Abstract
Differences in gene expression are thought to be an important source of phenotypic diversity, so dissecting the genetic components of natural variation in gene expression is important for understanding the evolutionary mechanisms that lead to adaptation. Gene expression is a complex trait that, in diploid organisms, results from transcription of both maternal and paternal alleles. Directly measuring allelic expression rather than total gene expression offers greater insight into regulatory variation. The recent emergence of high-throughput sequencing offers an unprecedented opportunity to study allelic transcription at a genomic scale for virtually any species. By sequencing transcript pools derived from heterozygous individuals, estimates of allelic expression can be directly obtained. The statistical power of this approach is influenced by the number of transcripts sequenced and the ability to unambiguously assign individual sequence fragments to specific alleles on the basis of transcribed nucleotide polymorphisms. Here, using mathematical modelling and computer simulations, we determine the minimum sequencing depth required to accurately measure relative allelic expression and detect allelic imbalance via high-throughput sequencing under a variety of conditions. We conclude that, within a species, a minimum of 500–1000 sequencing reads per gene are needed to test for allelic imbalance, and consequently, at least five to 10 millions reads are required for studying a genome expressing 10 000 genes. Finally, using 454 sequencing, we illustrate an application of allelic expression by testing for cis-regulatory divergence between closely related Drosophila species.
Keywords: cis-regulation, Drosophila melanogaster, Drosophila simulans, gene expression, hybrids
A major challenge in evolutionary biology today is understanding the genetic and molecular mechanisms that give rise to phenotypic differences within and between species. Such differences can arise from mutations affecting the function of gene products (i.e. proteins or RNAs) or mutations that affect expression of these genes. Historically, researchers have looked almost exclusively for (and often found) changes in protein coding regions that appeared to contribute to phenotypic evolution; however, during the last decade, there has been a dramatic increase in the number of studies showing that changes affecting gene regulation can also bring about diversity in ecologically relevant traits that affect behaviour, physiology and morphology (e.g. Duda & Remigio 2008; Giger et al. 2008; Voelckel et al. 2008; see also for reviews Wray 2007; Hoekstra & Coyne 2007; Stern & Orgogozo 2008; Pennisi 2008; Wolf et al. 2010).
Studies of gene expression have become routine with the development of techniques that quantify transcript abundance in a high-throughput way. Microarray studies, in particular, have produced valuable catalogues of differences in transcript levels between individuals (Oleksiak et al. 2002; Whitehead & Crawford 2006), between species in diverse taxa (Rifkin et al. 2003) and between ecological conditions (Reymond et al. 2000; Carsten et al. 2005; Derome et al. 2006). Such studies also show that inter-individual differences in gene expression are often highly heritable (Wayne et al. 2004; Gibson & Weir 2005; Hughes et al. 2006; Lemos et al. 2008; Ayroles et al. 2009).
Because of this heritability, quantitative trait locus (QTL) mapping can be combined with microarray analysis to investigate the genetic basis of variable gene expression (Vasemagi & Primmer 2005). When a QTL affecting a gene’s transcription maps close to the affected gene it can be classified as cis-acting, while a QTL that maps further away on the same chromosome, or to another chromosome, can be classified as trans-acting (Brem et al. 2002). However, strictly speaking, ‘cis’ describes mutations that affect expression of only the allele on the same chromosome as the mutation, whereas ‘trans’ describes mutations that affect allelic expression on both homologous chromosomes. Examples of cis-acting sequences include promoters and enhancers, which are typically located close to the gene that they regulate, while examples of trans-acting regulators include genes that encode transcription factors, which may be located anywhere in the genome. Classifications of expression QTLs as cis- or trans-acting based solely on their proximity to the affected gene are therefore only an approximation – and one that comes with many caveats (Rockman & Kruglyak 2006).
Nevertheless, studies mapping expression QTLs suggest that both cis- and trans-regulatory mutations contribute to transcriptional variation, with a preponderance of expression QTLs appearing to be cis-acting (Wayne et al. 2004; Hughes et al. 2006; Osada et al. 2006; Bergen et al. 2007; Genissel et al. 2008; Gilad et al. 2008; Price et al. 2008; Lemos et al. 2008; but see Morley et al. 2004), although this methodology generally has less statistical power to detect trans-acting than cis-acting variants (Cookson et al. 2009). In addition, QTL mapping studies of variable gene expression require microarrays suitable for studying the species of interest, molecular markers that cover its complete genome, and resources for genotyping these markers in a segregating population. The lack of any one of these things can be a significant impediment for mapping expression QTLs outside well-established genetic model systems.
An alternative strategy for studying regulatory variation uses allelic transcript abundance and the fact that cis-regulatory mutations have allele-specific effects on gene expression while trans-regulatory mutations affect expression of both alleles in a diploid cell (Cowles et al. 2002; Wittkopp et al. 2004). One or more transcribed differences in nucleotide sequence are used to discriminate between transcripts produced by each allele. Asymmetric expression of two alleles, also known as allelic imbalance (AI) that is observed between alleles present in the same cell (i.e. exposed to the same trans-regulatory environment) provides direct evidence of cis-regulatory differences. Expression differences observed between individuals homozygous for two different alleles that are not also observed between these same alleles in heterozygotes are attributed to trans-regulatory differences (Wittkopp et al. 2004).
This allele-specific approach has now been used to decompose variable gene expression into its cis- and trans-regulatory component parts for flies (e.g. Wittkopp et al. 2008a,b), humans (e.g. Pant et al. 2006; Serre et al. 2008), plants (e.g. de Meaux et al. 2005; Guo et al. 2008) and yeast (Tirosh et al. 2009). With the exception of Tirosh et al. (2009), who developed custom microarrays, the methods used to measure allelic expression in these studies are not readily scalable to an entire genome. Furthermore, methods used in these studies, including Tirosh et al. (2009), require polymorphic sites that differentiate alleles to be known a priori. For these reasons, studying allelic expression genome wide has been impractical for nonmodel (as well as most model) species.
Next generation sequencing technologies have the potential to revolutionize studies of allelic expression. Because they obviate the need for a priori sequence information, molecular markers, and locus-specific genotyping assays, next generation sequencing methods can measure allelic abundance at a genomic level in virtually any species. Only transcribed nucleotide differences between alleles and sufficient sequencing depth for detecting AI are required. For these reasons, we expect measurements of allelic expression based on next generation sequencing will soon be acquired by many researchers, not only to disentangle cis- and trans-regulatory variation, but also to quantify the heritability of gene expression, examine dominance among regulatory alleles, evaluate their contribution to morphological, physiological, or behavioural changes, and reveal patterns of allelic variation within and between species.
Not surprisingly, the benefits of next generation sequencing come with a price – and often a high one. A single ‘run’ of high-throughput sequencing can provide up to hundreds of millions of sequences, but currently costs thousands of dollars. The precise cost per base differs among technologies, as does the length of each sequenced fragment and the total number of sequences collected. Because of this cost, careful experimental design that maximizes the data per dollar for allelic expression studies using next generation sequencing is critical. Optimal experimental design is particularly paramount for studies in molecular ecology that seek to examine allelic expression in multiple individuals, species or environmental conditions.
