using immunohistochemistry on tissue microarrays representing an independent set of 225 prostate tumors. Positive staining for MUC1, a gene highly expressed in the subgroups with “aggressive” clinicopathological features, was associated with an elevated risk of recurrence (P = 0.003), whereas strong staining for AZGP1, a gene highly expressed in the other subgroup, was associated with a decreased risk of recurrence (P = 0.0008). In multivariate analysis, MUC1 and AZGP1 staining were strong predictors of tumor recurrence independent of tumor grade, stage, and preoperative prostate-specific antigen levels. Our results suggest that prostate tumors can be usefully classified according to their gene expression patterns, and these tumor subtypes may provide a basis for improved prognostication and treatment stratification.
Worldwide, prostate cancer is the third most common cancer and the cause of 6% of cancer deaths in men (1). Its incidence and mortality vary in different parts of the world and are highest in Western countries (2). In the United States, it is the most frequently diagnosed and the second leading cause of cancer death in men (3). Despite these high death rates, prostate cancer is often an indolent disease, and patients can remain asymptomatic for years. The widespread use of serum prostate-specific antigen (PSA) screening has led to identification of an increasing number of asymptomatic low-stage tumors in younger men (4, 5). An important clinical question has become whether and how aggressively to treat such patients with localized prostate cancer.
Currently, prognostication and treatment stratification at the time of diagnosis are based on clinical stage, biopsy Gleason grade (a measure of tumor differentiation), and serum PSA levels. In cases treated by radical prostatectomy, prognosis can be refined by using pathological stage and grade. However, these prognostic indicators do not accurately predict clinical outcome for individual patients. Improved markers are needed to determine which patients might benefit from a more aggressive treatment, and which patients might be spared unnecessary and potentially harmful interventions.
The observed clinical heterogeneity of prostate cancer is likely to reflect underlying molecular heterogeneity among tumors, which, although largely invisible under the light microscope, might be captured by profiling gene expression using DNA microarrays. Indeed, microarray profiling studies have identified clinically relevant gene-expression subtypes in leukemia (6, 7), lymphoma (8), breast cancer (9, 10), and lung cancer (11–13). Although DNA microarray studies of prostate cancer have identified genes differentially expressed in tumor compared to nontumor samples (14–18) and genes whose expression correlates with tumor grade, metastasis, and disease recurrence (14, 17, 19, 20), to date, tumor subtypes based on gene expression have not been appreciated.
Here we report a cDNA microarray-based study in prostate cancer leading to the identification of biologically and clinically relevant gene-expression tumor subtypes. Furthermore, we demonstrate that the protein expression levels for two genes, serving as surrogate markers for tumor subtypes, are strong predictors of tumor recurrence, independent of known risk factors. Our results support the existence of distinct gene expression subtypes in prostate cancer, and their potential use in disease diagnosis and management.
Materials and Methods
Gene Expression Profiling. Freshly frozen prostate surgical specimens were obtained from Stanford University, Karolinska Institute, and Johns Hopkins University, with institutional review board approval from the involved centers (see Supporting Note 1, which is published as supporting information on the PNAS web site). In total, we selected for study 62 primary prostate tumors (61 adenocarcinomas and one adenoid cystic tumor), 41 matched normal prostate tissues (from the noncancerous region of the prostate), and nine unmatched (i.e., different patient) pelvic lymph node metastases. Detailed pathological and clinical data for specimens are provided in Table 2, which is published as supporting information on the PNAS web site. Gene expression profiling was performed essentially as described (9), by using cDNA microarrays containing 26,260 different human genes (UniGene clusters). More detailed information, including data selection and manipulation methods, is available in Supporting Note 2, which is published as supporting information on the PNAS web site.
Tissue Microarrays. A tissue arrayer (Beecher Instruments, Sun Prarie, WI) was used to construct a prostate cancer tissue microarray comprising an independent set of 225 formalin-fixed, paraffin-embedded primary prostate tumor cases selected from diagnostic radical prostatectomy specimens collected at Stanford University, with institutional review board approval. Duplicate 0.6-mm tumor cores represented each case, and the series was associated with a minimum clinical follow-up of 5 years and a median follow-up of 8 years. Primary antibodies directed against MUC1 (SC-7313, Santa Cruz Biotechnology) and AZGP1 (SC-11242, Santa Cruz Biotechnology) were used for immunohistochemical staining. More detailed information is available in Supporting Note 3, which is published as supporting information on the PNAS web site.
