Abstract
Legionella pneumophila is an intracellular pathogen that multiplies in a specialized vacuole within host cells. Biogenesis of this vacuole requires the Dot/Icm type IV protein translocation system. By using a Cre/loxP-based protein translocation assay, we found that proteins translocated by the Dot/Icm complex across the host phagosomal membrane can also be transferred from one bacterial cell to another. The flexibility of this system allowed the identification of several families of proteins translocated by the Dot/Icm complex. When analyzed by immunofluorescence microscopy, a protein identified by this procedure, SidC, was shown to translocate across the phagosomal membranes to the cytoplasmic face of the L. pneumophila phagosome. The identification of large numbers of these substrates, and the fact that the absence of any one substrate rarely results in strong defects in intracellular growth, indicate that there is significant functional redundancy among the Dot/Icm translocation targets.
Keywords: bacterial pathogenesis, protein translocation, plasmid conjugation
Active modification of host cellular functions is essential for a bacterial pathogen to establish a successful infection. Such modification often is mediated by injecting effectors into the host cytoplasm through specialized protein secretion systems (1). Among these systems, conjugation-adaptive transporters, also called type IV secretion systems (TFSS), have been identified in a number of bacterial pathogens (2). Many of these TFSS are dedicated DNA transfer apparatuses, whereas others allow Gram-negative bacterial pathogens to translocate protein substrates directly into the cytosol of eukaryotic cells. Only a few protein substrates of TFSS that are translocated into host cells have been identified (2).
Legionella pneumophila is an intracellular pathogen that causes Legionnaire's disease. After being phagocytosed by macrophages, the bacteria multiply within a specialized vacuole that is initially isolated from the endocytic pathway (3, 4), possibly by intercepting early secretory vesicles (5). Biogenesis of this replicative phagosome requires the TFSS transporter called Dot/Icm (6, 7). Substrates transported by this apparatus are believed to directly promote the targeting pathway of the bacterial vacuole (8). Two of these substrates, RalF and LidA, have been identified (9, 10). RalF is a guanine nucleotide exchange factor for multiple Arf proteins (9), whereas the biochemical activity of LidA is unknown (10). There are clearly other unidentified substrates of Dot/Icm, as mutations that specifically eliminate LidA cause negligible defects in intracellular growth and mutations in ralF are proficient for intracellular replication (9, 10). A comprehensive analysis of the identity and function of effectors translocated by the Dot/Icm apparatus is crucial in determining how this bacterium establishes a replication vacuole. We report here that proteins translocated from bacteria to host cells can also be transferred between bacterial cells, allowing identification of a large cohort of proteins transferred by the Dot/Icm apparatus.
Materials and Methods
Bacterial Strains and Growth Conditions. All L. pneumophila strains used in this study are derivatives of the wild-type strain Lp02 (thyA, hsdR, and rpsL) (11). Lp03 is an isogenic dotA– mutant (12). All strains were grown on casamino acids yeast extract thymidine (CYET) plates or in N-(2-acetamido)-2-aminoethanesulfonic acid yeast extract (AYE) broth (11). Bone marrow-derived macrophages were prepared as described (11). To assay for intracellular growth within macrophages from A/J mice (11) or within the amoebal host Dictyostelium discoideum (13), L. pneumophila strains were grown to postexponential phase, as judged by bacterial motility and cell density (OD600 = 3.3–3.7).
Plasmid Constructions. A derivative of pBRR1MCS mob– (Cmr) (14), which was suitable for reporting interbacterial protein translocation (called pZL184; Fig. 1), was constructed in a cloning process involving multiple steps. Sequences of oligonucleotides used, sources of gene cassettes, and details of cloning are found in Supporting Methods, which is published as supporting information on the PNAS web site. To express the Cre fusions, we first constructed pZL180 (Ampr and thyA+), which contains cre on the transfer deficient RSF1010 plasmid pJB908 (J. Vogel, Washington University School of Medicine, St. Louis), and fusions were constructed as follows: Whole ORFs were amplified by PCR and were fused to the 3′ end of cre on pZL180 for genes smaller than 2 kb, whereas for genes larger than 2 kb, the 3′ regions encoding 500 or 700 amino acids of each ORF was fused to cre.
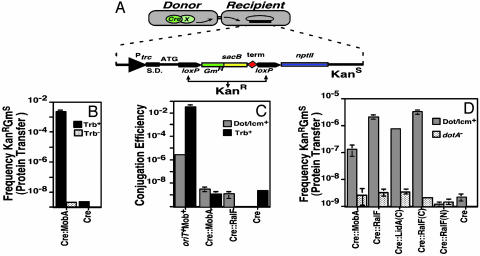
Fig. 1.