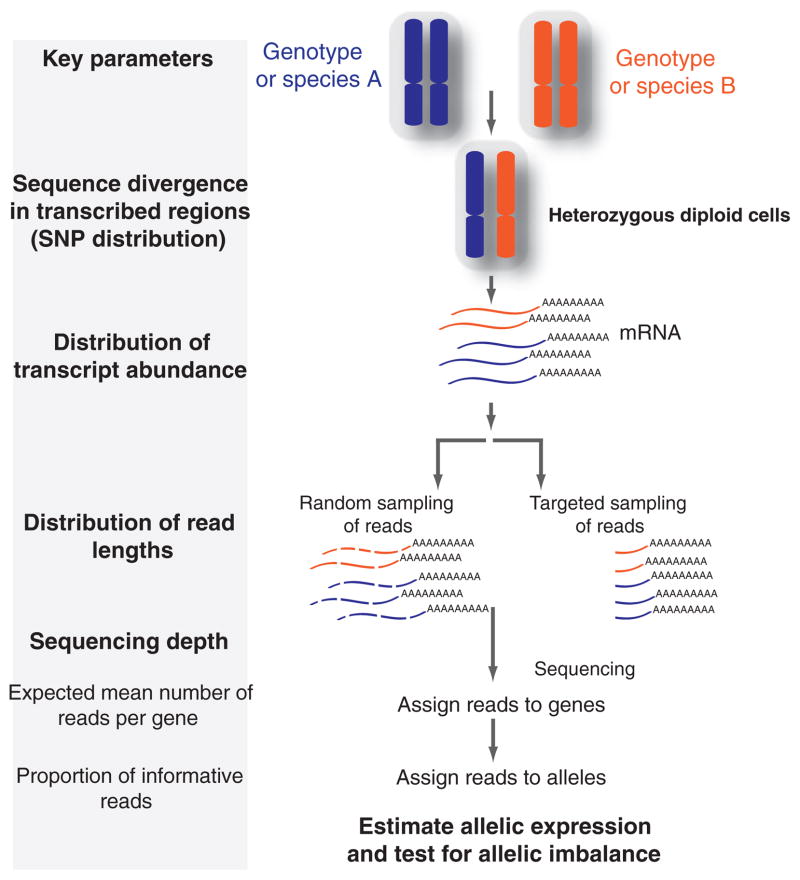
In this study, we use mathematical modelling and computer simulations to identify critical parameters affecting measurements of allelic expression and the detection of AI with high-throughput sequencing. We show that the statistical power of this method depends upon four crucial parameters (Fig. 1): sequence divergence between alleles, the relative transcript abundance, the average read length (i.e. amount of transcript sequenced) and sequencing depth (i.e. average number of reads per gene). The latter two parameters determine the number of sequencing reads expected to map to each gene. The former two parameters determine the proportion of sequence reads per gene that are informative for allelic expression [i.e. contain one or more single nucleotide polymorphisms (SNPs) that allow reads to be unambiguously assigned to an allele]. We show that this probability is strongly affected by the location of SNPs within an mRNA as well as by the way in which the cDNA library is prepared for sequencing. Here, we derive a mathematical model that determines the minimum number of reads required to test for significant AI given various levels of sequence divergence, read lengths, and distributions of relative transcript abundance, and we compare these results with simulations. Finally, to illustrate the potential of this approach, we describe an empirical study using measurements of allelic expression in F1 hybrids between Drosophila melanogaster and Drosophila simulans obtained using 454 sequencing (Roche 454 Life Sciences).
Fig. 1.
High-throughput sequencing technology allows measurement of relative allelic expression genome wide. The schematic representation shown illustrates the steps require to collect allelic expression data. Key parameters associated with each step that ultimately affect the statistical power for detecting significant allelic imbalance (AI) are also shown.
Materials and methods
Fly strains, rearing and crosses
F1 hybrids were produced by crossing 4-day-old virgin Drosophila melanogaster Canton S females with Drosophila simulans C167.4 males. Each mating vial contained 10 females and 10 males. Flies were reared on standard cornmeal medium at 20 °C under a 10/14-h light/dark cycle. Hybrid females were collected at emergence and were stored for 5–6 days at room temperature and then snap frozen in liquid nitrogen.
Preparation of cDNA libraries and 454 sequencing
Total RNA was extracted by homogenizing ~500 hybrid females in 4 mL of TRIZOL Reagent (Invitrogen). We isolated mRNA using an Oligotex Direct mRNA Mini Kit (Qiagen). One milligram of mRNA was used for the first-strand cDNA synthesis (Superscript II; Invitrogen). Reverse transcriptase reactions were performed with biotinylated polyT primer. Second-strand synthesis reactions contained 20 units of DNA ligase, 5 units of RNase H and 30 units of DNA polymerase I (New England Biolabs). The second-strand reactions were randomly sheared by sonication. The cDNA fragment was blunt-ended with T4 polymerase (New England Biolabs). Finally, 3′-end cDNA fragments containing the biotinylated polyT primers were removed from cDNA fragment pool by using Dynabeads (Invitrogen). Sequencing was performed on a GS FLX Instrument following standard protocols (454 Life Science Roche Diagnostics). Sequencing beads containing less than 30 bases with high quality score (>20) were discarded. All sequences are accessible in GenBank (genome project ID 41715).
Data handling and analysis of 454 sequences
A custom Perl script was used (i) to BLAST 454 reads against genic and intergenic sequences from D. melanogaster (Flybase release 4.1, http://flybase.org/) and against the complete genome of D. simulans (Apr. 2005 assembly, UCSC Genome Bioinformatics, http://genome.ucsc.edu/), (ii) to assign to each 454 read to the most probable species (i.e. D. melanogaster or D. simulans) as well as to a specific gene or intergenic region and (iii) to extract descriptive information from each read: number of basepairs (i.e. sequence length), extent of homology identified by BLAST, genomic position in the D. melanogaster and D. simulans genomes, number of gaps in the sequence alignments, number of shared SNPs, and number of species-specific SNPs. The first five of these six parameters describe the quality of 454 reads, while the final parameter was used to classify each sequence read as derived from D. melanogaster or D. simulans or to classify it as having an undetermined origin, which was most common for sequence reads that matched either nonpolymorphic or extremely polymorphic regions. All statistical analyses and simulations described in this work were performed using R (R Development Core Team, 2005).
Pyrosequencing data collection and analysis
To validate measures of allelic expression based on 454 sequencing, pyrosequencing assays were performed for 14 genes (Table S2, Supporting information) using protocols described in Wittkopp et al. (2008a,b). For each gene analysed, a custom pyrosequencing assay consisting of three unique primers was developed and tested for specificity (primer sequences available upon request). Using these assays, we quantified AI in each of eight replicate cDNA samples, each independently synthesized from new mRNA extracted from flies collected at the same time as those used for 454 sequencing. Genomic DNA was also extracted from flies obtained in the initial collection and analysed in quadruplicate using pyrosequencing. From each pyrosequencing reaction, the ratio of D. melanogaster to D. simulans alleles was calculated as described in Wittkopp et al. (2008a,b). For each gene, the ratio of relative allelic expression (i.e. D. melanogaster/D. simulans) was log (base 2) transformed, and the average value from replicate genomic DNA samples was subtracted from the average value of replicate cDNA samples, effectively correcting for any bias in PCR-amplification between alleles (Wittkopp et al. 2004).
Quantifying allelic expression using short-read sequencing: expectations and statistical power
Despite significant improvements in read length since their release, current high-throughput sequencing technologies (e.g. Illumina Solexa, Roche 454 Life Sciences, ABI Solid) remain ‘short-read’ (i.e. <500 bp) sequencing methods. The length of sequences is particularly important for studying allelic expression because, to be informative, a sequencing read must include one or more SNPs that discriminate between alleles. If the distance between heterozygous sites within a transcript is greater than the average sequence length, many reads will include only invariant sites and thus be uninformative for allelic expression. A single ‘run’ of next generation sequencing generates hundreds of thousands to millions of sequencing reads, but only the subset of reads that are informative contribute to estimates of allelic expression. The relative frequency of informative and uninformative reads has a major impact on the statistical power for detecting AI in a given experiment.
In the following section, we derive the probability of obtaining informative and uninformative sequences depending on the number of total sequencing reads, the read length and the sequence divergence between alleles. The model assumes that informative reads could be assigned unambiguously to alleles: it implies that allelic reference sequences are known and read mapping procedures are without errors. For very short reads, alignments against reference genome can be sensitive to mismatches but a large number of new algorithms have been recently released to deal with this issue (see Bateman & Quackenbush 2009; Kofler et al. 2009). Nevertheless, in our model, confidence of allele assignment can be controlled for by the number of SNPs required (see below). We then relax the model assumptions and discuss possibilities of measuring allelic expression without reference genomes or a priori sequences. We show that the total number of sequence reads, in combination with the distribution of transcript abundance among genes, determines the sequencing depth (i.e. number of reads) needed per gene. The read length and sequence divergence define the likelihood of sampling one or more transcribed SNPs, which in turn determines the proportion of informative reads. We explore a wide range of parameter values that should encompass most biological comparisons within and between closely related species. These conditions also reflect the current and anticipated output of multiple high-throughput sequencing technologies. The ultimate goal of this work was to provide guidance for researchers designing allelic expression experiments in their favourite system.