Results
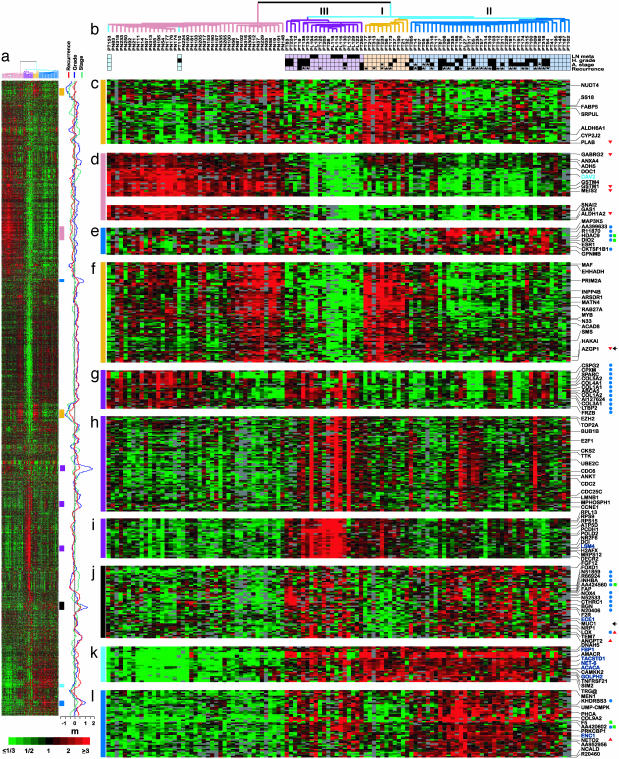
Identification of Prostate Tumor Subtypes. To survey the molecular variation among prostate tumors, we profiled gene expression in 112 prostate tissues, including 62 primary tumors, 41 matched normal prostate tissues, and nine unmatched lymph node metastases, by using cDNA microarray containing 26,260 different genes (see Materials and Methods). To explore the relationship among samples and underlying features of gene expression, we applied an unsupervised two-way hierarchical clustering method using the 5,153 cDNAs whose expression varied most across samples (Fig. 1a). Overall, samples divided into two major clusters (Fig. 1b), one representing tumors and the other, with two exceptions, representing the normal prostate samples. One of these exceptions was an adenoid cystic tumor (sample PT153), a rare neoplasm that shares features of basal epithelial cells (21), which are present in normal prostate glands but absent in prostate adenocarcinoma. The other exception (PT110) may reflect an unexpectedly high level of normal tissue contamination. Although tumor specimens in general had a higher epithelial cell fraction compared with normal prostate samples, tumor-normal gene expression distinctions were not merely reflective of varied epithelial cell content (see Supporting Note 4, which is published as supporting information on the PNAS web site). The gene expression “feature” representing genes more-highly expressed in prostate adenocarcinoma (Fig. 1k) included AMACR and TACSTD1, both previously described (18, 22, 23), as well as tumor necrosis factor receptor superfamily, member 21 (TNFRSF21), golgi phosphoprotein 2 (GOLPH2), net-6, and acetyl-Coenzyme A carboxylase α (ACACA), the latter which, like α-methylacyl-CoA racemase (AMACR), is involved in fatty acid metabolism. These genes, and others within this feature, may provide a basis for improved diagnosis or therapy. A larger cluster of genes had consistently lower expression in tumors than in normal prostate samples (partially displayed in Fig. 1d).
Fig. 1.