Interbacterial protein translocation by the Dot/Icm system in the absence of DNA transfer. (A) Assay for interbacterial protein transfer. Translocation of Cre hybrid protein from a donor bacterial strain is measured by removal of a floxed transcriptional terminator located between the trc promoter and the npt II (kanR) gene on plasmid pZL184 harbored by a recipient bacterial strain. Bacteria harboring the intact reporter are unable to grow on media containing kanamycin and sucrose. The translocation of Cre hybrid protein into the recipient strain leads to the excision, through recombination at the loxP sites, of the DNA fragment that confers sucrose sensitivity (sacB) and reconstitution of a functional loxP-npt II translational fusion. S.D., Shine–Dalgarno sequence; npt II, neomycin phosphotransferase; red diamond, transcriptional terminator. Arrows indicate the trc promoter and loxP sites. (B) Interbacterial transfer of a fusion derived from the RSF1010 mobA gene. The mobA gene was fused to cre, and RP4-dependent protein translocation into a recipient E. coli strain was measured by using E. coli S17–1 (15) as the donor selecting kanamycin resistance and screening for gentamicin sensitivity. Black bars, S17–1 Trb+ donor; stippled bars, E. coli DH5α Trb– donor. (C). Plasmids harboring Cre fusions cannot be transferred to recipient cells. Plasmids expressing the designated proteins were harbored in either E. coli S17–1 (Trb+) or L. pneumophila Lp02 (Dot/Icm+; ref. 11), and the efficiency of plasmid transfer was measured by using either recipient E. coli or L. pneumophila strains, respectively. As a positive control, the identical plasmids having an intact oriT and mob system were used to demonstrate transfer proficiency of donor strains. Black bars, donor strain E. coli S17–1 (Trb+); gray bars, donor strain L. pneumophila Lp02 (Dot/Icm+; ref. 11). (D) Transfer of translocated Dot/Icm substrates between bacterial cells. Protein transfer was performed as described in Materials and Methods, by using either Lp02 (Dot/Icm+) or Lp03(dotA–) expressing the designated protein fusions as the donor strains. Gray bars, donor strain L. pneumophila Lp02 (Dot/Icm+); stippled bars, donor strain L. pneumophila Lp03 (dotA–).
Bacterial Two-Hybrid Screening. First, ralF was translationally fused to the 3′ end of fragment T18 of the Bordetella pertussis cya gene, encoding adenylate cyclase, on pUT18 (15). Sau3AI-generated genomic DNA fragments from L. pneumophila strain Lp02 (11), ranging from 800 base pairs to 5 kb, were inserted into pKT25 (15), resulting in a library of L. pneumophila proteins fused to the C terminus of B. pertussis adenylate cyclase T25 fragment. Escherichia coli strain BTH101 (15), expressing cya::ralF(C), was used to identify RalF-interacting proteins by screening either on LB medium containing 40 μg/ml X-Gal or on the synthetic M63 medium (15) with lactose as the sole carbon source. Strains with functional adenylate cyclase proteins were identified based on the presence of detectable Lac+ phenotypes on these media. Proteins that interact with DotF were identified in a similar manner by using either DotF or DotF (28–123) as the bait.
Bacterial Matings and Intercellular Protein Translocation. For RP4 Trb-mediated translocation of the Cre::MobA hybrid, ≈2.5 × 108 cells from saturated cultures of S17–1 (Trb+) (16) or DH5α (Trb–) expressing the fusion grown in LB broth were mixed with a 15-fold excess of recipient strain XL1Blue carrying pZL184. The mixtures were spotted onto 0.45-μm nitrocellulose filters, placed on LB plates, and incubated at 37°C for 3 h. The excisants were selected on LB medium containing 5% sucrose and 30 μg/ml kanamycin.
For L. pneumophila to L. pneumophila translocation, Lp02 (11) or Lp03dotA– (12) containing plasmids expressing the appropriate Cre fusions were grown to postexponential phase in AYE broth. For matings, 0.45-μm nitrocellulose membranes were first placed onto CYET medium containing 100 nM isopropyl β-d-thiogalactoside (IPTG) and the plates were incubated at 37°C for 1 h before spotting with a mixture of ≈3.5 × 108 donor and a 15-fold excess of recipient Lp03(pZL184). The mating plates were incubated at 37°C for 14 h and excisants were selected on CYET –5% sucrose, 30 μg/ml kanamycin, which kills both parents. Each mating was performed in triplicate and repeated at least three times. Transfer frequencies were expressed as number of excisants (resistant to kanamycin and sucrose but sensitive to gentamicin) per donor bacterium and the number of donor bacteria was determined by counting colonyforming units derived from appropriately diluted donor cells plated onto CYET medium before mating.
For plasmid transfer, transconjugants were detected by plating the protein translocation mating mixture onto LB containing 100 μg/ml ampicillin and 30 μg/ml chroramphenicol (for E. coli) or onto charcoal yeast extract containing 5 μg/ml chloramphenicol (selecting Thy+ and CmR) for transfer in L. pneumophila). As a positive control, the wild-type RSF1010 derivative pKB5 (11) was used in both E. coli and L. pneumophila matings (6).
Construction of In-Frame Deletions and Complementation of the Deletion Mutants. In-frame deletions of L. pneumophila genes were performed by a two-step allelic exchange strategy as described (17). In each case, the deletion construct was designed such that the intact gene was replaced by an ORF predicted to express a 20-aa polypeptide consisting of the first 10 amino acids and the last 10 amino acids of the gene. To perform complementation studies on deletion mutants, the gene of interest was amplified by PCR and inserted into pJB908 or pBBRMCS2 (14).