The relationship between sequence divergence and read length
When quantifying total levels of transcript abundance by sequencing cDNA, reads from anywhere within the transcript are informative as long as they are long enough to map unambiguously to a single site in the genome (Torres et al. 2008; Wang et al. 2009). By contrast, when measuring allelic expression, only the subset of these reads containing one or more SNPs that distinguish transcripts derived from different alleles of the same gene are informative. Therefore, read length is a critical parameter for sequencing-based studies of allelic expression: longer reads increase the probability of sequencing a polymorphic site. Sequence divergence between the two alleles under study must also be considered: greater divergence increases the probability of sequencing polymorphic sites for a given read length.
More formally, the probability of sampling at least x SNPs in a read of length l can be approximated by:
| (1) |
where d is the sequence divergence (i.e. the probability of observing a SNP at each nucleotide position). The probability of obtaining exactly n informative reads with at least x SNPs in s randomly sampled reads from a heterozygous gene is given by the probability mass function:
| (2) |
The mean and the variance of this distribution are
| (3) |
and,
| (4) |
Consequently, Pr (X ≥ x) is the expected proportion of informative reads in a random sample.
If only one SNP is required to assign alleles unambiguously, eqn (1) becomes:
| (5) |
For this special case, the minimum read length required to observe a minimum proportion of Pr(X ≥ 1) reads with at least one SNP is
| (6) |
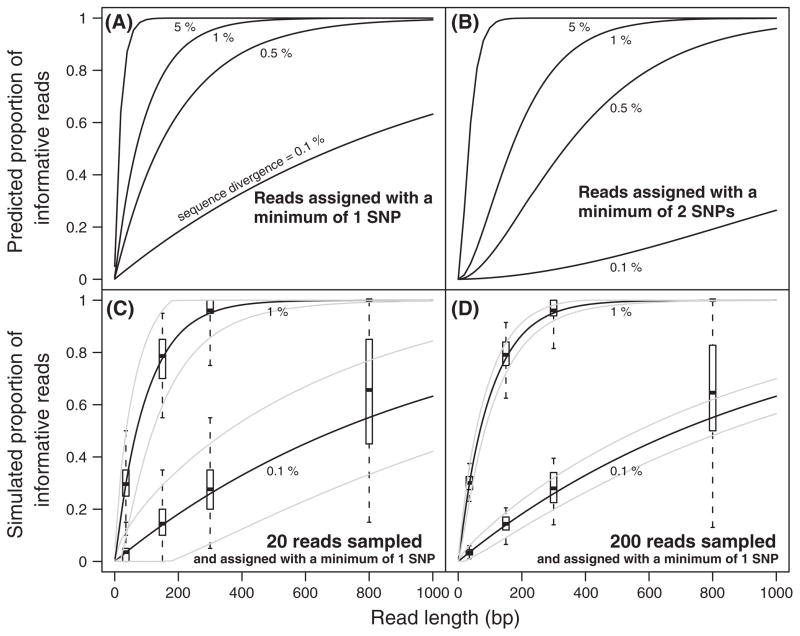
Hence, for mRNA sequences that differ at 0.5% of sites, sequence reads longer than 138 bp are needed in order to obtain ≥50% of reads with at least one SNP. However, in some cases, more than one SNP may be required to unequivocally discriminate between the two alleles. For example, depending on the specific experiment, recombination, homoplasy, or errors in transcription or sequencing could lower the confidence of assignments only based on a single SNP. Therefore, we also calculated the expected proportion of informative reads when more than one SNP is required for allele assignment. By applying eqn (2), we show that this parameter diminishes rapidly when the minimum number of required SNPs increases (Fig. 2A, B). Sequences with little genetic divergence (i.e. <1%) are particularly sensitive to the minimum number of SNPs required to discriminate between alleles.
Fig. 2.
The expected proportion of informative reads increases with genetic divergence and read length. (A, B) Black lines show expected proportions of informative reads (i.e. sequence fragments that could be unambiguously assigned to one allele) predicted by eqn (3) for transcribed sequences containing 0.1, 0.5, 1 or 5% sequence divergence, as indicated. Predictions are shown in which either one single nucleotide polymorphism (SNP) (A) or two SNPs (B) were required for a sequencing read to be informative for measuring allelic expression. (C, D) Predictions based on 0.1% and 1% sequence divergence and requiring only one SNP to be informative are shown again, as they were in (A). Results from simulated data sets are also shown. Each simulation contained either 20 (C) or 200 (D) reads that were generated using a virtual 2000 bp mRNA sequence, 0.1% or 1% sequence divergence, and sequencing reads of 35, 150, 300 and 800 bp. Each scenario was simulated 500 times, and is summarized by boxplots showing the median, lower and upper quartiles, as well as the 1.5 interquartile range. The gray lines are the 95% confidence intervals of the expected proportions based on binomial sampling (Clopper-Pearson interval on eqn (1), Clopper & Pearson 1934).
To determine how well this mathematical model predicts the proportion of informative reads, we randomly sampled reads in silico with lengths ranging from 35 to 800 bp from pairs of virtual mRNA sequences 2000 bp long that contained various levels of sequence divergence. SNPs were distributed uniformly across the length of each hypothetical mRNA sequence. In each simulation, we counted the number of informative reads, that is, the number containing one or more polymorphic sites. As shown in Fig. 2C, D, predictions from our model are most accurate for sequence reads that are ≤300 bp. For longer reads, our model underestimates the mean as well as the variance. Large means are inaccurately estimated because eqn (1) assumes that mRNA molecules have infinite length and consequently, that SNPs can be sampled with replacement. When read lengths converge to mRNA lengths, this assumption then becomes invalid and eqn (1) should be replaced by a hypergeometric form that includes a parameter for mRNA length (see legend for Fig. S1, Supporting information). Although this hypergeometric model is indeed more accurate (compare Fig. 2 and Fig. S1, Supporting information), it is also more complex mathematically. For most experimental design applications, we anticipate that the simpler binomial model will be sufficient.
Note that both the binomial (Fig. 2) and hypergeometric (Fig. S1, Supporting information) models underestimate the variance observed in the simulation study. This is because these models do not take into account the location of polymorphic sites within each mRNA. The probability of sampling a SNP dependent upon its position in the mRNA (Fig. S2, Supporting information) can be described as:
| (7) |
where i is the nucleotide position in the mRNA sequence, lg the mRNA length, and
The mean of this probability distribution is
| (8) |
and its variance is
| (9) |
Hence, considering the location of a SNP requires a much more complex model and only special cases can be easily derived. For instance, for the simple situation of an mRNA sequence containing one and only one SNP, the probability of sampling informative reads becomes
| (10) |
The model described above treats the read length l as a fixed value even though (after filtering for base quality) all sequencing technologies produce reads with a range of alignable lengths. To incorporate this element of the data, we represent the distribution of l as Pr(l), and incorporate this new assumption into eqn (1), resulting in
| (11) |
Incorporating variable read lengths into the model had little effect on the probability of sampling x SNPs in a read. For instance, assuming a Poisson distribution of read lengths, eqn (5) becomes
| (12) |
Finally, the proportion of genes with more than n informative reads can be estimated by:
| (13) |
where Pr(T = t) is the distribution of transcript levels t across the genome. This distribution can be empirically determined or approximated by either discrete decay or power law functions (e.g. Ogasawara et al. 2003). Assuming a geometric decay and using eqn (5), we can rewrite eqn (13) as:
| (14) |
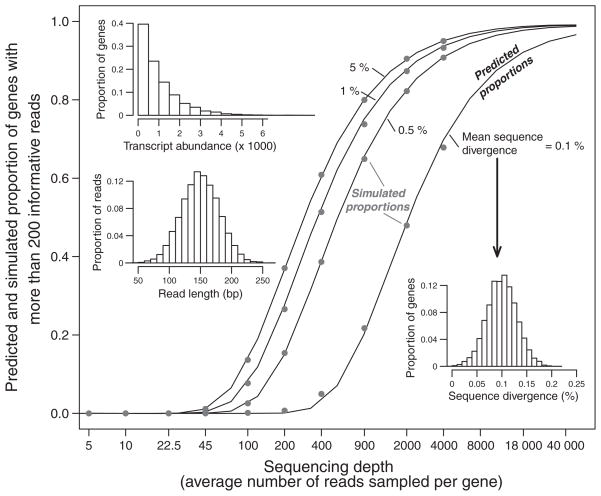
where T̄ is the mean number of transcripts per gene (i.e. the sequencing depth) and d̄ is the mean sequence divergence. Although eqn (14) lacks some of the variance in parameter values discussed above (i.e. read length and sequence divergence distributions as well as SNP location within a sequence), the model nevertheless does an excellent job of predicting the simulated proportion of genes with more than n informative reads (Fig. 3).
