Abstract
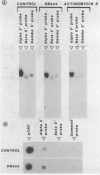
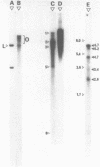
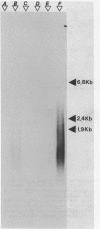
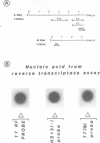
Sub-cellular fractions, isolated from cauliflower mosaic virus (CaMV)-infected turnip protoplasts, are capable of synthesising CaMV DNA in vitro on an endogenous template and of reverse transcribing oligo dT-primed cowpea mosaic virus RNA. The activity was not detected in mock-inoculated protoplasts. In vitro-labelled DNA hybridized to single-stranded M13 clones complementary to the putative origins of (-) and (+) strand CaMV DNA synthesis and to restriction endonuclease fragments encompassing more than 90% of the CaMV genome. The synthesis of (-) and (+) strand DNA appeared asymmetric. The template(s) for in vitro CaMV DNA synthesis are in a partially nuclease-resistant form. Fractions capable of in vitro CaMV DNA synthesis contained CaMV RNA both heterogeneous and as discrete species; they also contained a range of different sizes of CaMV DNA. Several lines of evidence indicate that this range of in vitro-labelled CaMV DNA, extending from 0.6kb to 8.0kb in length, represents elongating (-) strand DNA. These are discussed in relation to their role as possible replicative intermediates.
Full text
PDF




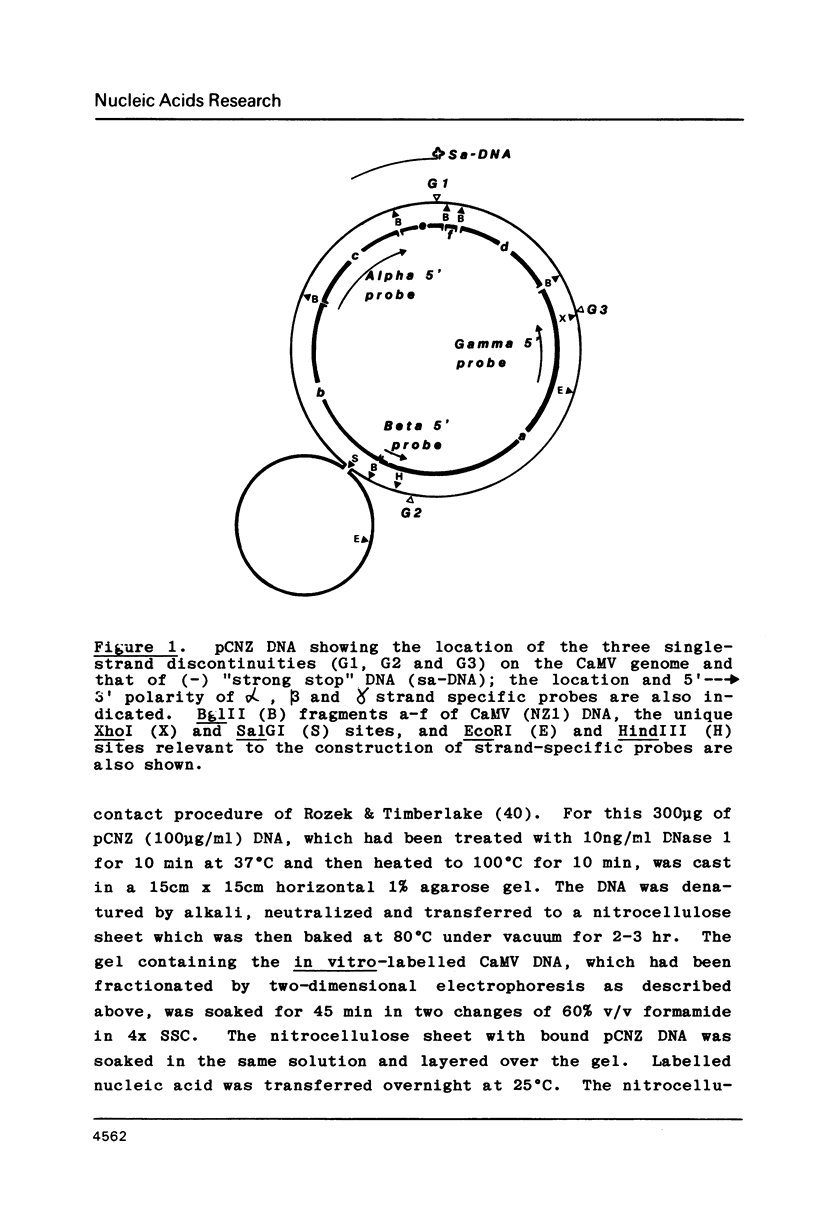