Protein Purification and Antibody Preparation. The predicted ORF of SidC was inserted into pQE30 and the resulting His6-SidC fusion protein was purified from E. coli by using Ninitrilotriacetic acid resin (Qiagen, Valencia, CA). Rabbit polyclonal serum was generated by the Pocono Rabbit Farm (Canadensis, PA; ref. 10). Antibodies were affinity-purified by using a matrix containing purified (His)6-SidC covalently coupled to Affigel-10 beads (Bio-Rad) (18).
Western Blot and Immunofluorescence Staining. Total L. pneumophila proteins separated by SDS/PAGE were transferred onto Immobilon-P membranes (Millipore) and probed by Western blotting, as described (16). Filters were probed with affinity-purified anti-SidC antibody (diluted 1:5,000), or anti-Bacillus subtilis isocitrate dehydrogenase polyclonal antibody (diluted 1:5,000; a kind gift from Dr L. Sonenshein, Tufts University School of Medicine). Immunofluorescence staining was performed with affinity-purified rabbit anti-(His)6-SidC antibodies followed by a Texas red-conjugated goat anti-rabbit antibody (Molecular Probes). Fixation and probing techniques were performed as described (10). Postnuclear L. pneumophila phagosomes were isolated as described (10).
Results
Interbacterial Protein Translocation by Dot/Icm. To identify substrates of the Dot/Icm translocator, a screening strategy was devised that allowed us to directly assay for protein translocation, using the Cre/loxP system from bacteriophage P1 (for review, see ref.19). Previous work (20) demonstrated that the presence of Cre does not interfere with the translocation of the TFSS substrates VirE2 and VirF in Agrobacterium tumefaciens. A reporter suitable for monitoring translocation of Cre fusion proteins between two prokaryotic cells was constructed in which expression of the npt II gene depended on the excision of a floxed cassette consisting of a gentamicin resistance gene, the sacB gene, and a transcriptional terminator (Fig. 1 A). By using this system, E. coli and L. pneumophila could transfer a hybrid protein in which cre was fused to the mobA gene of plasmid RSF1010 (Fig. 1B). When the Cre::MobA hybrid protein was expressed from a plasmid lacking its origin of transfer (oriT) in the E. coli strain S17–1 (16), transfer of the fusion protein into another E. coli strain could be detected based on excision of the stopper sequence and expression of kanamycin resistance (Fig. 1B, and Fig. 6, which is published as supporting information on the PNAS web site). No transfer occurred when the Trb– strain DH5α, which lacks conjugative transfer functions, was used as the donor, or when a plasmid having cre alone was tested (Fig. 1B). Moreover, there was no transfer of the plasmid to the recipient, indicating that the translocation of the fusion protein occurred in the absence of DNA transfer (Fig. 1C; Cre::MobA or Cre). L. pneumophila also was able to transfer the hybrid protein in a Dot/Icm-dependent manner (Fig. 1D; Cre::MobA), indicating that the Dot/Icm system is able to transfer proteins interbacterially in the absence of DNA transfer (Fig. 1C). Plasmid Mob proteins such as the MobA of RSF1010 are believed to be transferred to recipient cells as protein–single-stranded DNA complexes (21). However, our results demonstrate that conjugation systems such as Trb and Dot/Icm are able to transfer MobA (and presumably similar proteins) in the absence of DNA transfer. This result is similar to what had been observed with the ColIb-P9 SogL protein (22).
Two proteins known to be transferred from bacterial cells to mammalian cells could also be translocated interbacterially by using this strategy. Full-length ralF and the 3′ half of lidA [called lidA(C)] were each translationally fused to cre and the resulting hybrids were examined for bacterium-bacterium translocation. We found that wild type L. pneumophila, but not a Dot/Icmdeficient strain, could transfer Cre::RalF or Cre::LidA(C), based on the ability of recipient strain to excise the floxed DNA fragment and to express kanamycin resistance [Fig. 1D; Cre::RalF and Cre::LidA(C)]. However, Cre itself could not be translocated by wild-type L. pneumophila (Fig. 1D). The translocation proficiency of Cre::LidA(C) indicates that targeting information resides in the C terminus of the protein, at least for interbacterial transfer. We examined whether this finding was also true for RalF by testing Dot/Icm-mediated translocation of Cre::RalF(C) and Cre::RalF(N) respectively. Only Cre:RalF(C) was translocated at a detectable frequency (Fig. 1D). These observations are similar to findings regarding A. tumefaciens Vir proteins, in which secretion signals are localized to the C termini of the substrates (20, 23).