Fig. 3.
Predicted proportions of genes with more than 200 informative reads for a given sequencing depth are consistent with simulated data. Predicted values (lines) were obtained using eqn (14), assuming a mean read length of 150 bp and sequence divergence of 0.1%, .5%, 1%, and 5%, as indicated. Simulated data (points) used distributions of transcript abundance, read length, and sequence divergence, as shown for the insets. Two replicate simulations were performed and found to be highly correlated with each other (Spearman’s Rho >99%).
Therefore, for a given number of transcribed genes and total number of sequencing reads (from which T̄ is derived), as well as a particular mean sequence divergence and mean read length, the number of genes expected to have more than n informative reads can be robustly predicted. This number (n) is critical for designing allelic expression experiments using next generation sequencing because it directly determines the statistical power for detecting AI.
Allelic imbalance and statistical power
If we specifically define AI as the ratio of allelic transcript abundance (i.e. number of transcripts from allele 1 divided by the number of transcripts from allele 2), then the probability of observing na1 reads from allele 1 in a sample of n informative reads is given by:
| (15) |
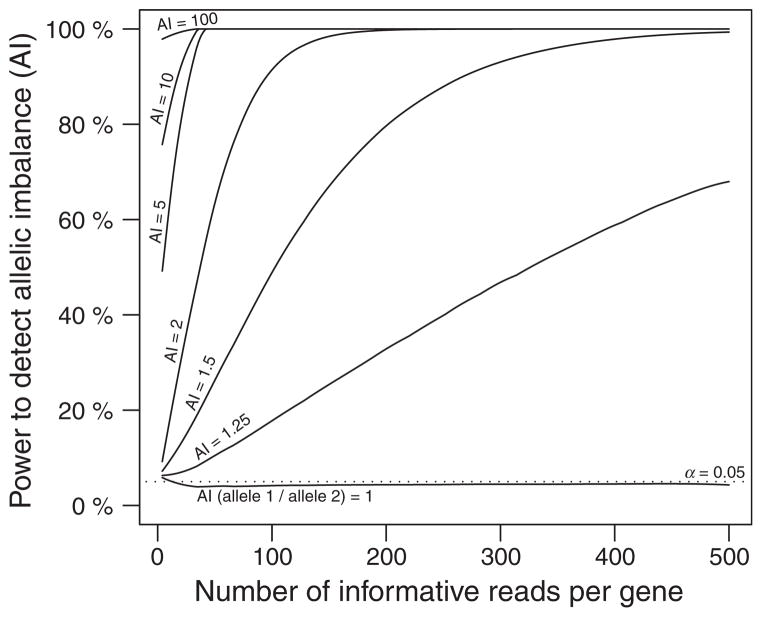
Using this equation, we can determine whether an observed AI value for a gene is significantly different from a null hypothesis of no difference in allelic expression (AI = 1). As shown in Fig. 4, statistical power rises quickly when the true value of AI is ≥2 (e.g. a sample of only 50 informative reads provides 60% statistical power). On the other hand, small imbalances (<1.25-fold) require more than 500 informative reads to reach this same power. Although statistical power to detect significant AI can be achieved easily with small samples, large samples are generally required to produce reasonably precise estimates of AI, especially when the true value of AI is very large (Fig. S3, Supporting information). Prior studies comparing allelic expression within and between Drosophila species observed a median AI of 1.4 for all genes and samples examined and a median AI of 1.7 for cases classified as having significant AI (Wittkopp et al. 2008a,b). This suggests that 200 or more informative reads will generally be required to detect a significant and precise AI values using high-throughput sequencing.
Fig. 4.
Detecting significant allelic imbalance (AI) for genes with small differences in allelic expression requires a large number of informative reads per gene. Statistical power for detecting significant AI for a type I error α = 5% under different conditions is shown. Each line shows the power to detect significant AI, assuming that the true value of AI is 1, 1.25, 1.5, 2, 7, 10 or 100.
Read sampling strategies
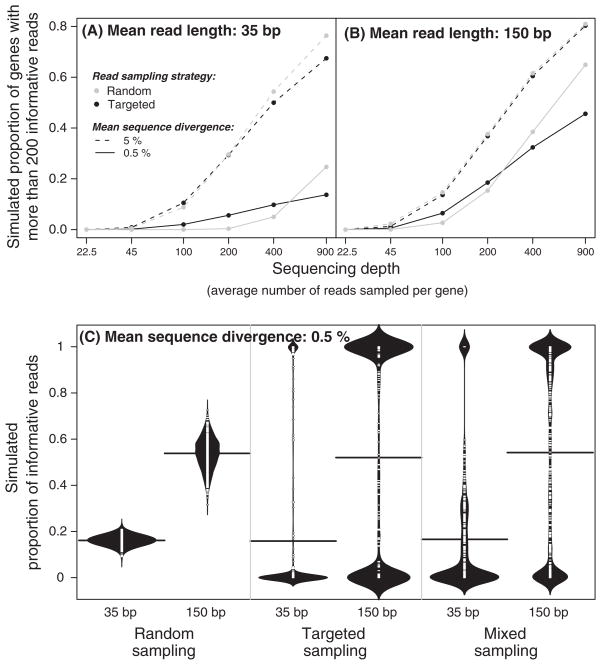
The number of transcribed genes and the mean sequence divergence are both dictated by the species and genotypes under study and cannot be altered. Furthermore, the researcher can affect the average length of sequence reads only in a limited way by choosing one next generation sequencing technology instead of another. The aspect of a sequencing-based allelic expression experiment that the researcher has the most control over is the preparation of cDNA (or equivalent) libraries used for sequencing. Two general types of cDNA libraries can be used for measuring allelic expression. The first is essentially a shotgun library, in which fragments are randomly sampled from the transcriptome. The second is more targeted, containing fragments only from a predetermined region of each transcript. For instance, the 5′- or 3′-end can be systematically sampled from each transcript (e.g. Gowda et al. 2006). The sampling strategy should be chosen carefully because, as shown in eqn (7) and Fig. S2 (Supporting information), the location of SNPs within the cDNA template used for sequencing affects the probability of collecting sequence reads informative for allelic expression.
For a given sequence divergence and read length, the sampling strategy does not affect the expected mean proportion of informative reads; however, it has a large effect on the variance among genes in the proportion of informative reads (Fig. 5C). With a targeted sampling approach all reads for a given gene will either be informative or uninformative, depending on the position of SNPs in the transcript. Consequently, targeted sampling maximizes the statistical power to detect AI for genes that have at least one polymorphic site in the targeted region, but provides no information about relative allelic expression for genes that lack variation in this region. With random sampling, the situation is reversed: estimates of allelic expression can be obtained for more genes, but the power to detect significant AI for any given gene is reduced. Additionally, under random sampling, gene length affects the relative estimated expression among genes (i.e. more reads should come from longer genes), but has a negligible effect on measures of relative allelic expression because transcripts from both alleles of a gene are usually the same length. (For this reason, we did not include gene length in the mathematical models described above.)
Fig. 5.