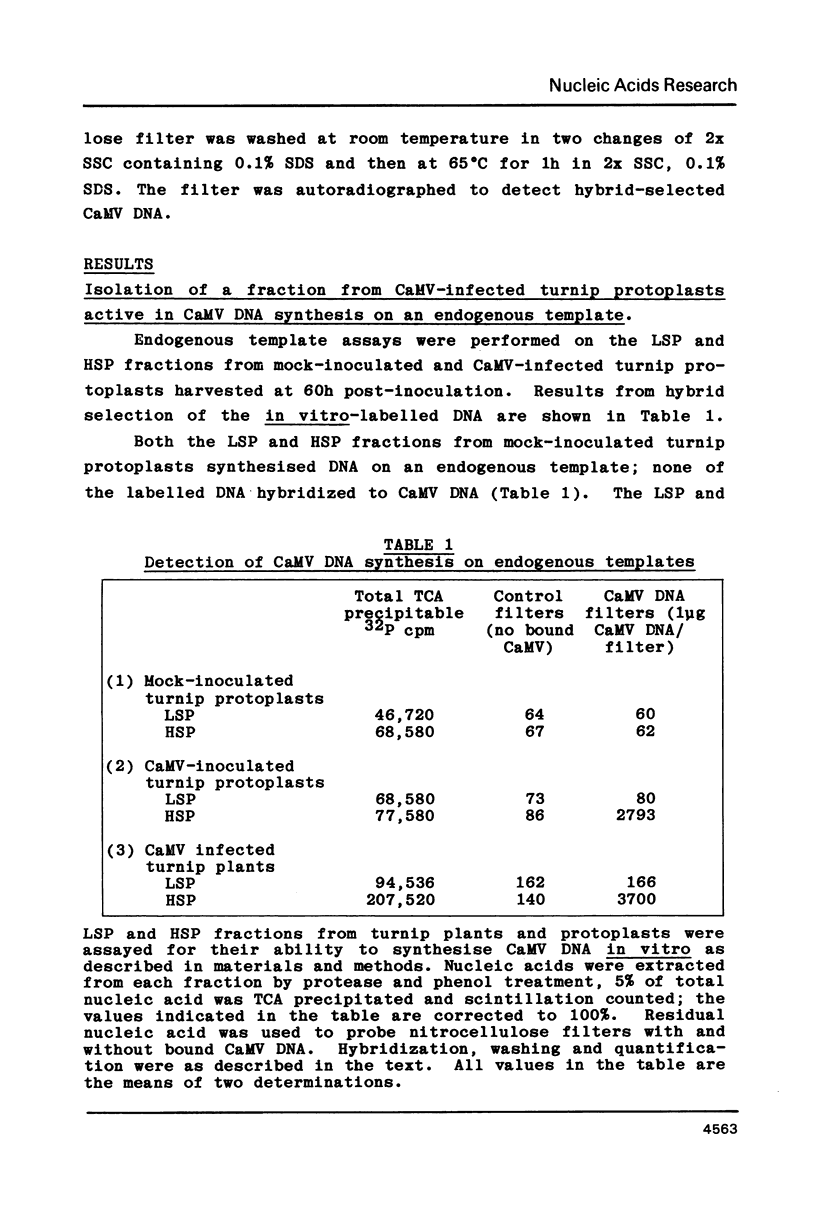

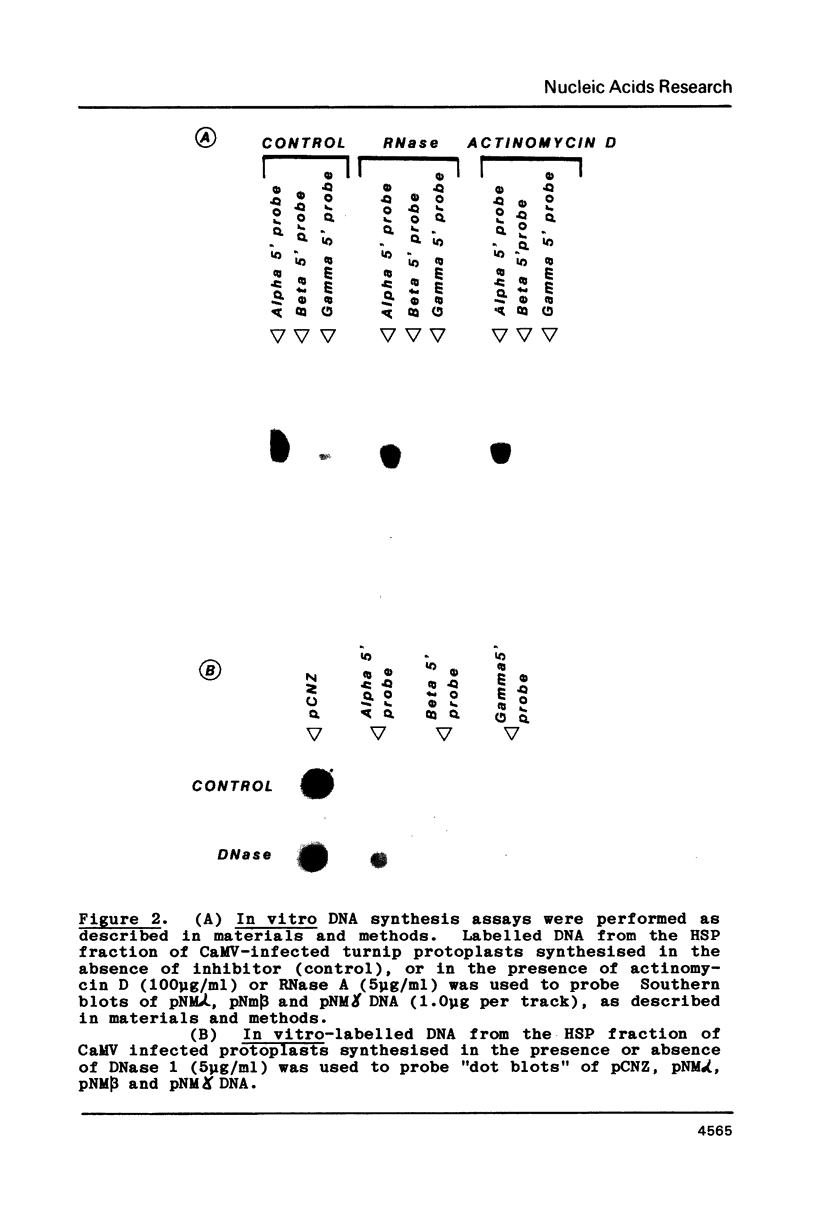


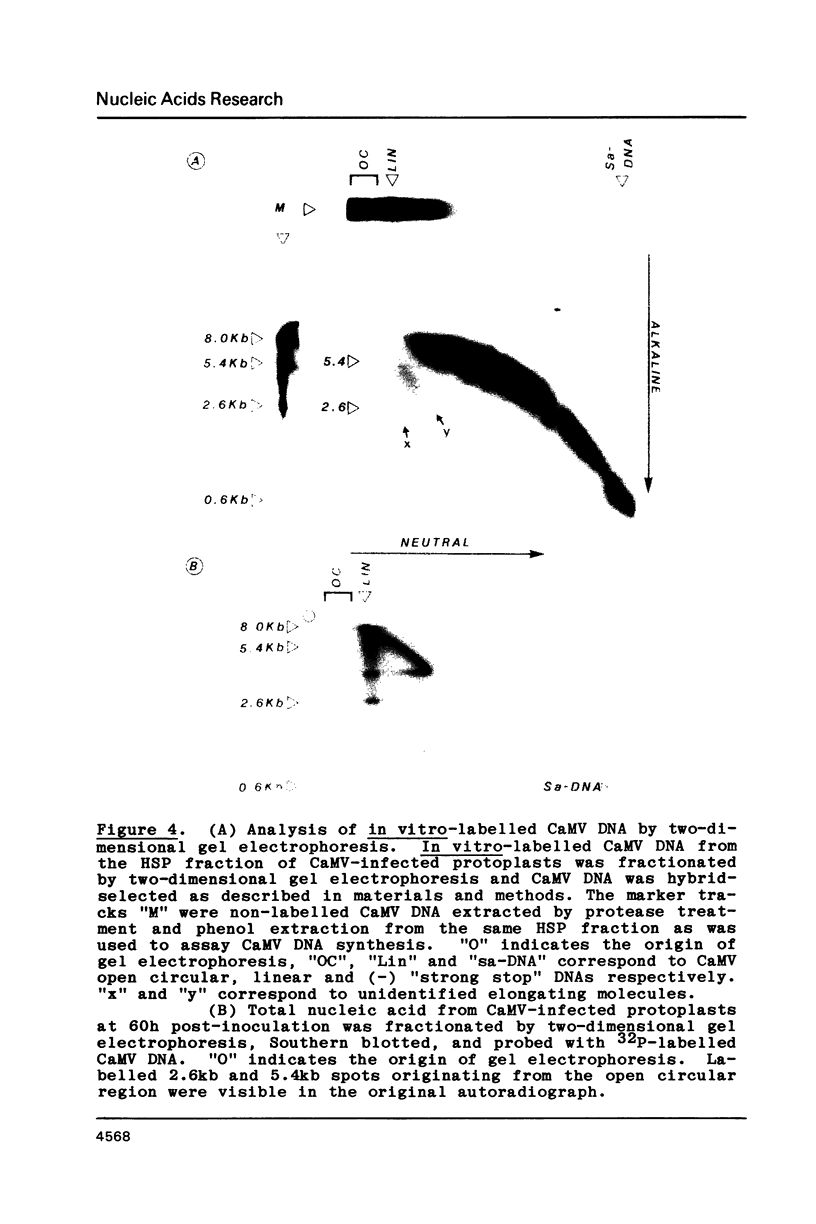

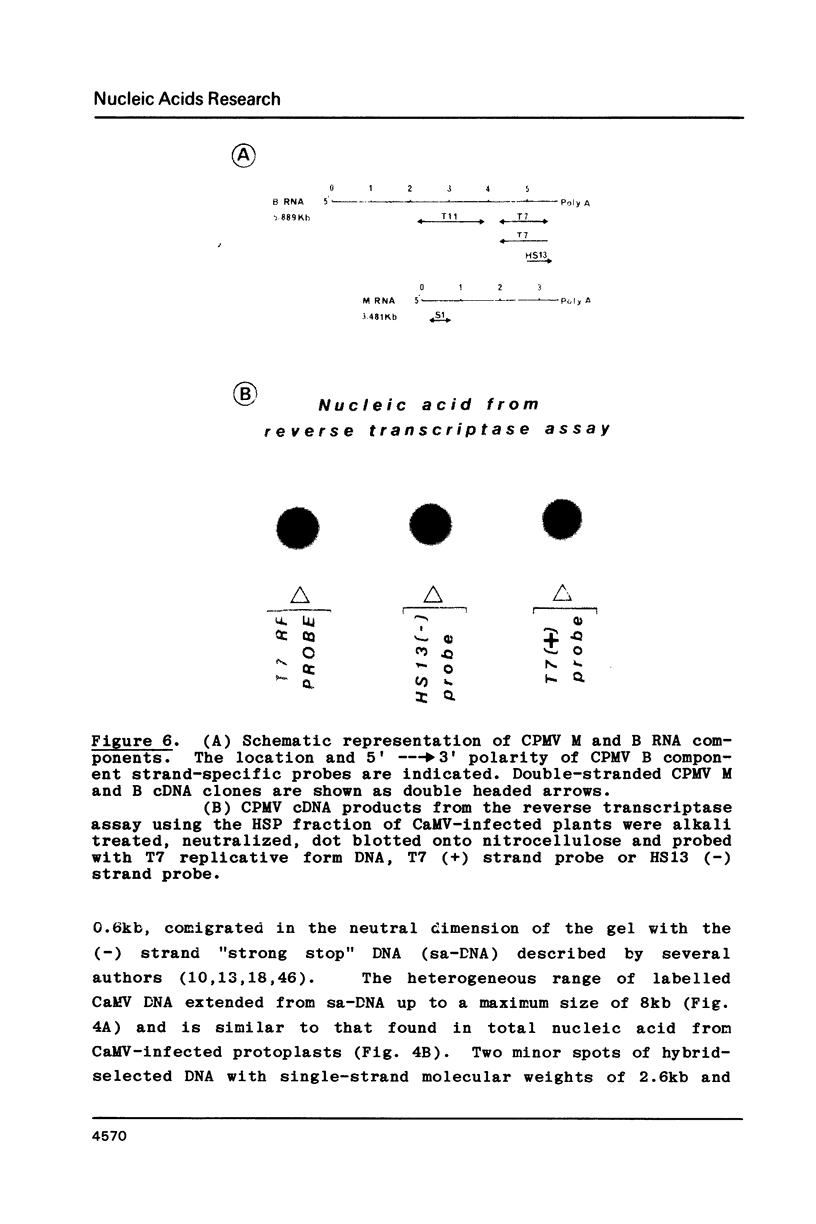






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balàzs E., Guilley H., Jonard G., Richards K. Nucleotide sequence of DNA from an altered-virulence isolate D/H of the cauliflower mosaic virus. Gene. 1982 Oct;19(3):239–249. doi: 10.1016/0378-1119(82)90013-0. [DOI] [PubMed] [Google Scholar]
- Blum H. E., Haase A. T., Harris J. D., Walker D., Vyas G. N. Asymmetric replication of hepatitis B virus DNA in human liver: demonstration of cytoplasmic minus-strand DNA by blot analyses and in situ hybridization. Virology. 1984 Nov;139(1):87–96. doi: 10.1016/0042-6822(84)90332-5. [DOI] [PubMed] [Google Scholar]
- Brown D. M., Frampton J., Goelet P., Karn J. Sensitive detection of RNA using strand-specific M13 probes. Gene. 1982 Dec;20(2):139–144. doi: 10.1016/0378-1119(82)90032-4. [DOI] [PubMed] [Google Scholar]
- Condit C., Hagen T. J., McKnight T. D., Meagher R. B. Characterization and preliminary mapping of cauliflower mosaic virus transcripts. Gene. 1983 Nov;25(1):101–108. doi: 10.1016/0378-1119(83)90172-5. [DOI] [PubMed] [Google Scholar]