Identification of Substrates of the Dot/Icm Transporter. Because the translocated proteins LidA and RalF could be transferred interbacterially, we used the bacterial translocation assay to identify proteins transferred by this transporter. To simplify the screening of a randomly generated fusion library, we chose to screen through a preselected pool of candidate substrates. We hypothesized that some translocated substrates may interact with specific components of the Dot/Icm complex. Therefore, a bacterial two-hybrid screen (15) was used to identify L. pneumophila proteins that specifically interacted with the carboxyl portion of RalF. From 41 positive clones sequenced, DotF, predicted to be an inner membrane component of the Dot/Icm complex, was identified nine times independently. Interestingly, in all cases, only the portion of DotF that spans amino acids 28–123 was obtained. The full-length DotF also gave a positive two-hybrid readout, although at a somewhat lower level (Fig. 2). A bacterial two-hybrid study was then performed by using Cya fusions to the 3′ ends of L. pneumophila genes and baits of DotF or its derivative, DotF (28–123). From ≈150,000 candidates, we isolated 148 clones that showed some level of interaction higher than background, and these were sequenced and analyzed. Although it was clear that some strains gave barely detectable β-galactosidase activity and were of questionable significance, this strategy greatly reduced the number of strains to be analyzed. Complete ORFs of these genes were retrieved from the L. pneumophila genome database (http://genome3.cpmc.columbia.edu/~legion) and 68 genes were identified. Of these genes, 20 candidates were eliminated based primarily on their high hydrophobicity, which may have caused nonspecific interactions with DotF. Forty-eight candidate genes were subsequently tested for interbacterial transfer (see Table 2, which is published as supporting information on the PNAS web site, for data on 17 such candidates from a single experiment). Table 1 shows eight clones that gave a positive signal by using the Cre/loxP reporter system. We designated these proteins substrate of Icm/Dot transporter (Sid) (Table 1). The efficiency of interbacterial transfer varied, ranging from 10–6 to 10–5 excisants per input donor cell. Moreover, transfer of all of these proteins depended on the Dot/Icm apparatus, because mating with dotA– strains expressing these fusions failed to cause the excision of the floxed DNA fragment and the subsequent expression of the kanamycin resistance marker (Table 1).
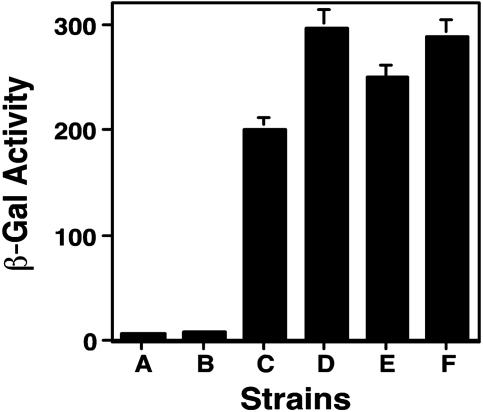
Fig. 2.
Interactions between DotF and the C termini of RalF and SidC, respectively. E. coli strain BTH101 (14) containing the indicated plasmids was grown overnight at 28°C in LB and the cultures were diluted 20-fold in the same medium containing 100 nM IPTG. Cultures were grown for 14 h before an appropriate volume was withdrawn for β-galactosidase assay. Tested strains are as follows: (column A) BTH101(pKT25dotF, pUT18C); (column B) BTH101(pKT25, pUT18CRalF(C); (column C) BTH101(pKT25dotF, pUT18CRalF(C); (column D) BTH101(pDotF (28–123), pKT25RalF(C); (column E) BTH101 [pDotF (28–123), pSidC300(C)]k; and (column F) the leucine zipper domain of the yeast GCN4 protein (14) was used as positive control in BTH101(pKT25Zip, pUT18CZip). β-galactosidase activity was expressed as Miller units. Experiments were performed in triplicate for three independent times. Data shown are from one representative experiment.
Table 1. L. pneumophila proteins identified in this study that are translocated between bacterial cells by the Dot/Icm system.
| Gene name | Accession no. | Protein size, aa | L. pneumophila paralogs | E values of paralogs | Orthologs in other species (E value) | Translocation frequency* |
|---|---|---|---|---|---|---|
| sidA | AY504668 | 474 | - | NA | - | 7.2 ± 1.2 × 10-6 |
| sidB | AY504669 | 417 | 4 | 7e-26 to 5e-09 | Rtx toxin/lipase (e-5) | 6.7 ± 0.2 × 10-6 |
| sidC | AY504673 | 918 | 2 | 0† | - | 3.3 ± 0.7 × 10-6 |
| sidD | AY504675 | 472 | - | NA | - | 2.3 ± 0.2 × 10-5 |
| sidE | AY504676 | 1,495 | 5 | 0-2e-05 | - | 1.2 ± 0.5 × 10-6 |
| sidF | AY504681 | 912 | NA | - | 6.2 ± 0.2 × 10-5 | |
| sidG | AY504682 | 965 | - | NA | - | 8.2 ± 0.3 × 10-6 |
| sidH | AY504683 | 2,225 | 3 | 7e-08 to 3e-04 | - | 3.2 ± 0.4 × 10-5 |
| SdeC‡ | AY504679 | 1,538 | 5 | 0 to 2e-05 | - | 2.2 ± 0.9 × 10-6 |
Translocation assay was performed as described in Materials and Methods. NA, not applicable.