Read sampling strategy affects the proportion of informative reads per gene and thus the number of genes for which significant allelic imbalance (AI) can be detected. (A, B) Simulated proportions of genes with more than 200 informative reads using a random or a targeted read sampling strategy are shown for mean read lengths of 35 bp (A) and 150 bp (B), with individual reads sampled from a Poisson distribution. See Fig. 2 for a more detailed description of the simulation parameters. (C) The proportion of informative reads per gene using random (left), targeted (middle) and mixed (right) sampling strategies are shown. Each beanplot represents the distribution (500 replicates) of the proportion of informative reads among 500 sampled reads. The horizontal bar shown on each beanplot indicates the mean of these distributions. For the mixed strategy, fragments with sequence lengths drawn from a Poisson distribution with a mean of 500 bp were anchored to a fixed, predetermined location (the 3′ end), and sequences of either 18 or 75 bp were taken from each end to simulate paired-end sequencing.
To compare the power of random and targeted sampling, we simulated the proportion of genes with more than 200 informative reads under different conditions. As described above, 200 informative reads per gene provides reasonable statistical power to detect reasonably small difference in allelic expression (see Fig. 4). Results from this simulation show that random sampling generally performs better than targeted sampling, except when sequence divergence and the number of sequences per gene are low, and especially when sequencing reads are short (Fig. 5A, B). As an alternative, a ‘mixed’ sampling scheme that included 50% targeted and 50% randomly located sequence reads produced results most similar to targeted sampling (Fig. 5C). Such a ‘mixed’ sampling strategy could be employed by paired-end sequencing a cDNA library containing fragments of variable length that all share the same 5′- or 3′-end.
Based on these results, we conclude that, for allelic expression experiments with at least moderate sequence depth in species with reference genomes available (that can be used to map sequencing reads to particular genes), a random sampling strategy will almost always provide the most information per unit cost. However, if no reference genome sequence is available, or if sequencing depth is limited, targeted sampling may have advantages that offset the loss of information on a genomic scale. For example, targeted sampling simplifies the process of determining which sequence reads come from the same gene. In the absence of a reference genome, this is especially important because sequence reads must be assembled into (hopefully, gene-specific) contigs de novo and all reads from the same gene generated by targeted sampling should overlap. Alternatively if a reference genome is available, but only low depth sequencing is possible, a targeted strategy would provide more accurate estimates of allelic expression for the subset of the genes with SNPs in the targeted regions than random sampling. Finally, although there may be some cases for which ‘mixed’ sampling would be the best choice, in the two scenarios considered above, reducing coverage in the targeted region and distributing some of the reads more evenly across the transcriptome does not offer any clear advantages.
Allelic expression in Drosophila hybrids
To illustrate one application of allelic expression measurements, we quantified allelic expression in cDNA pools derived from interspecific F1 hybrids and used these data to test for significant AI (i.e. differences in expression between the maternal and paternal alleles). F1 hybrids used for this study were produced by crossing Drosophila melanogaster females and Drosophila simulans males, collecting virgin female progeny, and aging them for 5–6 days. After extracting mRNA from these flies, a cDNA pool containing random fragments from the transcriptome was constructed and analysed using the high-throughput sequencing technology developed by 454 Life Sciences (Roche).
Analysis of sequencing statistics, informative reads and allelic expression
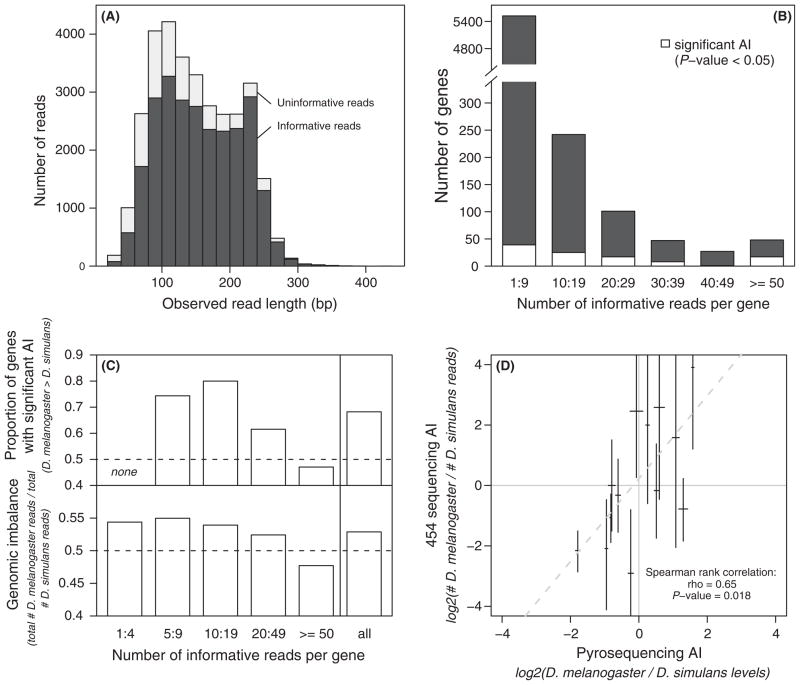
In all, 36 855 high-quality 454 sequencing reads were obtained from the random interspecific hybrid cDNA library (Table 1). The average length of these reads was 170 bp, with 74 and 250 bp for the 2.5 and 97.5 percentile respectively. Eighty-eight per cent of these reads had at least one homologous sequence in either the D. melanogaster or D. simulans genome (BLAST, E-value < 10−4), 5% of which showed homology to only one of the two parental genomes. These reads mapped to 5591 genes and 975 intergenic regions. Sequences derived from intergenic regions accounted for 7% of the mapped reads, and might have resulted from transcription of unannotated genes or exons, abnormal splicing that generated unexpected splice junctions, transposable elements or spurious transcription (e.g. Stolc et al. 2004). More than 10 reads matched each of 577 genes, while more than 50 reads matched each of 83 genes. Despite this relatively low sequencing depth, our data appear to correctly, albeit crudely, measure total levels of gene expression: estimates of transcript abundance in F1 hybrids measured using 454 sequencing correlated significantly (Spearman correlation, Rho = 0.45, P < 0.001) with expression levels reported for female D. melanogaster that were obtained using microarrays (Harbison et al. 2005).
Table 1.
Number of reads, mean read and BLAST hit length, and average number of discriminating SNPs for Drosophila melanogaster, Drosophila simulans and uninformative reads. Within brackets: 2.5 and 97.5 percentile. ‘Undetermined SNPs’ are polymorphic sites in 454 reads that do not match either reference genome
| No. reads | Mean reads length | Mean D. melanogaster BLAST hit length | Mean D. simulans BLAST hit length | No. D. melanogaster SNPs | No. D. simulans SNPs | No. unknown SNPs | |
|---|---|---|---|---|---|---|---|
| All reads | 36 855 | 173 (74;250) | 150 (45;259) | 149 (43;259) | 1.92 (0;5) | 1.61 (0;4) | 0.72 (0;2) |
| Nonattributed | 4502 | 192 (71;305) | |||||
| D. melanogaster BLAST hits only | 1429 | 160 (68;282) | 123 (22;261) | ||||
| D. simulans BLAST hits only | 271 | 138 (64;279) | 98 (23;257) | ||||
| D. melanogaster alleles (≥1 SNP) | 13 770 | 176 (81;279) | 160 (61;263) | 154 (52;259) | 4.7 (1;15) | 0.24 (0;2) | 0.78 (0;6) |
| D. simulans alleles (≥1 SNP) | 12 269 | 177 (80;280) | 156 (54;259) | 159 (58;262) | 0.39 (0;3) | 4.45 (1;14) | 1 (0;6) |
| Uninformative reads (≥1 SNP) | 4614 | 140 (64;265) | 110 (30;238) | 107 (28;236) | 0.28 (0;2) | 0.28 (0;2) | 0.76 (0;6) |
| D. melanogaster alleles (≥2 SNPs) | 10 984 | 182 (86;281) | 168 (72;264) | 161 (61;260) | 5.56 (2;15) | 0.24 (0;2) | 0.78 (0;6) |
| D. simulans alleles (≥2 SNPs) | 9521 | 184 (86;281) | 164 (63;261) | 168 (69;265) | 0.4 (0;3) | 5.35 (2;15) | 1.02 (0;6) |
| Uninformative reads (≥2 SNPs) | 10 148 | 147 (67;266) | 121 (34;246) | 118 (32;244) | 0.57 (0;3) | 0.56 (0;3) | 0.81 (0;6) |
SNP, single nucleotide polymorphism.