Hierarchical cluster analysis of prostate samples. (a) Thumbnail overview of the two-way hierarchical cluster of 112 prostate specimens (columns) and 5,153 variably expressed genes (rows). Mean-centered gene expression ratios are depicted by a log2 pseudocolor scale (ratio fold-change is indicated); gray denotes poorly measured data. The complete data set depicted here is available at http://microarray-pubs.stanford.edu/prostateCA. (b) Enlarged view of the sample dendrogram. Terminal branches for normal prostate samples are colored pink, and those for tumor samples are colored according to gene expression subgroups: III (purple), I (yellow), and II (dark blue). Two tumors clustering with normal samples (see text) are colored light blue. Clinicopathological features associated with individual tumor samples are indicated by black boxes below the dendrogram (asterisks indicate missing data). High grade indicates Gleason grade ≥4 + 3; advanced stage indicates pathological stage ≥T3; tumor recurrence indicates PSA rise after surgery or clinical metastasis. (c–l) Selected gene expression “features” extracted from cluster (locations indicated by vertical colored bars). Because of space limitations, only selected genes are indicated. Genes are annotated as indicated if associated in supervised analysis with high-grade (blue circles), advanced stage (green squares), short time to recurrence (red triangles), or long time to recurrence (red inverted triangles). Genes positively and negatively associated with epithelial cell content are indicated by colored text (dark blue and light blue, respectively; see Supporting Note 1). Genes characterized by immunohistochemistry are indicated with arrow. m, moving average (41-gene window) plots for the t test statistic (grade and stage) and Cox's proportional hazards partial likelihood score (recurrence-free survival) shown for the 5,153 genes in the cluster. Note that peaks (high grade, advanced stage, early recurrence) and valleys frequently correspond to gene expression features characterizing tumor subtypes.
Notably, unsupervised clustering also divided tumor samples into three major subgroups based on distinct patterns of gene expression (Fig. 1b). These subgroups were identified by using a variety of preclustering data filtering and sample selection criteria (Fig. 4, which is published as supporting information on the PNAS web site), and were also evident by principal component analysis (Fig. 5, which is published as supporting information on the PNAS web site), suggesting that they represent robust classes (also see Supporting Note 5 and Table 3, which are published as supporting information on the PNAS web site). Subtype III included primary tumors as well as most of the lymph node metastases, and the associated gene expression features (Fig. 1 g–i) included genes related to extracellular matrix (e.g., COL1A1, COL1A2, CSPG2, SPARC; Fig. 1g), cell proliferation (e.g., TOP2A, E2F1, CDC2, CDC25C; Fig. 1h), and increased metabolic activity reflected by energy production (e.g., ATP5D, DCI, DECR2; Fig. 1i) and protein synthesis (e.g., RPL13, RPS15, RPS9). Subtype I tumors were associated with two features of gene expression (Fig. 1 c and f), one of which (Fig. 1f) included genes also expressed in normal prostate epithelium, such as AZGP1 (24) and ARSDR1 (25). Subtype II represented the largest tumor subclass and, in addition to having a characteristic gene expression feature (Fig. 1e), also shared expression features with tumor subtype III (Fig. 1 j and l).
Tumor Subtypes Are Associated with Distinct Clinicopathological Features. As noted above, primary tumors within subgroup III shared an expression signature with unmatched lymph node metastases. To gain further insight into the biological and clinical relevance of new molecular subtypes, we examined the distribution of clinicopathological parameters among primary tumors (Fig. 1b). High grade and advanced stage tumors were more highly represented among tumor subgroups II and III (P = 0.04, χ2 test; Table 4, which is published as supporting information on the PNAS web site). Seven tumors associated with early recurrence in our dataset were also found to reside only within subgroups II and III. Consistent with these observations, a supervised analysis using the significance analysis of microarrays (SAM) method (ref. 26; see Supporting Note 2) identified sets of genes whose expression were associated with high-grade, advanced stage or early tumor recurrence (Figs. 1m and 2); these genes were predominantly located within gene expression features characterizing tumor subtypes II and/or III (Fig. 1 e, g, j, and l; also see Supporting Note 6 and Table 5, which are published as supporting information on the PNAS web site).
Fig. 2.
