Abstract
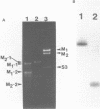
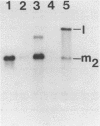
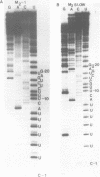
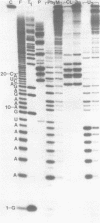
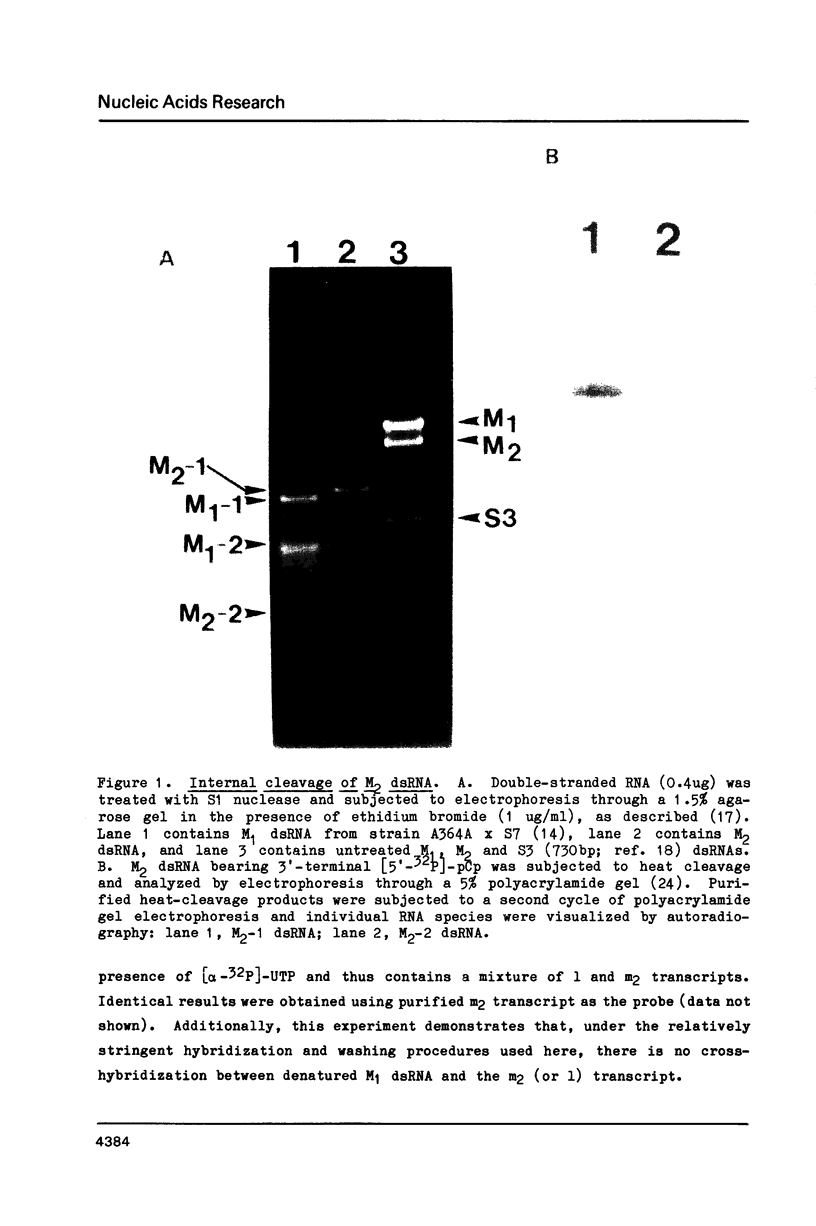
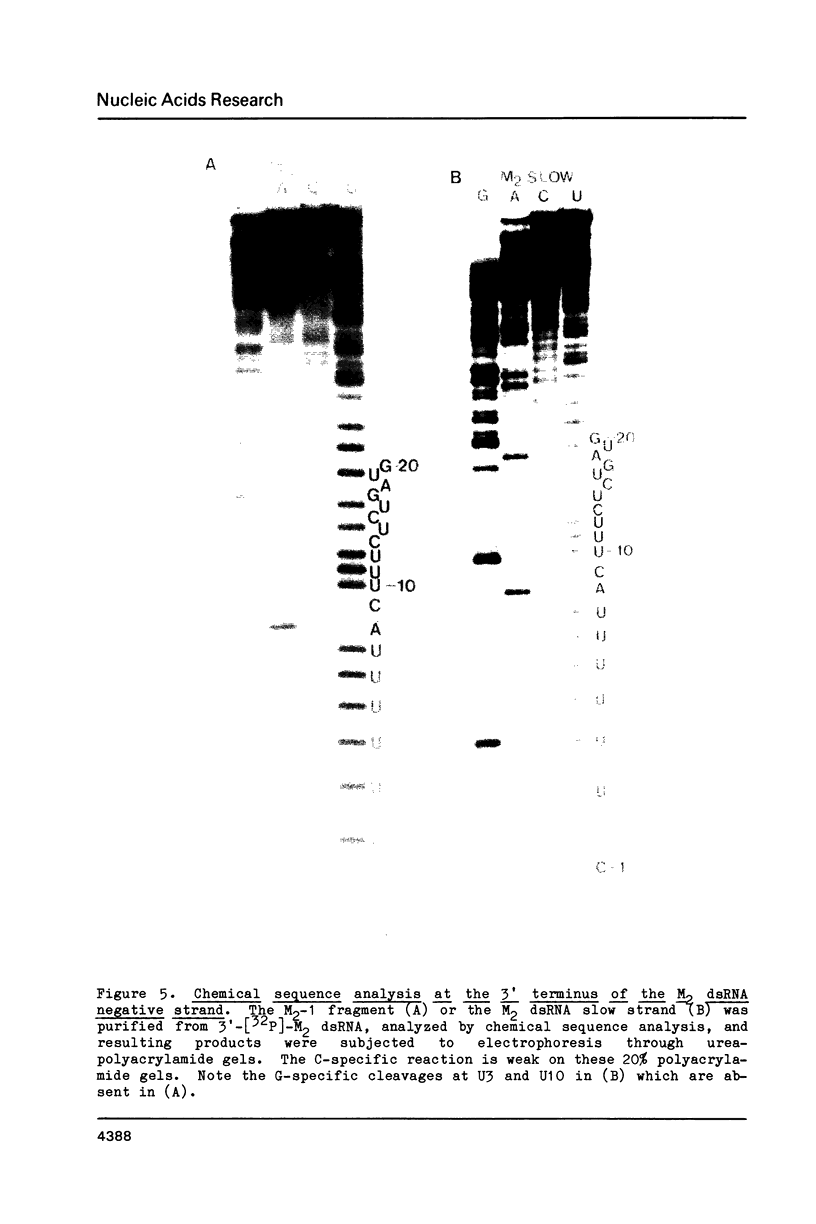
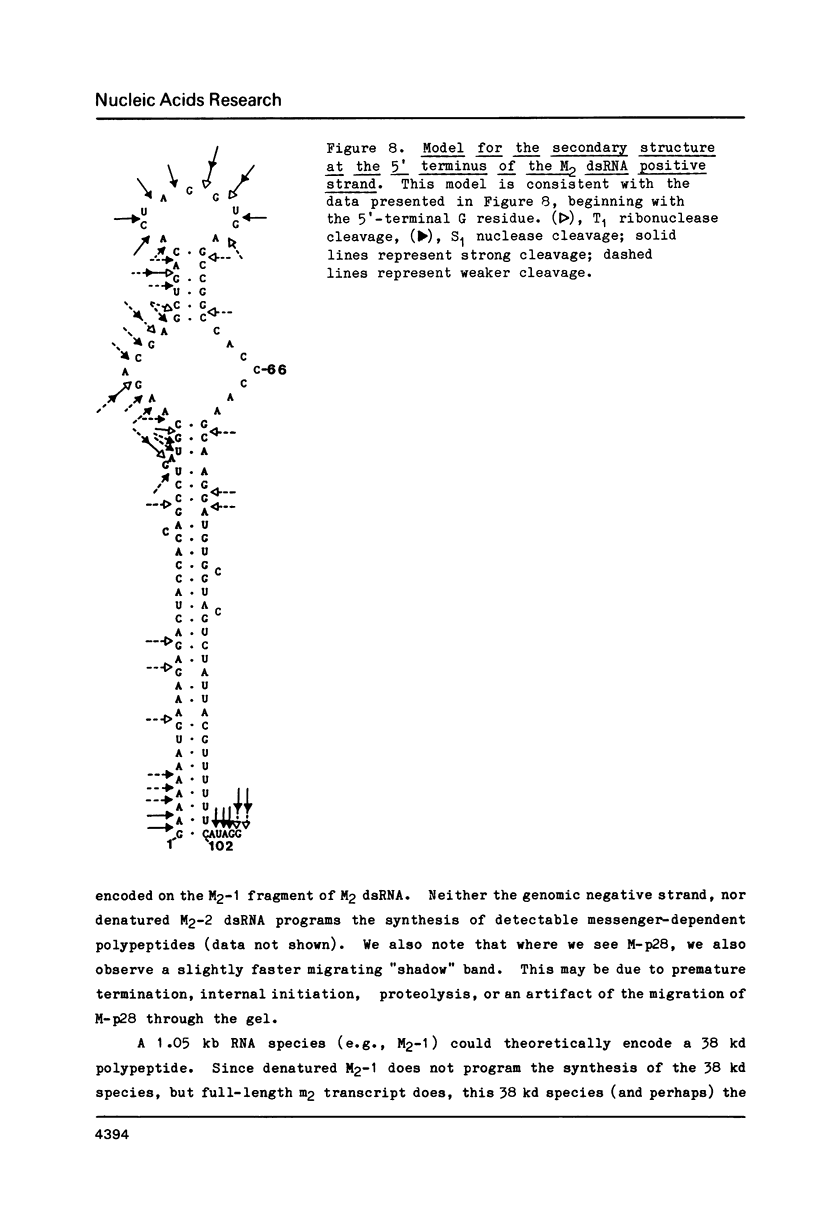
The M2 double-stranded (ds) RNA species encodes toxin and resistance functions in Saccharomyces cerevisiae strains with the K2 killer specificity. RNA sequence analysis reveals the presence of a large open reading frame on the larger heat-cleavage product of M2 dsRNA, which is translated in vitro to yield a 28 kd polypeptide as a major product. The postulated translation initiator AUG triplet is located within a stem and loop structure near the 5' terminus of the positive strand, which also contains plausible 18S and 5.8S ribosomal RNA binding sites. These features may serve to regulate the translation of the K2 toxin precursor. The M1 (from type 1 yeast killers) and M2 dsRNA species lack extensive sequence homology, although specific features are shared, which may represent structural elements required for gene expression and replication.
Full text
PDF
















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. N., McAllister W. T. Mapping of promoter sites utilized by T3 RNA polymerase on T3 DNA. Nucleic Acids Res. 1980 Nov 11;8(21):5071–5088. doi: 10.1093/nar/8.21.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball S. G., Tirtiaux C., Wickner R. B. Genetic Control of L-a and L-(Bc) Dsrna Copy Number in Killer Systems of SACCHAROMYCES CEREVISIAE. Genetics. 1984 Jun;107(2):199–217. doi: 10.1093/genetics/107.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bostian K. A., Burn V. E., Jayachandran S., Tipper D. J. Yeast killer dsRNA plasmids are transcribed in vivo to produce full and partial-length plus-stranded RNAs. Nucleic Acids Res. 1983 Feb 25;11(4):1077–1097. doi: 10.1093/nar/11.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostian K. A., Elliott Q., Bussey H., Burn V., Smith A., Tipper D. J. Sequence of the preprotoxin dsRNA gene of type I killer yeast: multiple processing events produce a two-component toxin. Cell. 1984 Mar;36(3):741–751. doi: 10.1016/0092-8674(84)90354-4. [DOI] [PubMed] [Google Scholar]