In order to assign each read to a specific parental allele, we determined the number of variable sites among 454 reads for a particular gene (i.e. SNPs) that were identical to the D. melanogaster and D. simulans reference genomes. This analysis was performed only for reads that showed a significant alignment with both reference genomes. Sequences were assigned to the species with the highest number of identical SNPs. For instance, if three SNPs within a given read were identical to the D. melanogaster reference genome and two (different) SNPs were identical to the D. simulans reference genome, the read was assumed to be derived from the D. melanogaster allele. Reads were expected to differ from the reference genomes because the strains of D. melanogaster and D. simulans used to generate the F1 hybrids analysed by 454 sequencing were not the same as the strains sequenced to assemble these reference genomes. As mentioned above, shared recombination, homoplasy, or errors in transcription or sequencing might also complicate the assignment of individual reads to one species or the other. In all, 26 039 reads, which is 71% of all high-quality reads, were assigned to one species or the other. At least 10 informative reads were identified for each of 465 genes, with 58 of these genes having more than 50 informative reads each (Fig. 6B). As shown in Fig. 4, genes with 50 informative reads had 60% power to detect significant AI of at least twofold, while genes with 10 reads had only 20% power to detect changes of the same magnitude. Consistent with both intuitive and mathematical predictions, long reads were more often informative for allele-specific expression than short reads (Fig. 6A), with the average length of uninformative reads only ~70% that of informative reads (Table 1).
Fig. 6.
Analysis of allelic expression and allelic imbalance (AI) in Drosophila F1 hybrids. (A) The distribution of sequence fragment lengths for both informative and uninformative reads is shown. (B) The number of genes in different gene expression level classes (as measured by the abundance of informative reads) are shown along with the number of genes in each class that showed significant AI. (C) The top panel shows the proportion of genes with significant AI (see Table S1, Supporting information) for which the D. melanogaster allele is most abundant. The bottom panel shows the proportion of informative reads in a given expression level class that were assigned to D. melanogaster. In both panels, the dotted line corresponds to a balanced proportion (50%). (D) The relationship between relative allelic expression as measured by 454 sequencing and by pyrosequencing is shown. For pyrosequencing, the average of eight replicates is plotted and the 95% confidence intervals are indicated by the horizontal bars. For 454 sequencing, the relative number of informative reads is shown, with vertical bars indicating the Clooper-Pearson 95% confidence intervals derived from binomial sampling (see Supplementary Fig. S3). The dotted line indicates the slope of the nonparametric regression.
Intriguingly, more informative reads (53%) were assigned to the D. melanogaster allele than to the D. simulans allele (13 770 and 12 269 reads, respectively; binomial test: P < 0.001). The excess of D. melanogaster alleles was greater among sequences from genomic regions annotated as genic than those annotated as intergenic (53.0% vs. 50.3%), although this difference was not significant (χ2 = 3, P = 0.08) and may be an artefact of the much smaller number of reads classified as intergenic (93% vs. 7%). The overabundance of D. melanogaster alleles was observed not only across the whole transcriptome analysed but also for subsets of genes with different expression levels (Fig. 6C). The one (surprising) exception to this was the subset of genes with the highest overall expression levels, which showed an excess of D. simulans alleles (Fig. 6C). This shows that the greater abundance of D. melanogaster reads observed in F1 hybrids cannot be explained by higher expression of the D. melanogaster allele of only a few highly expressed genes. In addition, this pattern is unlikely to result from poor quality sequences or alignments because (i) BLAST hit lengths are on average identical in both reference genomes, (ii) there were on average 4.5 discriminating SNPs per informative read, in both species, which makes nearly all assignments unambiguous and (iii) the trend remained after applying a higher stringency cutoff (≥2 SNPs, see Table 1). Based on all of these observations, we conclude that the preferential expression of D. melanogaster alleles observed in F1 hybrids is a real (biological) property of these flies and likely results from a process that affects allelic transcription on a genomic scale. This expression bias does not appear to be caused by a particular tissue, a particular biological function, or a particular chromosomal location (data not shown); however, deeper sequence coverage is required to test these hypotheses definitively.
Allelic imbalance in Drosophila hybrids
As described in the Introduction, quantifying AI in F1 hybrids provides a direct readout of relative cis-regulatory activity. We tested for significant AI of the 891 genes that show more than six informative reads because it is impossible to reach significance (P < 0.05) with a binomial test and less than six observations. One hundred seven of these 891 genes (12%) showed significant AI based on our data (Table S1, Supporting information). For this analysis, we estimate the false discovery rate to be c. 17% by resampling our observed distribution of informative read abundance assuming observed AIs are true or equal to 1. On the other hand, we estimated c. 13% of false negative AI. Consequently, the analysis suggests that more than 190 genes could show true AI (21% of tested genes). In fact, despite the low level of sequencing depth, we detected significant AI for 35% of genes with more than 50 informative reads and for 14% of genes with more than 10 informative reads. On average, the more abundant allele of a gene was observed five times more than the less abundant allele, with a minimum of 1.6 and a maximum of 15 (Fig. S4, Supporting information). Finally and perhaps not surprisingly given the overall excess of reads most similar to D. melanogaster, the D. melanogaster allele was more abundant than the D. simulans allele for 73 (68%) of these 107 genes (binomial test: P < 0.001, Fig. 6C).
454 sequencing and pyrosequencing produce similar estimates of allelic expression
To determine whether 454 sequencing produces accurate estimates of allelic expression, we used pyrosequencing to independently quantify AI in 14 genes and compared the results to estimates obtained using 454 sequencing. The 14 genes selected for this analysis had allelic expression differences that spanned the range of AI values observed with 454 sequencing (compare Fig. 6D and Fig. S4). Pyrosequencing permits quantifications of allelic expression for individual genes and produces estimates of AI that are consistent with those obtained using both quantitative real-time PCR and microarrays (Wittkopp et al. 2006). Pyrosequencing allows high levels of replication for a modest cost, thus very precise estimates of AI can be obtained. Despite the small number of informative 454 sequencing reads for some of the genes analysed (Table S2, Supporting information), we found a significant correlation between estimates of allelic expression obtained using these two techniques (Fig. 6D; Spearman’s Rho = 0.65, P < 0.05). The effect of sequencing depth for individual genes on the precision of allelic expression estimates is readily visible by examining the gene-specific binomial sampling confidence intervals shown in Fig. 6D. Because precision improves with deeper sequencing coverage, we also examined the correlation between measures of allelic expression for the eight genes with at least 20 informative reads each. This subset of the data showed a stronger correlation (Spearman’s Rho = 0.74, P < 0.05), as expected.
Discussion
Less than a decade ago, Gibson (2002) predicted that microarrays would have a tremendous impact on molecular and genetic research at the interface of organismal and population biology. Since then, this methodology has proven to be a formidable tool for examining the extent of gene expression polymorphism within species and divergence between species, as well as for studying the evolutionary processes that generate and act upon regulatory variation. Microarray studies have revealed extensive inter-individual variation in gene expression and showed that this variation is often heritable and its segregation within and between species is primarily affected by neutral drift and stabilizing selection (see for review Whitehead & Crawford 2006). Perhaps surprisingly, very little adaptive regulatory variation has been identified. This may be because it is uncommon or because we do not yet have the correct theoretical framework for distinguishing the effects of neutral and non-neutral evolution (Fay & Wittkopp 2008).
High-throughput sequencing technologies are now poised to replace microarrays for measuring gene expression on a genomic scale, especially for evolutionary and ecological studies that require analysis of many species and/or individuals with divergent genotypes. (Elmer et al., 2010) Compared to microarrays, a high-throughput sequencing approach is more complex technologically, yet much simpler statistically and methodologically. Furthermore, it does not require taxon-specific probes or any a priori sequence information, making it suitable for measuring expression profiles of virtually any species. Arguably, its greatest advantage relative to microarrays is that both total and allelic measures of gene expression are obtained simultaneously. With measures of allelic expression, there is a closer relationship between an allele’s sequence and its activity, making it easier to study the inheritance of gene regulation phenotypes within and between species.