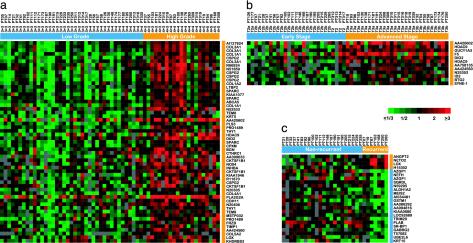
Genes associated with high grade, advanced stage, and tumor recurrence. Genes identified in a supervised analysis using the significance analysis of microarrays (SAM) method (see Supporting Note 2) are ordered by rank value of their SAM score; samples are grouped by clinicopathological parameter and ordered by rank value within groups. Gene expression ratios are depicted by a log2 pseudocolor scale (ratio fold-change is indicated). (a) Forty-one genes (represented by 55 cDNAs), positively associated with high grade, with a FDR of 2%; note that, at this FDR, no negatively associated genes were identified. (b) Eleven genes (represented by 12 cDNAs) positively associated with advanced stage (FDR 8%); at this FDR, no negatively associated genes were identified. (c) Four genes positively and 19 genes negatively associated with short time interval to tumor recurrence (FDR 16%). Orange bars indicate samples and genes associated with high grade (a), advanced stage (b), or early tumor recurrence (c).
In all, we identified 41 genes associated with high grade (Fig. 2a) and 11 genes associated with advanced stage (Fig. 2b), with false discovery rates (FDR) of 2% and 8%, respectively. Among the genes associated with high grade, COL1A2, SPARC, ABCA5 and BGN were reported by Singh et al. (17) to correlate with tumor grade; however, that same study had not identified genes correlated with tumor stage. We also identified four genes positively and 19 genes negatively associated with early recurrence (Fig. 2c), with a FDR of 16%; the relatively high FDR is likely attributable to the short period of clinical follow-up. Although there was no overlap between this gene set and five outcome predictor genes reported by Singh et al. (17), our set of 23 genes correlating with outcome accurately predicted recurrence for patients in their study (Fig. 6, which is published as supporting information on the PNAS web site).
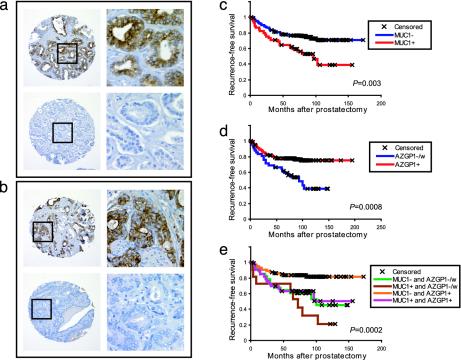
Surrogate Markers for Tumor Subtypes Predict Recurrence. To further characterize the clinical relevance of tumor subtypes, we evaluated as surrogate markers two genes differentially expressed among tumor subtypes, by using immunohistochemistry on tissue microarrays comprising an independent set of 225 primary prostate tumors with a minimum clinical follow-up of 5 years and a median follow-up of 8 years. The two genes were selected based on their differential expression across subtypes and the availability of specific antibodies. MUC1, encoding the mucin 1 transmembrane protein, resided within a gene expression feature that characterized tumor subtypes II and III (Fig. 1j), both associated with more “aggressive” clinicopathological features. MUC1 protein expression, determined by immunohistochemical staining on the tissue microarray, was variable across tumors (Fig. 3a); in Kaplan–Meier survival analysis, positive MUC1 staining was associated with significantly shorter time to recurrence (P = 0.003; Fig. 3c). In contrast, AZGP1, encoding zinc-α-2-glycoporotein, resided within a feature characterizing tumor subtype I (Fig. 1f), and strong immunostaining of AZGP1 was associated with significantly prolonged time to recurrence (P = 0.0008; Fig. 3d). The widest separation in recurrence-free survival curves was attained by combining immunostaining data on MUC1 and AZGP1 (Fig. 3e), suggesting an additive value for prognostication.
Fig. 3.
Expression of MUC1 and AZGP1 predict prostate tumor recurrence. (a and b) Immunohistochemical staining of prostate cancer tissue microarray. Representative positively and negatively staining cores are shown for MUC1 (a) and AZGP1 (b). Original magnifications are ×200 and ×400 (Inset). (c–e) Kaplan–Meier recurrence-free survival analysis based on immunostaining for MUC1 (c, 173 scoreable cases), AZGP1 (d, 170 scoreable cases), or both (e, 160 scoreable cases). MUC1 expression is stratified by positive vs. negative staining. AZGP1 expression is stratified by strong vs. weak/negative staining. P values (log rank test) are indicated.