- Bostian K. A., Hopper J. E., Rogers D. T., Tipper D. J. Translational analysis of the killer-associated virus-like particle dsRNA genome of S. cerevisiae: M dsRNA encodes toxin. Cell. 1980 Feb;19(2):403–414. doi: 10.1016/0092-8674(80)90514-0. [DOI] [PubMed] [Google Scholar]
- Bostian K. A., Jayachandran S., Tipper D. J. A glycosylated protoxin in killer yeast: models for its structure and maturation. Cell. 1983 Jan;32(1):169–180. doi: 10.1016/0092-8674(83)90507-x. [DOI] [PubMed] [Google Scholar]
- Bostian K. A., Sturgeon J. A., Tipper D. J. Encapsidation of yeast killer double-stranded ribonucleic acids: dependence of M on L. J Bacteriol. 1980 Jul;143(1):463–470. doi: 10.1128/jb.143.1.463-470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan V. E., Field L., Cizdziel P., Bruenn J. A. Sequences at the 3' ends of yeast viral dsRNAs: proposed transcriptase and replicase initiation sites. Nucleic Acids Res. 1981 Aug 25;9(16):4007–4021. doi: 10.1093/nar/9.16.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn J., Bobek L., Brennan V., Held W. Yeast viral RNA polymerase is a transcriptase. Nucleic Acids Res. 1980 Jul 11;8(13):2985–2997. doi: 10.1093/nar/8.13.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey H. Physiology of killer factor in yeast. Adv Microb Physiol. 1981;22:93–122. doi: 10.1016/s0065-2911(08)60326-4. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Fink G. R. Electron microscopic heteroduplex analysis of "killer" double-stranded RNA species from yeast. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4224–4228. doi: 10.1073/pnas.75.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gordon K. H., Symons R. H. Satellite RNA of cucumber mosaic virus forms a secondary structure with partial 3'-terminal homology to genomal RNAs. Nucleic Acids Res. 1983 Feb 25;11(4):947–960. doi: 10.1093/nar/11.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K., Cruz P., Tinoco I., Jr, Jovin T. M., van de Sande J. H. 'Z-RNA'--a left-handed RNA double helix. Nature. 1984 Oct 11;311(5986):584–586. doi: 10.1038/311584a0. [DOI] [PubMed] [Google Scholar]