Indeed, using allelic expression rather than total gene expression not only facilitates studies exploring the evolutionary consequences of naturally occurring regulatory variation, but also simplifies mapping of regulatory mutations variation, which is necessary to elucidate the genetic architecture of gene expression. Such studies allow researchers to more efficiently identify loci, genes and molecular processes that play important roles in adaptation. We fully anticipate that studies of allelic expression will soon produce novel insights for the field of molecular ecology, as well as many other fields. We hope that the theoretical and empirical information provided by this study helps researchers design experiments that test specific hypotheses efficiently and cost effectively.
To this end, we have developed a mathematical model that allows researchers to estimate the minimum sequencing depth needed to detect significant AI, based on the average sequence read length of their chosen next generation sequencing technology, the average genetic divergence between alleles under study, the approximate number of transcribed genes, and the anticipated distribution of transcript levels across the genome. For instance, eqn (14) shows that, for an experiment that uses reads averaging 150 bp and compares allelic expression in a polymorphic species with 0.1% sequence divergence, more than 4000 sequence reads per gene are needed to achieve 60% statistical power for detecting significantly AI larger than 1.5-fold (see Figs 3 and 4). For a genome containing 10 000 transcribed genes (and assuming an exponential decay distribution for expression levels), such a study would require c. 40 million reads (Fig. 3). If the average sequence length were increased to 500 bp, the number of sequence reads required would be decreased fourfold to c. 10 million reads. With only three million reads, a statistical power of ~20% would be achieved.
Caveats and considerations
We stress that these predictions should be treated as approximate guidelines only because a number of features of real transcriptomes violate our model and cause our equations to over- or underestimate the requisite number of sequence reads. In the following paragraphs, we discuss four such features and their impact on study design.
We modelled the distribution of transcript levels using a standard decay function, yet the distribution of transcript abundances in real organisms will virtually never fit such as function perfectly. In particular, strong deviations in distribution tails, such as many more genes with extremely high or low expression levels, will cause the model to underestimate the required minimum sequencing depth. If the precise distribution of transcript levels is known a priori for a particular organism, however, this information can easily be incorporated into eqn (13) to improve the accuracy of the predictions.
Another potential caveat comes from the imprecise construction of cDNA pools. We found that the way sequences are sampled from mRNA transcripts affects the statistical power for a given set of parameters (Fig. 5). We modelled cases only in which cDNA pools contain completely random fragments and perfectly targeted fragments; however, one or more of the technical steps used to create cDNA libraries for sequencing may often introduce imperfections. For example, an unexpected bias may arise during the production of ‘random’ cDNA libraries caused by factors such as the preferential ligation of adapters used for sequencing. Alternatively, targeted libraries may include unwanted sequence fragments that result from reads in untargeted regions. However, our analysis of the ‘mixed’ sampling strategy suggests that small errors in the ultimate composition of cDNA libraries will have little effect on the power to detect AI for most genes. Paired-end sequencing, which is an option now available for some technologies, has no clear advantages for AI analyses, although it could be used to evaluate read and sample quality.
Yet another factor to consider carefully is which genes (with respect to expression level) are of utmost interest. For example, if highly transcribed genes are of primary concern, shallow sequencing may be sufficient; increasing sequencing depth has little impact on the statistical power to detect significant AI in these genes. By contrast, if lowly transcribed genes are of most interest, sequencing the entire transcriptome may require such high coverage as to make the experiment impractical. Removing transcripts from genes with the highest expression by subtraction or normalization is one way to increase coverage of the more lowly expressed genes; however, care must be taken not to alter the relative abundance of alleles for the genes of interest. Methods such as quantitative PCR or pyrosequencing may be much more cost-effective and reliable for measuring allelic expression of a few lowly expressed genes.
Finally, computer simulations showed that the position of SNPs within a transcript increases variance in the expected number of informative reads per gene. The model assumes, however, that SNPs are distributed randomly across the length of each mRNA. In reality, polymorphic sites are rarely distributed randomly within a transcript. For instance, they are known to be more frequent in 5′ or 3′ UTRs than nonsynonymous sites (Andolfatto 2005). Differences also exist in the amount of sequence polymorphism and divergence among genes. In general, regions of transcripts with greater sequence variation are more likely to produce informative reads than regions with fewer polymorphic or divergent sites. Finally, our model assumes that transcript abundance and sequence divergence are independent, but in fact, they are often correlated (Subramanian & Kumar 2004).
Properties of allelic expression in Drosophila hybrids
Interspecific hybrids between D. melanogaster and D. simulans show extensive misexpression relative to either parental species (Ranz et al. 2004). The primary cause of this misexpression is unknown, but it is generally attributed to incompatibilities between genomes that disrupt gene regulatory networks (Landry et al. 2007). Consistent with this idea, prior work suggests that the co-evolution of cis- and trans-regulatory factors may contribute to misexpression when divergent alleles meet in interspecific hybrids (Landry et al. 2005).
To compare the activity of homologous cis-regulatory sequences between species, we used estimates of allelic expression in interspecific F1 hybrids obtained using 454 sequencing. We measured allelic expression for over 5500 of the most highly expressed genes and we were able to test AI in ~900 of these genes. Overall, we detected significant AI for 12% of these genes; among genes with the most informative reads (i.e. ≥50 reads per gene), 35% of genes showed significant AI. Our study almost certainly underestimates the percentage of genes with differences in expression between species-specific alleles in F1 hybrids because even 50 informative reads per gene provides only 25% and 60% statistical power to detect significant AI for genes with true allelic expression differences of 1.5- and twofold respectively. Consistent with this interpretation, prior studies of relative cis-regulatory activity between D. melanogaster and D. simulans reported much higher percentages of genes with significant AI (Lemos et al. 2008; Wittkopp et al. 2008a,b).
Genomic imbalance in interspecific hybrids
Widespread misexpression of genes is common in inter-specific hybrids, and the genomic imbalance that we observed in F1 hybrids between D. melanogaster and D. simulans alleles may provide insight into the molecular mechanisms responsible for this misexpression. Drosophila melanogaster alleles are repeatedly overexpressed in F1 hybrids despite the fact that both alleles are expected to be regulated by the same pool of maternal and paternal trans-acting factors. Long-range chromatin effects that favour expression of D. melanogaster chromosomes could contribute to this genomic imbalance, as could interactions between the nuclear genome and cytoplasmic components (especially mitochondria), which were inherited from D. melanogaster in the hybrids examined. It will be interesting to see whether these same expression differences exist between the two alleles if they are compared in cells containing only trans-acting factors from D. melanogaster or D. simulans. Such a comparison would provide a direct test for complex interactions among divergent trans-acting factors and cis-regulatory sequences. Although the prevalence of such interactions remains an open question, a small scale study of D. melanogaster found no evidence of such cis-by-trans interactions (Wittkopp et al. 2008a,b).
Concluding remarks
Comparing the activity of orthologous cis-regulatory alleles, as described here for interspecific Drosophila hybrids, is but one application of sequence-based measures of allelic expression. Quantifying heritability of gene expression, examining dominance and imprinting among regulatory alleles, or revealing patterns of allelic variation within and between species are some of additional applications for quantitative measures of relative allelic expression. By identifying parameters critical for such experiments and exploring their impact on possible outputs, we anticipate that results from this study will help researchers optimize their available resources and generate data sets that are best suited for addressing their primary question(s) of interest.
Supplementary Material
Acknowledgments
We thank the FAS Center for Systems Biology for the use of their Facility and the Sequencing Platform at the Broad Institute. PF was supported by a Marie Curie Outgoing Fellowship from the European Commission and postdoctoral fellowships from the Swiss National Science Foundation. CRL was supported by the National Sciences and Engineering Research Council of Canada (NSERC) and a Frank Knox Memorial Fellowship at Harvard University, and a Canadian Institute of Health Research (CIHR) Fellowship at University of Montreal and is now a CIHR New Investigator at Laval University. Funding for this work was provided by NIH grants GM068465 and GM065169 to DLH and a March of Dimes Basil O’Connor Starter Scholar award to PJW (5-FY07-181). P.J.W. is an Alfred P. Sloan Research Fellow.