Translocation frequency was expressed as numbers of excisants (resistant to both kanamycin and sucrose and sensitive to gentamicin) per donor cell. No translocation occurred when the dotA- strain Lp03 was used as the donor. The number of donor cells was determined by plating the appropriate dilutions of the donor culture onto solid CYE medium. The rates of spontaneous mutants resistant to kanarnycin and sucrose of the reporter strain appeared on the selective medium used to select excisants were approximately -2.5 × 10-9 per recipient.
An E value of 0 indicates that the paralogs are almost identical proteins.
Paralogs are named for the gene identified in DotF two-hybrid assays. For instance, paralog A of sidE is called sdeA.
Characteristics of sid Genes. Sequence analyses revealed that with the exception of sidB, the sid genes have no significant orthologs present in the GenBank NR database (Table 1). SidB contains a putative active site found in some lipases and shows a region similar to a portion of the Rtx toxin from Vibrio cholerae (24). In contrast, many of these proteins have one or more paralogs present in the L. pneumophila genome (Table 1). The similarities among the paralogs range from almost identical predicted proteins (E = 0) to rather loose similarity (E = 5e–2 to 2e–8; Table 1). Interestingly, in some cases, subsets of paralogs are organized into contiguous ORFs. For instance, sidC and its homolog sdcA are separated by only 150 base pairs, whereas sdeA, sdeB, and sdeC are closely clustered (Fig. 3). In addition, a paralog of sidE (sdeD) is located directly upstream from what appears to be the operon encoding sidC (Fig. 3B). Interestingly, two putative genes, orf1 and orf3, located near the sidA and sdeC regions are highly similar (E = 0; Fig. 3 A and B), although these genes were not identified in the original two-hybrid assay.
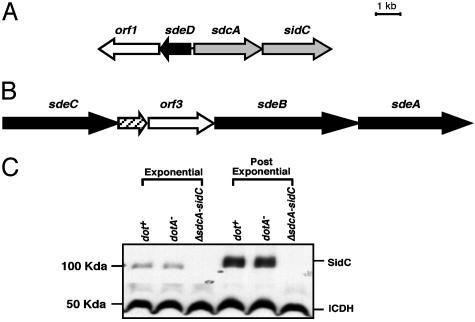
Fig. 3.
Clustering of sid genes, their paralogs into operon-like structures, and growth phase regulation of sidC.(A) sidC and its homolog sdcA are separated by only 150 bp, and the two genes are closely linked to a paralog of sidE(sdeD). (B) Three paralogs of sidE are part of a contiguous region of the chromosome that contains five significant ORFs. (C) Growth phase regulation of sidC. Bacteria were grown in AYE broth to either an OD600 = 1.8 (exponential) or 3.7 (postexponential), harvested, and analyzed by SDS/PAGE and immunoblotting with affinity-purified anti-(His)6-SidC. Displayed are Lp02(dot/icm intact; ref. 11), Lp03 (dotA–; ref. 12), and Lp02(ΔsdcA, ΔsidC), an Lp02 derivative deleted for sidC and its upstream paralog. The isocitrate dehydrogenase (ICDH) protein was used as a loading control by probing with antiserum raised against B. subtilis ICDH.
SidC Is Translocated by Dot/Icm Into the Cytosol of Host Cells. To verify that proteins identified in our assay are translocated into mammalian cells, affinity-purified antiserum against His6-SidC was used to analyze expression and translocation of SidC by L. pneumophila. As predicted, a protein of ≈110 kDa was detected in both a dot/icm+ strain and a dotA– mutant of L. pneumophila, but not in a mutant missing sidC and its paralog sdcA [Δ(sdcA-sidC)] (Fig. 3C). Moreover, expression of SidC was induced ≈7-fold in cells grown to post exponential phase (Fig. 3C). The translocated substrates RalF and LidA also show induced expression in post exponential phase (ref. 9 and G. M. Conover and R.R.I., unpublished observations), and a large body of evidence indicates that factors critical for intracellular survival of L. pneumophila are similarly regulated (25, 26). When infected macrophages were probed for SidC by indirect immunofluorescence microscopy, we found that 1 h after uptake, ≈86% of L. pneumophila phagosomes stained positively for SidC (Fig. 4A), with the protein localizing about the phagosomal membrane (Fig. 4 C and D). In some cases, there was asymmetric diffusion of the protein emanating from the phagosome surface, with most infected cells showing protein only in the region near the phagosome (Fig. 4D).
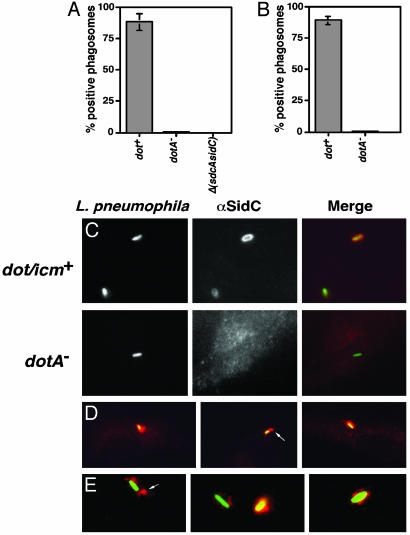
Fig. 4.