- Hannig E. M., Thiele D. J., Leibowitz M. J. Saccharomyces cerevisiae killer virus transcripts contain template-coded polyadenylate tracts. Mol Cell Biol. 1984 Jan;4(1):101–109. doi: 10.1128/mcb.4.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzeman R. A., Hagie F. E., Levine H. L., Goeddel D. V., Ammerer G., Hall B. D. Expression of a human gene for interferon in yeast. Nature. 1981 Oct 29;293(5835):717–722. doi: 10.1038/293717a0. [DOI] [PubMed] [Google Scholar]
- Hopper J. E., Bostian K. A., Rowe L. B., Tipper D. J. Translation of the L-species dsRNA genome of the killer-associated virus-like particles of Saccharomyces cerevisiae. J Biol Chem. 1977 Dec 25;252(24):9010–9017. [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Lolle S., Skipper N., Bussey H., Thomas D. Y. The expression of cDNA clones of yeast M1 double-stranded RNA in yeast confers both killer and immunity phenotypes. EMBO J. 1984 Jun;3(6):1383–1387. doi: 10.1002/j.1460-2075.1984.tb01981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumova G. I., Naumova T. I. Sravitel'naia genetika drozhzhei. Soobshchenie XIII. Sravitel'noe izuchenie sakharomitsetov-ubiits iz razlichnykh kollektsii. Genetika. 1973 Nov;9(11):140–145. [PubMed] [Google Scholar]
- Pavlakis G. N., Lockard R. E., Vamvakopoulos N., Rieser L., RajBhandary U. L., Vournakis J. N. Secondary structure of mouse and rabbit alpha- and beta-globin mRNAs: differential accessibility of alpha and beta initiator AUG codons towards nucleases. Cell. 1980 Jan;19(1):91–102. doi: 10.1016/0092-8674(80)90391-8. [DOI] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Ridley S. P., Sommer S. S., Wickner R. B. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol. 1984 Apr;4(4):761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoncsits A., Brownlee G. G., Brown R. S., Rubin J. R., Guilley H. New rapid gel sequencing method for RNA. Nature. 1977 Oct 27;269(5631):833–836. doi: 10.1038/269833a0. [DOI] [PubMed] [Google Scholar]