- Covey S. N., Turner D., Mulder G. A small DNA molecule containing covalently-linked ribonucleotides originates from the large intergenic region of the cauliflower mosaic virus genome. Nucleic Acids Res. 1983 Jan 25;11(2):251–264. doi: 10.1093/nar/11.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favali M. A., Bassi M., Conti G. G. A quantitative autoradiographic study of intracellular sites for replication of cauliflower mosaic virus. Virology. 1973 May;53(1):115–119. doi: 10.1016/0042-6822(73)90470-4. [DOI] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Franck A., Guilley H., Jonard G., Richards K., Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980 Aug;21(1):285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Gardner R. C., Howarth A. J., Hahn P., Brown-Luedi M., Shepherd R. J., Messing J. The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucleic Acids Res. 1981 Jun 25;9(12):2871–2888. doi: 10.1093/nar/9.12.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilley H., Dudley R. K., Jonard G., Balàzs E., Richards K. E. Transcription of Cauliflower mosaic virus DNA: detection of promoter sequences, and characterization of transcripts. Cell. 1982 Oct;30(3):763–773. doi: 10.1016/0092-8674(82)90281-1. [DOI] [PubMed] [Google Scholar]
- Guilley H., Richards K. E., Jonard G. Observations concerning the discontinuous DNAs of cauliflower mosaic virus. EMBO J. 1983;2(2):277–282. doi: 10.1002/j.1460-2075.1983.tb01417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn T., Richards K., Geneviève-Lebeurier Cauliflower mosaic virus on its way to becoming a useful plant vector. Curr Top Microbiol Immunol. 1982;96:194–236. [PubMed] [Google Scholar]