SidC is translocated by the L. pneumophila Dot/Icm system to the host cell and is localized about the phagosomal membrane. (A) Bone marrow-derived macrophages from A/J mice were infected with Lp02(dot/icm intact), Lp03(dotA–), or Lp02(ΔsdcA-ΔsidC) strain expressing GFP, respectively. One hour after infection, cells were fixed as described (10), and SidC was probed with anti-(His)6-SidC antibodies and Texas red-labeled secondary antibodies. Stained macrophages were scored for translocation of SidC by counting phagosomes that stained positively with anti-(His)6-SidC. Data shown are from two independent experiments performed in triplicate in which at least 100 phagosomes were scored per coverslip. (B) SidC staining on PNS prepared from L. pneumophila-infected U937 cells in the absence of permeabilization. Sample preparation, immunostaining, and data collection were performed as described in ref. 10 or in A. (C) DotA-dependent translocation of SidC. (Left) Bacteria expressing GFP associated with bone marrow-derived macrophage. Strains used were Lp02(dot/icm intact; Upper) and Lp03 (dotA–; Lower). (Center) Immunoprobing of infected cells with anti-(His)6-SidC. (Right) Merged images of GFP and anti-(His)6-SidC staining. (D) Limited diffusion of SidC from L. pneumophila phagosome. Shown are images of Lp02(dot/icm intact) with murine bone borrow-derived macrophage. (E) SidC is translocated across the phagosomal membrane. Shown are images of PNSs of Lp02(dot/icm intact)-infected macrophages. Bacteria and SidC are probed as above, with bacteria marked by GFP and anti-SidC marked in red.
Because the above strategy only detects secretion of SidC and does not demonstrate translocation across the phagosomal membrane, a second approach was pursued (10). To demonstrate SidC translocation across the phagosomal membrane by the Dot/Icm transporter, we prepared postnuclear supernatants (PNS) from macrophages incubated with L. pneumophila and probed intact phagosomes for SidC in the absence of permeabilization reagents. Approximately 85% of these phagosomes stained positively for SidC (Fig. 4 B and E), whereas <5% stained positively with anti-L. pneumophila serum, demonstrating that the phagosomal membranes were intact. Less than 0.3% of macrophages containing a L. pneumophila dotA– strain stained positively for SidC (Fig. 4 A and C), which was consistent with our data showing that interbacterial translocation of this protein requires the Dot/Icm apparatus. Similarly, in PNS, no isolated phagosome containing a dotA– strain stained positively for SidC (Fig. 4B). The antibody reactivity was specific, because no macrophages harboring a mutant lacking sidC and its upstream paralog stained positively for this protein [Fig. 4A; Δ(sdcA-sidC)], nor did macrophages infected with a simple deletion of sidC (data not shown). These results demonstrate that during infection, SidC is translocated into mammalian cells by the Dot/Icm system, and that translocation occurs across the phagosomal membrane. Because the transfer frequencies of other proteins in our Cre/loxP system were comparable to that of SidC, we postulate that these proteins are also targeted to the host cell by the Dot/Icm transporter.
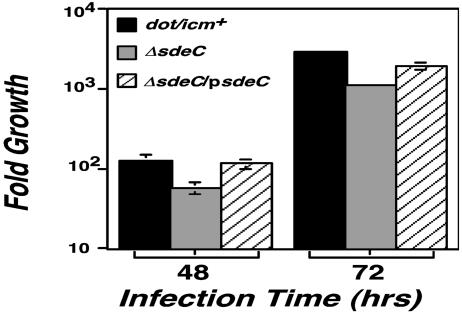
SdeC Is Required for Efficient Intracellular Growth by L. pneumophila. To examine the importance of the Sid proteins in L. pneumophila pathogenesis, we constructed in-frame deletions in some of these genes and tested the resulting mutants for intracellular growth in bone marrow-derived macrophages (10). Individual deletions of sidA, sidD, sidF, or sidG, which have no detectable paralogs in the available L. pneumophila database, resulted in strains with intracellular growth properties that were difficult to distinguish from the wild-type strain. Furthermore, a mutant lacking sidC and its upstream paralog sdcA grew proficiently (data not shown). Finally, a quadruple mutant lacking sidB and its three paralogs has a defect in intracellular growth, but this result was not significantly different from a mutant missing sdbA alone (data not shown), suggesting that functional redundancy extends beyond specific substrate families. We then constructed in-frame deletion mutants lacking individual paralogs of sidE and examined intracellular growth of these mutants in bone marrow-derived or in D. discoideum. Unexpectedly, only deletion of the single paralog sdeC had a detectable growth defect (Fig. 5). In D. discoideum, by using L. pneumophila strains harboring the plasmid vector, the yield of viable counts after 48 h after incubation was depressed 3-fold for the mutant lacking sdeC (Fig. 5). Defective growth could be complemented in trans by a single ORF containing sdeC (Fig. 5). A similar growth defect of this mutant also was observed in macrophage (data not shown). In contrast, mutants missing sidE, sdeA, or sdeB, respectively, had no detectable growth defect in bone marrow-derived macrophages (data not shown). As expected, a Cre::SdeC fusion was found to be translocated between bacterial cells (Table 1).