- Skipper N., Thomas D. Y., Lau P. C. Cloning and sequencing of the preprotoxin-coding region of the yeast M1 double-stranded RNA. EMBO J. 1984 Jan;3(1):107–111. doi: 10.1002/j.1460-2075.1984.tb01769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. S., Wickner R. B. Co-curing of plasmids affecting killer double-stranded RNAs of Saccharomyces cerevisiae: [HOK], [NEX], and the abundance of L are related and further evidence that M1 requires L. J Bacteriol. 1982 May;150(2):545–551. doi: 10.1128/jb.150.2.545-551.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. S., Wickner R. B. Double-stranded RNAs that encode killer toxins in Saccharomyces cerevisiae: unstable size of M double-stranded RNA and inhibition of M2 replication by M1. Mol Cell Biol. 1984 Sep;4(9):1747–1753. doi: 10.1128/mcb.4.9.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. S., Wickner R. B. Yeast L dsRNA consists of at least three distinct RNAs; evidence that the non-Mendelian genes [HOK], [NEX] and [EXL] are on one of these dsRNAs. Cell. 1982 Dec;31(2 Pt 1):429–441. doi: 10.1016/0092-8674(82)90136-2. [DOI] [PubMed] [Google Scholar]
- Thiele D. J., Hannig E. M., Leibowitz M. J. Genome structure and expression of a defective interfering mutant of the killer virus of yeast. Virology. 1984 Aug;137(1):20–31. doi: 10.1016/0042-6822(84)90004-7. [DOI] [PubMed] [Google Scholar]
- Thiele D. J., Hannig E. M., Leibowitz M. J. Multiple L double-stranded RNA species of Saccharomyces cerevisiae: evidence for separate encapsidation. Mol Cell Biol. 1984 Jan;4(1):92–100. doi: 10.1128/mcb.4.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele D. J., Leibowitz M. J. Structural and functional analysis of separated strands of killer double-stranded RNA of yeast. Nucleic Acids Res. 1982 Nov 11;10(21):6903–6918. doi: 10.1093/nar/10.21.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele D. J., Wang R. W., Leibowitz M. J. Separation and sequence of the 3' termini of M double-stranded RNA from killer yeast. Nucleic Acids Res. 1982 Mar 11;10(5):1661–1678. doi: 10.1093/nar/10.5.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tipper D. J., Bostian K. A. Double-stranded ribonucleic acid killer systems in yeasts. Microbiol Rev. 1984 Jun;48(2):125–156. doi: 10.1128/mr.48.2.125-156.