- Howell S. H., Hull R. Replication of cauliflower mosaic virus and transcription of its genome in turnip leaf protoplasts. Virology. 1978 May 15;86(2):468–481. doi: 10.1016/0042-6822(78)90086-7. [DOI] [PubMed] [Google Scholar]
- Hull R., Covey S. N. Characterisation of cauliflower mosaic virus DNA forms isolated from infected turnip leaves. Nucleic Acids Res. 1983 Mar 25;11(6):1881–1895. doi: 10.1093/nar/11.6.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R., Howell S. H. Structure of the cauliflower mosaic virus genome. II. Variation in DNA structure and sequence between isolates. Virology. 1978 May 15;86(2):482–493. doi: 10.1016/0042-6822(78)90087-9. [DOI] [PubMed] [Google Scholar]
- Hull R., Shepherd R. J. The structure of cauliflower mosaic virus genome. Virology. 1977 Jun 1;79(1):216–230. doi: 10.1016/0042-6822(77)90346-4. [DOI] [PubMed] [Google Scholar]
- Kamei T., Rubio-Huertos M., Matsui C. Thymidine-3H uptake by X-bodies associated with cauliflower mosaic virus infection. Virology. 1969 Mar;37(3):506–508. doi: 10.1016/0042-6822(69)90241-4. [DOI] [PubMed] [Google Scholar]