Despite the association we noted between tumor subtypes and tumor grade and stage, no significant association was identified between MUC1 or AZGP1 protein expression and tumor grade or stage (Table 6, which is published as supporting information on the PNAS web site). To determine whether MUC1 and AZGP1 expression added prognostic information over and above known prognostic factors, we performed multivariate proportional hazards analysis. MUC1 and AZGP1 staining were found to be strong predictors of tumor recurrence [odds ratios = 2.4 (1.3–4.2) and 0.38 (0.21–0.69), respectively; P < 0.001], independent of tumor Gleason grade, stage and preoperative serum PSA (Table 1).
Table 1. Multivariate proportional hazards analysis.
| Variable | Hazard ratio (95% confidence interval) | P value* |
|---|---|---|
| Gleason grade, ≥4 + 3 vs. ≤3 + 4 | 3.11 (1.70-5.70) | 0.0002 |
| Pathological stage, ≥T3 vs. ≤T2 stage | 2.80 (1.46-5.35) | 0.002 |
| Preoperative serum PSA, per ng/ml† | 1.03 (1.01-1.05) | 0.0004 |
| MUC1 staining, positive vs. negative | 2.35 (1.30-4.24) | 0.0005 |
| AZGP1 staining, strong vs. weak/negative | 0.38 (0.21-0.69) | 0.002 |
Wald test.
Serum PSA was used as a continuous variable.
Discussion
The main objective of our study was to survey the molecular variation of prostate cancer, to gain new insight into the underlying biology of this clinically heterogeneous disease. We used unsupervised two-way hierarchical clustering to discover that primary prostate tumors stratify into three robust subtypes based on distinct patterns of gene expression. Moreover, the distribution of clinicopathological features, as well as the performance of surrogate immunohistochemical markers for these subtypes on an independent set of samples, suggests that these subtypes are associated with distinct biological and clinical behavior.
We have characterized subtype I tumors as the clinically least aggressive subclass. Indeed, one of the two gene expression features defining subtype I (Fig. 1f) includes genes expressed in normal prostate, suggesting that this subgroup may represent more highly differentiated tumors. We had selected AZGP1 expression as a surrogate marker for this tumor subtype, and strong immunostaining was associated with longer recurrence-free survival, independent of tumor grade and stage. AZGP1 has previously been reported to be expressed in primary prostate tumors, and to a lesser extent in metastases (27, 28). Hale et al. (28) found an inverse association with tumor stage and grade, whereas Gagnon et al. (27), as did we, found no such association. Given that subgroup I, although predominantly comprising low-grade tumors, also included higher-grade tumors, we speculate that expression profiling may identify a molecular signature of differentiation not apparent by histology.
We have determined subtype III, along with subtype II, to represent a clinically aggressive tumor subclass. Notably, primary tumors within subtype III shared features of gene expression with unmatched lymph node metastases (Fig. 1 g–i). Distinct gene expression signatures between primary prostate tumors and metastases have been reported (14, 19). Interestingly, in both of these studies, a small proportion of primary tumors also clustered together with metastases. Recently, Ramaswamy et al. (20) reported a gene expression signature of metastasis present in a subset of primary solid tumors, including prostate cancer. Our findings are consistent with their conclusion that a metastatic phenotype may preexist within the bulk tumor population for a subset of primary tumors. Importantly, however, because both tumor subtypes II and III are associated with tumor recurrence, our data suggest that at least in prostate cancer, this metastatic signature may represent only one of at least two distinct signatures associated with poor outcome (also see Supporting Note 7, which is published as supporting information on the PNAS web site).
Subtype II tumors represent the second clinically aggressive tumor subclass, and the gene expression feature that characterizes this subgroup included several genes identified in supervised analysis to be associated with both high grade and advanced stage, such as HDAC9 and DIO2 (Fig. 1e). Other genes associated with high grade (e.g., NOX4) and advanced stage (e.g., F5) resided within two features of gene expression also shared with subtype III (Fig. 1 j and l). The role of these genes in the development or progression of prostate cancer remains to be determined. Nevertheless, it is worth noting that nearly half of the tumors within subgroup II are low grade and early stage, suggesting again that gene expression features may represent molecular signatures of biological processes relevant to tumor progression that are not appreciable by pathological analysis.