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi A. K., Wickner R. B., Tabor C. W., Tabor H. Specificity of polyamine requirements for the replication and maintenance of different double-stranded RNA plasmids in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1149–1153. doi: 10.1073/pnas.81.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert G., Fiers W. A micromethod for base analysis of 32P-labeled oligoribonulcleotides. Anal Biochem. 1977 Nov;83(1):222–227. doi: 10.1016/0003-2697(77)90530-9. [DOI] [PubMed] [Google Scholar]
- Welsh D., Leibowitz M. J. Transcription of killer virion double-stranded RNA in vitro. Nucleic Acids Res. 1980 Jun 11;8(11):2365–2375. doi: 10.1093/nar/8.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J. D., Leibowitz M. J. Localization of genes for the double-stranded RNA killer virus of yeast. Proc Natl Acad Sci U S A. 1982 Feb;79(3):786–789. doi: 10.1073/pnas.79.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J. D., Leibowitz M. J., Wickner R. B. Virion DNA-independent RNA polymerase from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Jun 11;8(11):2349–2363. doi: 10.1093/nar/8.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Genetic control of replication of the double-stranded RNA segments of the killer systems in Saccharomyces cerevisiae. Arch Biochem Biophys. 1983 Apr 1;222(1):1–11. doi: 10.1016/0003-9861(83)90496-4. [DOI] [PubMed] [Google Scholar]
- Wickner R. B. Killer systems in Saccharomyces cerevisiae: three distinct modes of exclusion of M2 double-stranded RNA by three species of double-stranded RNA, M1, L-A-E, and L-A-HN. Mol Cell Biol. 1983 Apr;3(4):654–661. doi: 10.1128/mcb.3.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B. Plasmids controlled exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell. 1980 Aug;21(1):217–226. doi: 10.1016/0092-8674(80)90129-4. [DOI] [PubMed] [Google Scholar]
- Wickner R. B., Toh-e A. [HOK], a new yeast non-Mendelian trait, enables a replication-defective killer plasmid to be maintained. Genetics. 1982 Feb;100(2):159–174. doi: 10.1093/genetics/100.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]