- Lomonossoff G. P., Shanks M. The nucleotide sequence of cowpea mosaic virus B RNA. EMBO J. 1983;2(12):2253–2258. doi: 10.1002/j.1460-2075.1983.tb01731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco Y., Howell S. H. Intracellular forms of viral DNA consistent with a model of reverse transcriptional replication of the cauliflower mosaic virus genome. Nucleic Acids Res. 1984 Feb 10;12(3):1517–1528. doi: 10.1093/nar/12.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason W. S., Aldrich C., Summers J., Taylor J. M. Asymmetric replication of duck hepatitis B virus DNA in liver cells: Free minus-strand DNA. Proc Natl Acad Sci U S A. 1982 Jul;79(13):3997–4001. doi: 10.1073/pnas.79.13.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule A. J., Hull R., Donson J. The application of spot hybridization to the detection of DNA and RNA viruses in plant tissues. J Virol Methods. 1983 Apr;6(4):215–224. doi: 10.1016/0166-0934(83)90048-4. [DOI] [PubMed] [Google Scholar]
- Menissier J., Laquel P., Lebeurier G., Hirth L. A DNA polymerase activity is associated with Cauliflower Mosaic Virus. Nucleic Acids Res. 1984 Dec 11;12(23):8769–8778. doi: 10.1093/nar/12.23.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miller R. H., Marion P. L., Robinson W. S. Hepatitis B viral DNA-RNA hybrid molecules in particles from infected liver are converted to viral DNA molecules during an endogenous DNA polymerase reaction. Virology. 1984 Nov;139(1):64–72. doi: 10.1016/0042-6822(84)90330-1. [DOI] [PubMed] [Google Scholar]
- Ménissier J., de Murcia G., Lebeurier G., Hirth L. Electron microscopic studies of the different topological forms of the cauliflower mosaic virus DNA: knotted encapsidated DNA and nuclear minichromosome. EMBO J. 1983;2(7):1067–1071. doi: 10.1002/j.1460-2075.1983.tb01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P., Hohn T. Involvement of reverse transcription in the replication of cauliflower mosaic virus: a detailed model and test of some aspects. Cell. 1983 Jul;33(3):781–789. doi: 10.1016/0092-8674(83)90020-x. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rozek C. E., Timberlake W. E. Restriction endonuclease mapping by crossed contact hybridization: the ribosomal RNA genes of Achlya ambisexualis. Nucleic Acids Res. 1979 Nov 24;7(6):1567–1578. doi: 10.1093/nar/7.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Summers J., Mason W. S. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982 Jun;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Volovitch M., Modjtahedi N., Yot P., Brun G. RNA-dependent DNA polymerase activity in cauliflower mosaic virus-infected plant leaves. EMBO J. 1984 Feb;3(2):309–314. doi: 10.1002/j.1460-2075.1984.tb01801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]