Footnotes
Research interests: PF studies gene expression evolution and mechanisms of molecular evolution. CRL studies gene expression regulatory networks and protein-protein interaction networks. PJW studies genetic changes responsible for phenotypic differences within and between species. JDG is exploring fundamental characteristics of gene expression evolution. CN and CR are responsible for development, implementation and application of new sequencing technologies at the Broad Institute. DLH’s research interests include evolutionary genomics, molecular evolution and population genetics.
Conflicts of interest
The authors have no conflict of interest to declare and note that the funders of this research had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, et al. Systems genetics of complex traits in Drosophila melanogaster. Nature Genetics. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Quackenbush J. Bioinformatics for next generation sequencing. Bioinformatics. 2009;25:429. doi: 10.1093/bioinformatics/btp037. [DOI] [PubMed] [Google Scholar]
- Bergen AW, Baccarelli A, McDaniel TK, et al. cis sequence effects on gene expression. BMC Genomics. 2007;8:296. doi: 10.1186/1471-2164-8-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- Carsten LD, Watts T, Markow TA. Gene expression patterns accompanying a dietary shift in Drosophila melanogaster. Molecular Ecology. 2005;14:3203–3208. doi: 10.1111/j.1365-294X.2005.02654.x. [DOI] [PubMed] [Google Scholar]
- Clopper CJ, Pearson ES. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nature Reviews Genetics. 2009;10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. Detection of regulatory variation in mouse genes. Nature Genetics. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- Derome N, Duchesne P, Bernatchez L. Parallelism in gene transcription among sympatric lake whitefish (Coregonus clupeaformis Mitchill) ecotypes. Molecular Ecology. 2006;15:1239–1249. doi: 10.1111/j.1365-294X.2005.02968.x. [DOI] [PubMed] [Google Scholar]
- Duda TF, Remigio EA. Variation and evolution of toxin gene expression patterns of six closely related venomous marine snails. Molecular Ecology. 2008;17:3018–3032. doi: 10.1111/j.1365-294X.2008.03804.x. [DOI] [PubMed] [Google Scholar]
- Elmer KR, Fan S, Gunter HM, Jones JC, Boekhoff S, Kuraku S, Meyer A. Rapid evolution and selection inferred from the transcriptomes of sympatric crater lake cichlid fishes. Molecular Ecology. 2010;19 (Suppl 1):196–210. doi: 10.1111/j.1365-294X.2009.04488.x. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity. 2008;100:191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- Genissel A, McIntyre LM, Wayne ML, Nuzhdin SV. Cis and trans regulatory effects contribute to natural variation in transcriptome of Drosophila melanogaster. Molecular Biology and Evolution. 2008;25:101–110. doi: 10.1093/molbev/msm247. [DOI] [PubMed] [Google Scholar]
- Gibson G. Microarrays in ecology and evolution: a preview. Molecular Ecology. 2002;11:17–24. doi: 10.1046/j.0962-1083.2001.01425.x. [DOI] [PubMed] [Google Scholar]
- Gibson G, Weir B. The quantitative genetics of transcription. Trends in Genetics. 2005;21:616–623. doi: 10.1016/j.tig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Giger T, Excoffier L, Amstutz U, et al. Population transcriptomics of life-history variation in the genus Salmo. Molecular Ecology. 2008;17:3095–3108. doi: 10.1111/j.1365-294X.2008.03820.x. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends in Genetics. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda M, Li HM, Alessi J, et al. Robust analysis of 5′-transcript ends (5′-RATE): a novel technique for transcriptome analysis and genome annotation. Nucleic Acids Research. 2006;34:e126. doi: 10.1093/nar/gkl522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Yang S, Rupe M, et al. Genome-wide allele-specific expression analysis using Massively Parallel Signature Sequencing (MPSS (TM)) reveals cis- and trans-effects on gene expression in maize hybrid meristem tissue. Plant Molecular Biology. 2008;66:551–563. doi: 10.1007/s11103-008-9290-z. [DOI] [PubMed] [Google Scholar]
- Harbison ST, Chang S, Kamdar KP, Mackay TFC. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biology. 2005;6:R36. doi: 10.1186/gb-2005-6-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hughes KA, Ayroles JF, Reedy MM, et al. Segregating variation in the transcriptome: Cis regulation and additivity of effects. Genetics. 2006;173:1347–1364. doi: 10.1534/genetics.105.051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Torres TT, Lelley T, Schlotterer C. PanGEA: identification of allele specific gene expression using the 454 technology. BMC Bioinformatics. 2009;10:143. doi: 10.1186/1471-2105-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, et al. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Hartl DL, Ranz JM. Genome clashes in hybrids: insights from gene expression. Heredity. 2007;99:483–493. doi: 10.1038/sj.hdy.6801045. [DOI] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proceedings of the National Academy of Sciences, USA. 2008;105:14471–14476. doi: 10.1073/pnas.0805160105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Meaux J, Goebel U, Pop A, Mitchell-Olds T. Allele-specific assay reveals functional variation in the chalcone synthase promoter of Arabidiopsis thaliana that is compatible with neutral evolution. Plant Cell. 2005;17:676–690. doi: 10.1105/tpc.104.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M, Molony CM, Weber TM, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara O, Kawamoto S, Okubo K. Zipf’s law and human transcriptomes: an explanation with an evolutionary model. Comptes Rendus Biologies. 2003;326:1097–1101. doi: 10.1016/j.crvi.2003.09.031. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nature Genetics. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Osada N, Kohn MH, Wu CI. Genomic inferences of the cis-regulatory nucleotide polymorphisms underlying gene expression differences between Drosophila melanogaster mating races. Molecular Biology and Evolution. 2006;23:1585–1591. doi: 10.1093/molbev/msl023. [DOI] [PubMed] [Google Scholar]
- Pant PV, Tao H, Beilharz EJ, et al. Analysis of allelic differential expression in human white blood cells. Genome Research. 2006;16:331–339. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Deciphering the genetics of evolution. Science. 2008;321:760–763. doi: 10.1126/science.321.5890.760. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson N, Hancks DC, et al. Effects of cis and trans genetic ancestry on gene expression in African Americans. Plos Genetics. 2008;4:e1000294. doi: 10.1371/journal.pgen.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Namgyal K, Gibson G, Hartl DL. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Research. 2004;14:373–379. doi: 10.1101/gr.2019804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nature Genetics. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nature Reviews Genetics. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Serre D, Gurd S, Ge B, et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic Cis-acting mechanisms regulating gene expression. Plos Genetics. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- Torres TT, Metta M, Ottenwalder B, Schlotterer C. Gene expression profiling by massively parallel sequencing. Genome Research. 2008;18:172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasemagi A, Primmer CR. Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Molecular Ecology. 2005;14:3623–3642. doi: 10.1111/j.1365-294X.2005.02690.x. [DOI] [PubMed] [Google Scholar]
- Voelckel C, Heenan PB, Janssen B, et al. Transcriptional and biochemical signatures of divergence in natural populations of two species of New Zealand alpine Pachycladon. Molecular Ecology. 2008;17:4740–4753. doi: 10.1111/j.1365-294X.2008.03933.x. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne ML, Pan YJ, Nuzhdin SV, McIntyre LM. Additivity and trans-acting effects on gene expression in male Drosophila simulans. Genetics. 2004;168:1413–1420. doi: 10.1534/genetics.104.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Variation within and among species in gene expression: raw material for evolution. Molecular Ecology. 2006;15:1197–1211. doi: 10.1111/j.1365-294X.2006.02868.x. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Parent-of-origin effects on mRNA expression in Drosophila melanogaster not caused by genomic imprinting. Genetics. 2006;173:1817–1821. doi: 10.1534/genetics.105.054684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Independent effects of cis- and trans-regulatory variation on gene expression in Drosophila melanogaster. Genetics. 2008a;178:1831–1835. doi: 10.1534/genetics.107.082032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nature Genetics. 2008b;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- Wolf JBW, Bayer T, Haubold B, Schilhabel M, Rosenstiel P, Tautz D. Nucleotide divergence versus gene expression differentiation: comparative transcriptome sequencing in natural isolates from the carrion crow and its hybrid zone with the hooded crow. Molecular Ecology. 2010;19 (Suppl 1):162–175. doi: 10.1111/j.1365-294X.2009.04471.x. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.