Fig. 5.
sdeC is required for efficient intracellular growth. D. discoideum cells were infected with a multiplicity of infection of 0.05, and growth of bacteria was monitored as described (13). Total bacterial cells were washed from individual microtiter wells at designated times, and the appropriate dilutions were plated on charcoal yeast extract plates to obtain the colony-forming units. Fold of growth was obtained by dividing colony-forming units at a given time point by the input bacterial cell numbers. Strains tested are as follows: black bars, Lp02(intact dot/icm); gray bars, Lp02(ΔsdeC); striped bars, Lp02(ΔsdeC) harboring pZL192 that carries the ORF of sdeC. Data shown are from two independent experiments performed in triplicate.
Discussion
By developing a genetic assay for monitoring protein translocation using the cre/loxP system, we conclusively demonstrated that the Dot/Icm TFSS transporter can perform interbacterial transfer of proteins known to be targeted to mammalian cells, an observation that we have exploited to identify a large number of translocated proteins. The majority of the proteins we identified have no significant orthologs in the database, suggesting that biogenesis of the L. pneumophila replication compartment may involve mechanisms that differ from other organisms, such as Chlamydia trachomatis and Mycobacterium spp., which reside in similar intracellular vacuoles. Alternatively, proteins with similar functions in different microbial species may have evolved independent of each other, and show little sequence similarity to each other. Many of the translocated proteins have paralogs within L. pneumophila, often encoded in chromosomal regions devoted to expression of translocated substrates of Dot/Icm (Fig. 3 A and B). These genes may have been organized to ensure proper expression during exposure to environmental conditions that are optimal for initiating intracellular growth.
The lack of intracellular growth defects observed in L. pneumophila mutants lacking translocated substrates is reminiscent of a L. pneumophila ralF mutant, which is indistinguishable from the parental wild-type strain in regards to intracellular growth (9). Nevertheless, a ralF mutant is unable to recruit Arf1 to the surface of the replicative phagosome, a property that may play some unknown role in the lifestyle of the organism (27). It is possible that elimination of some of the genes here identified similarly results in the formation of phagosomes with alterations in either morphology or host protein content that have little consequence with respect to growth in cultured cells. Alternatively, the roles played by these proteins may be substituted by other substrates of Dot/Icm, or some of these proteins may be important for growth in some untested host cell.
It is common that a bacterial pathogen codes for numerous effectors but the presence of multiple paralogs of a specific translocated effector in the same organism is only occasionally found (28–30). The close similarity among proteins in a family points to functional redundancy, which may provide an explanation for the failure to identify these proteins in previous genetic screens for bacterial mutants defective in intracellular growth. In the case of the sidB family, however, functional redundancy clearly extends beyond the identified paralogs, and proteins of little similarity in sequence or function may be redundant. It is plausible that each paralog may be adapted to promote growth only in specific host cells. Because L. pneumophila is a versatile pathogen that interacts with very diverse hosts in the environment, the establishment of a successful intracellular replication niche may require only a subset of translocated substrates, with a single subset adapted for a particular host cell type.
Sorting out the minimal complement of translocated proteins necessary for intracellular replication will be a challenge for the future, because the number of such proteins may be quite large, based on the data displayed in Table 1. In addition to the products of the genes identified in the original Cre/loxP assay and sdeC, we have tested four paralogs of Sid proteins for interbacterial transfer, and all can promote transfer of Cre (data not shown). Based on this finding, we speculate that it is likely that all of the paralogs are capable of translocation. Furthermore, in another study we found an additional family of four proteins that can be transferred (S. M. VanRheenen, Z.-Q.L., and R.R.I., unpublished results). Altogether, this brings the minimum number of translocated proteins encoded by L. pneumophila to 24. In fact, there are probably many more proteins translocated by Dot/Icm, as the DotF interaction screen was not carried to saturation. The gene bank required in-frame fusions to a Sau3AI restrictions site, which may not be present in all genes encoding translocated substrates. Furthermore, the screen described here focused on only the subset of DotF interactors, and it appears that not all translocated substrates bind DotF at detectable levels in this assay.
The other salient feature of the Dot/Icm translocation system that was uncovered here is the large size of the proteins that were translocated. Many of the proteins identified in this study are >90 kDa, with one predicted to be >240 kDa. It appears likely that TFSS have evolved to transport large molecules, such as DNA–protein complexes, and these transfer systems may be the preferred translocators for high molecule weight substrates.
In summary, the L. pneumophila TFSS is a transporter system of striking flexibility. It is capable of promoting bacterial conjugation and translocation of proteins from bacteria into mammalian cells, as well as transporting these same proteins between bacterial strains. Although the efficiency of interbacterial protein transfer appears to be many orders of magnitude lower than the transfer from bacteria to macrophages (Table 1 and Figs. 1D and 4 A and B), this property has allowed us to identify substrates of the Dot/Icm system. Understanding the functions of these proteins should allow elucidation of the mechanisms underlying the biogenesis of the L. pneumophila-replicative phagosome. Finally, the methods we described here may be generalized to identify mammalian effectors transferred by all TFSS.