One expression feature of particular interest (Fig. 1j), its expression shared by subgroups II and III, included several genes involved in cellular invasion and/or angiogenesis, such as F2R, MUC1, NRP1, LOX, ANGPT2, and TEM7 (29–35). We had selected MUC1 expression as a surrogate marker for this feature, characterizing the clinically aggressive subgroups II and III, and found positive immunostaining to be associated with shorter recurrence-free survival, independent of tumor grade and stage. Increased expression of MUC1 has previously been associated with poor prognosis in other types of carcinoma (36–39). It has been proposed that overexpression of MUC1 increases the metastatic potential of cancer cells by reducing E-cadherin and intergrin-mediated cell adhesion (34, 35). However, a role of MUC1 in prostate cancer has not been established. One study reported MUC1 expression to correlate with prostate cancer grade and stage (40), whereas another found an association with intratumoral angiogenesis but not with grade (41). Interestingly, coexpression of MUC1 along with multiple angiogenic factors has been observed in non-small cell lung tumors (42). In our data set, MUC1 was coexpressed along with several genes involved in cellular invasion and angiogenesis, and this expression feature may represent the signature of these biological processes, which are important for prostate cancer progression.
In this study, we have further characterized the expression and prognostic value of two genes, functioning as surrogate markers for our newly identified tumor subtypes. In an independent set of 225 prostate tumors assessed by immunohistochemistry on tissue microarrays, MUC1 (a surrogate for subtypes II and III) and AZGP1 (subtype I) were found to be strong predictors of tumor recurrence. Importantly, these genes were found in multivariate analysis to add additional prognostic information over and above the known risk factors of tumor grade, stage, and preoperative PSA. Interestingly, these genes also provided prognostic value independent of one another, suggesting that using two genes improves the accuracy of tumor subtyping and prognostication. It remains to be determined whether adding yet additional genes might further improve prognostication, and, of course, it will be important to validate our findings prospectively and on preoperative tumor biopsy samples (the specimens most relevant for treatment stratification). Nonetheless, our results suggest that prostate tumors can be usefully classified according to their gene expression patterns, and that these tumor subtypes may provide a basis for improved prognostication and treatment stratification.
Supplementary Material
Acknowledgments
We thank laboratory members for many helpful comments and suggestions. We also thank Janet Mitchell and the Stanford Tissue Bank for collection of tissues, Mike Fero and the staff of the Stanford Functional Genomics Facility for providing high-quality cDNA microarrays, and Gavin Sherlock and the Stanford Microarray Database group for providing outstanding database support. We are grateful to Helen Fedor, Gerrun March, and Marcella Southerland for assisting with tissue procurement and frozen sectioning at Johns Hopkins University. This work was supported by National Institutes of Health Grant U01CA85129, the Howard Hughes Medical Institute, the Grove Foundation, the Swedish Cancer Society, the Cancer Society in Stockholm, the Konung Gustaf V:s Jubileumsfond, Wallström and Osterman Foundations in Karolinska Institutet, and the Swedish Medical Society. The Johns Hopkins tissue bank is funded in part by the National Cancer Institute/National Institutes of Health Specialized Programs of Research Excellence (prostate) P50CA58236. J.L. was supported in part by a fellowship from the Canadian Institute of Health Research. P.O.B. is an investigator of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PSA, prostate-specific antigen; FDR, false discovery rate.
References
- 1.Parkin, D. M., Bray, F. I. & Devesa, S. S. (2001) Eur. J. Cancer 37, S4–S66. [DOI] [PubMed] [Google Scholar]
- 2.Hsing, A. W., Tsao, L. & Devesa, S. S. (2000) Int. J. Cancer 85, 60–67. [DOI] [PubMed] [Google Scholar]
- 3.Jemal, A., Murray, T., Samuels, A., Ghafoor, A., Ward, E. & Thun, M. J. (2003) CA Cancer J. Clin. 53, 5–26. [DOI] [PubMed] [Google Scholar]
- 4.Farkas, A., Schneider, D., Perrotti, M., Cummings, K. B. & Ward, W. S. (1998) Urology 52, 444–448; discussion, 448–449. [DOI] [PubMed] [Google Scholar]
- 5.Han, M., Partin, A. W., Piantadosi, S., Epstein, J. I. & Walsh, P. C. (2001) J. Urolol. 166, 416–419. [PubMed] [Google Scholar]
- 6.Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., Caligiuri, M. A., et al. (1999) Science 286, 531–537. [DOI] [PubMed] [Google Scholar]
- 7.Yeoh, E. J., Ross, M. E., Shurtleff, S. A., Williams, W. K., Patel, D., Mahfouz, R., Behm, F. G., Raimondi, S. C., Relling, M. V., Patel, A., et al. (2002) Cancer Cell 1, 133–143. [DOI] [PubMed] [Google Scholar]
- 8.Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503–511. [DOI] [PubMed] [Google Scholar]
- 9.Perou, C. M., Sørlie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., Rees, C. A., Pollack, J. R., Ross, D. T., Johnsen, H., Akslen, L. A., et al. (2000) Nature 406, 747–752. [DOI] [PubMed] [Google Scholar]
- 10.Sørlie, T., Perou, C. M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H., Hastie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beer, D. G., Kardia, S. L., Huang, C. C., Giordano, T. J., Levin, A. M., Misek, D. E., Lin, L., Chen, G., Gharib, T. G., Thomas, D. G., et al. (2002) Nat. Med. 8, 816–824. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharjee, A., Richards, W. G., Staunton, J., Li, C., Monti, S., Vasa, P., Ladd, C., Beheshti, J., Bueno, R., Gillette, M., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 13790–13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garber, M. E., Troyanskaya, O. G., Schluens, K., Petersen, S., Thaesler, Z., Pacyna-Gengelbach, M., van de Rijn, M., Rosen, G. D., Perou, C. M., Whyte, R. I., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 13784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhanasekaran, S. M., Barrette, T. R., Ghosh, D., Shah, R., Varambally, S., Kurachi, K., Pienta, K. J., Rubin, M. A. & Chinnaiyan, A. M. (2001) Nature 412, 822–826. [DOI] [PubMed] [Google Scholar]
- 15.Luo, J., Duggan, D. J., Chen, Y., Sauvageot, J., Ewing, C. M., Bittner, M. L., Trent, J. M. & Isaacs, W. B. (2001) Cancer Res. 61, 4683–4688. [PubMed] [Google Scholar]
- 16.Magee, J. A., Araki, T., Patil, S., Ehrig, T., True, L., Humphrey, P. A., Catalona, W. J., Watson, M. A. & Milbrandt, J. (2001) Cancer Res. 61, 5692–5696. [PubMed] [Google Scholar]
- 17.Singh, D., Febbo, P. G., Ross, K., Jackson, D. G., Manola, J., Ladd, C., Tamayo, P., Renshaw, A. A., D'Amico, A. V., Richie, J. P., et al. (2002) Cancer Cell 1, 203–209. [DOI] [PubMed] [Google Scholar]
- 18.Welsh, J. B., Sapinoso, L. M., Su, A. I., Kern, S. G., Wang-Rodriguez, J., Moskaluk, C. A., Frierson, H. F., Jr., & Hampton, G. M. (2001) Cancer Res. 61, 5974–5978. [PubMed] [Google Scholar]
- 19.LaTulippe, E., Satagopan, J., Smith, A., Scher, H., Scardino, P., Reuter, V. & Gerald, W. L. (2002) Cancer Res. 62, 4499–4506. [PubMed] [Google Scholar]
- 20.Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. (2003) Nat. Genet. 33, 49–54. [DOI] [PubMed] [Google Scholar]
- 21.Grignon, D. J., Ro, J. Y., Ordonez, N. G., Ayala, A. G. & Cleary, K. R. (1988) Hum. Pathol. 19, 1425–1433. [DOI] [PubMed] [Google Scholar]