Supplementary Material
Acknowledgments
We thank M. Tang for assistance in protein purification; S. Farrand, D. Ladant, A. Vergunst, and J. Vogel for supplying plasmids; the Isberg laboratory for helpful discussions; and Drs. Carol Kumamoto, Michael Malamy, Susan VanRheeven, Marion Shonn, Isabelle Derre, and Matthias Machner for review of the text. This work was supported by the Howard Hughes Medical Institute. Z.-Q.L. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation, and R.R.I. is a Howard Hughes Medical Institute Investigator.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TFSS, type IV secretion systems; Sid, substrate of Icm/Dot transporter.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. AY504668–AY504685).
References
- 1.Staskawicz, B. J., Mudgett, M. B., Dangl, J. L. & Galan, J. E. (2001) Science 292, 2285–2289. [DOI] [PubMed] [Google Scholar]
- 2.Christie, P. J. (2001) Mol. Microbiol. 40, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horwitz, M. A. (1983) J. Exp. Med. 158, 2108–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturgill-Koszycki, S. & Swanson, M. S. (2000) J. Exp. Med. 192, 1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kagan, J. C. & Roy, C. R. (2002) Nat. Cell Biol. 4, 945–954. [DOI] [PubMed] [Google Scholar]
- 6.Vogel, J. P., Andrews, H. L., Wong, S. K. & Isberg, R. R. (1998) Science 279, 873–876. [DOI] [PubMed] [Google Scholar]
- 7.Segal, G., Purcell, M. & Shuman, H. A. (1998) Proc. Natl. Acad. Sci. USA 95, 1669–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel, J. P. & Isberg, R. R. (1999) Curr. Opin. Microbiol. 2, 30–34. [DOI] [PubMed] [Google Scholar]
- 9.Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A. & Roy, C. R. (2002) Science 295, 679–682. [DOI] [PubMed] [Google Scholar]
- 10.Conover, G. M., Derre, I. I., Vogel, J. P. & Isberg, R. R. (2003) Mol. Microbiol. 48, 305–321. [DOI] [PubMed] [Google Scholar]
- 11.Berger, K. H. & Isberg, R. R. (1993) Mol. Microbiol. 7, 7–19. [DOI] [PubMed] [Google Scholar]
- 12.Berger, K. H., Merriam, J. J. & Isberg, R. R. (1994) Mol. Microbiol. 14, 809–822. [DOI] [PubMed] [Google Scholar]
- 13.Solomon, J. M., Rupper, A., Cardelli, J. A. & Isberg, R. R. (2000) Infect. Immun. 68, 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M. & Peterson, K. M. (1995) Gene 166, 175–176. [DOI] [PubMed] [Google Scholar]
- 15.Karimova, G., Pidoux, J., Ullmann, A. & Ladant, D. (1998) Proc. Natl. Acad. Sci. USA 95, 5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon, R., Priefer, U. & Puhler, A. (1983) Bio/Technology 1, 37–45. [Google Scholar]
- 17.Dumenil, G. & Isberg, R. R. (2001) Mol. Microbiol. 40, 1113–1127. [DOI] [PubMed] [Google Scholar]
- 18.Harlow, E. & Lane, D. (1999) in Using Antibodies, A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 311–343.
- 19.Van Duyne, G. D. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 87–104. [DOI] [PubMed] [Google Scholar]
- 20.Vergunst, A. C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C. M., Regensburg-Tuink, T. J. & Hooykaas, P. J. (2000) Science 290, 979–982. [DOI] [PubMed] [Google Scholar]
- 21.Christie, P. J. (1997) J. Bacteriol. 179, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkins, B. M. & Thomas, A. T. (2000) Mol. Microbiol. 38, 650–657. [DOI] [PubMed] [Google Scholar]
- 23.Simone, M., McCullen, C. A., Stahl, L. E. & Binns, A. N. (2001) Mol. Microbiol. 41, 1283–1293. [DOI] [PubMed] [Google Scholar]
- 24.Lin, W., Fullner, K. J., Clayton, R., Sexton, J. A., Rogers, M. B., Calia, K. E., Calderwood, S. B., Fraser, C. & Mekalanos, J. J. (1999) Proc. Natl. Acad. Sci. USA 96, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrne, B. & Swanson, M. S. (1998) Infect. Immun. 66, 3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammer, B. K. & Swanson, M. S. (1999) Mol. Microbiol. 33, 721–731. [DOI] [PubMed] [Google Scholar]
- 27.Roy, C. R. & Tilney, L. G. (2002) J. Cell Biol. 158, 415–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartman, A. B., Venkatesan, M., Oaks, E. V. & Buysse, J. M. (1990) J. Bacteriol. 172, 1905–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stender, S., Friebel, A., Linder, S., Rohde, M., Mirold, S. & Hardt, W. D. (2000) Mol. Microbiol. 36, 1206–1221. [DOI] [PubMed] [Google Scholar]
- 30.Miao, E. A., Scherer, C. A., Tsolis, R. M., Kingsley, R. A., Adams, L. G., Baumler, A. J. & Miller, S. I. (1999) Mol. Microbiol. 34, 850–864. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.