- 22.Rubin, M. A., Zhou, M., Dhanasekaran, S. M., Varambally, S., Barrette, T. R., Sanda, M. G., Pienta, K. J., Ghosh, D. & Chinnaiyan, A. M. (2002) J. Am. Med. Assoc. 287, 1662–1670. [DOI] [PubMed] [Google Scholar]
- 23.Luo, J., Zha, S., Gage, W. R., Dunn, T. A., Hicks, J. L., Bennett, C. J., Ewing, C. M., Platz, E. A., Ferdinandusse, S., Wanders, R. J., et al. (2002) Cancer Res. 62, 2220–2226. [PubMed] [Google Scholar]
- 24.Tada, T., Ohkubo, I., Niwa, M., Sasaki, M., Tateyama, H. & Eimoto, T. (1991) J. Histochem. Cytochem. 39, 1221–1226. [DOI] [PubMed] [Google Scholar]
- 25.Lin, B., White, J. T., Ferguson, C., Wang, S., Vessella, R., Bumgarner, R., True, L. D., Hood, L. & Nelson, P. S. (2001) Cancer Res. 61, 1611–1618. [PubMed] [Google Scholar]
- 26.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gagnon, S., Tetu, B., Dube, J. Y. & Tremblay, R. R. (1990) Am. J. Pathol. 136, 1147–1152. [PMC free article] [PubMed] [Google Scholar]
- 28.Hale, L. P., Price, D. T., Sanchez, L. M., Demark-Wahnefried, W. & Madden, J. F. (2001) Clin. Cancer Res. 7, 846–853. [PubMed] [Google Scholar]
- 29.Carson-Walter, E. B., Watkins, D. N., Nanda, A., Vogelstein, B., Kinzler, K. W. & St. Croix, B. (2001) Cancer Res. 61, 6649–6655. [PubMed] [Google Scholar]
- 30.Even-Ram, S. C., Maoz, M., Pokroy, E., Reich, R., Katz, B. Z., Gutwein, P., Altevogt, P. & Bar-Shavit, R. (2001) J. Biol. Chem. 276, 10952–10962. [DOI] [PubMed] [Google Scholar]
- 31.Kawakami, T., Tokunaga, T., Hatanaka, H., Kijima, H., Yamazaki, H., Abe, Y., Osamura, Y., Inoue, H., Ueyama, Y. & Nakamura, M. (2002) Cancer 95, 2196–2201. [DOI] [PubMed] [Google Scholar]
- 32.Kirschmann, D. A., Seftor, E. A., Fong, S. F., Nieva, D. R., Sullivan, C. M., Edwards, E. M., Sommer, P., Csiszar, K. & Hendrix, M. J. (2002) Cancer Res. 62, 4478–4483. [PubMed] [Google Scholar]
- 33.Sfiligoi, C., De Luca, A., Cascone, I., Sorbello, V., Fuso, L., Ponzone, R., Biglia, N., Audero, E., Arisio, R., Bussolino, F., et al. (2003) Int. J. Cancer 103, 466–474. [DOI] [PubMed] [Google Scholar]
- 34.Wesseling, J., van der Valk, S. W., Vos, H. L., Sonnenberg, A. & Hilkens, J. (1995) J. Cell Biol. 129, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wesseling, J., van der Valk, S. W. & Hilkens, J. (1996) Mol. Biol. Cell 7, 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guddo, F., Giatromanolaki, A., Koukourakis, M. I., Reina, C., Vignola, A. M., Chlouverakis, G., Hilkens, J., Gatter, K. C., Harris, A. L. & Bonsignore, G. (1998) J. Clin. Pathol. 51, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGuckin, M. A., Walsh, M. D., Hohn, B. G., Ward, B. G. & Wright, R. G. (1995) Hum. Pathol. 26, 432–439. [DOI] [PubMed] [Google Scholar]
- 38.Nakamori, S., Ota, D. M., Cleary, K. R., Shirotani, K. & Irimura, T. (1994) Gastroenterology 106, 353–361. [DOI] [PubMed] [Google Scholar]
- 39.Takao, S., Uchikura, K., Yonezawa, S., Shinchi, H. & Aikou, T. (1999) Cancer 86, 1966–1975. [DOI] [PubMed] [Google Scholar]
- 40.Kirschenbaum, A., Itzkowitz, S. H., Wang, J. P., Yao, S., Eliashvili, M. & Levine, A. C. (1999) Mol. Urol. 3, 163–168. [PubMed] [Google Scholar]
- 41.Papadopoulos, I., Sivridis, E., Giatromanolaki, A. & Koukourakis, M. I. (2001) Clin. Cancer Res. 7, 1533–1538. [PubMed] [Google Scholar]
- 42.Giatromanolaki, A., Koukourakis, M. I., Sivridis, E., O'Byrne, K., Cox, G., Thorpe, P. E., Gatter, K. C. & Harris, A. L. (2000) Clin. Cancer Res. 6, 1917–1921. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.