Abstract
The Keap1-Nrf2 interaction plays important roles in regulation of Nrf2 activity and induction of chemopreventive enzymes. To better understand the interaction and to determine the minimal Nrf2 sequence required for Keap1 binding, we synthesized a series of Nrf2 peptides containing ETGE motif and determined their binding affinities to the Kelch domain of Keap1 in solution using a surface plasmon resonance (SPR)-based competition assay. The equilibrium dissociation constant for the interaction between 16mer Nrf2 peptide and Keap1 Kelch domain in solution (KDsolution) was found to be 23.9 nM, which is 10× lower than the surface binding constant (KDsurface) of 252 nM obtained for the direct binding of Keap1 Kelch domain to the immobilized 16mer Nrf2 peptide on a SPR sensor chip surface. The binding affinity of Nrf2 peptides to Keap1 Kelch domain was not lost until after deletion of 8 residues from the N-terminus of the 16mer Nrf2 peptide. The 9mer Nrf2 peptide has a moderate binding affinity with a KDsolution of 352 nM and the affinity was increased 15× upon removal of the positive charge at the peptide N-terminus by acetylation. These results suggest that the minimal Nrf2 peptide sequence required for Keap1 binding is the 9mer sequence of LDEETGEFL.
Keywords: Keap1, Nrf2, protein-protein interaction, surface plasmon resonance, solution binding assay
The human body has developed intrinsic protection mechanisms to upregulate a number of antioxidative and cytoprotective enzymes in response to oxidative stresses.1–3 These enzymes include not only the enzymes involved in phase 2 biotransformation reactions such as glutathione S-transferases (GST)4 and UDP-glucuronyl transferase5 but also several enzymes that catalyze reactions in the phase 1 biotransformation reactions such as NAD(P)H:quinone oxidoreductase 1 (NQO1)3 and epoxide hydrolase6 as well as enzymes with anticarcinogenic and antioxidant activities such as glutamate cysteine ligase,7 thioredoxin,4 catalase,8 superoxide dismutase,8 glutathione reductase,8 and heme oxygenase 1 (HO1).9 Induction of these enzymes by a variety of chemical agents was shown to protect cells against mutagenesis, oxidative stress, and inflammation and is believed to be an effective approach to prevent carcinogenesis.10,11 The regulation of the enzymes’ expression is largely controlled by Keap1-Nrf2-ARE system. The antioxidant response element (ARE) is a cis-acting, upstream regulatory transcriptional enhancer sequence commonly present on these enzyme genes in single or multiple copies.12 Nuclear factor erythroid 2 (Nf-E2)-related factor 2 (Nrf2) is the principal transcription factor in the nucleus for transactivation of enzyme expression through binding to ARE element,13 while Kelch ECH-associated protein 1 (Keap1) is a cytosolic repressor protein that binds to Nrf2 and inhibits Nrf2 transcriptional activity.14
Nrf2 is a 66 kDa protein originally found to interact with Nf-E2 DNA binding motif;15 both Nrf2 and Nf-E2 belong to basic leucine zipper-type (bZIP) Cap n′ Collar (CNC) protein family. Six highly conserved Nrf2-ECH homologous domains (Neh1–6) were identified in Nrf2 across different species.14 As a bZIP CNC protein, Neh1 domain has a commonly shared C-terminal basic zipper that is required for binding to DNA and dimerization with other transcriptional factors,16 along with a nuclear localization signal (NLS) region known to facilitate its localization from cytoplasm to nucleus.17 The Neh2 domain at Nrf2 N-terminus binds to the cytoplasmic Keap1 protein and is responsible for ubiquitin conjugation, which leads to proteasomal degradation of Nrf2.14,18 The C-terminal Neh3 and a large central domain comprising Neh4 and Neh5 recruit transcriptional co-activators to ARE motifs in the promoter regions of ARE genes.19,20
Keap1 contains two conserved domains, namely, BTB domain and Kelch domain, which are shared with other BTB-Kelch proteins. The N-terminal broad complex, tramtrack, bric-a-brac (BTB) domain is a protein-protein interaction motif required for homodimerization21 and interaction with Cullin3,18 while the C-terminal Kelch domain is comprised of six Kelch repeats that form the β-propeller structure and binds to the Neh2 domain of Nrf2.22 Through these two major domains, Keap1 functions as a substrate adaptor protein that brings Nrf2 to Keap1-Cullin3 based E3-ligase ubiquitination complex for ubiquitination and subsequent proteasomal degradation.18,23 Keap1 also contains a cysteine-rich linker region between BTB and Kelch domains which is believed to serve as a “sensor” regulating the induction of cytoprotective enzymes against carcinogens and oxidants.24,25
Under basal conditions, Nrf2 is constantly targeted by Keap1 for ubiquination and proteasomal degradation and is, thus, kept in the cytoplasm and maintained at low levels; under stressful conditions such as oxidative stress and inflammation or by external induction, Nrf2 is liberated from Keap1-dependent degradation and translocated into the nucleus to transcriptionally activate downstream ARE genes.18,23,26,27 Experiments using gene-knockout mouse models have shown that mice with disrupted Nrf2 are more sensitive to toxicological effects of carcinogens, drugs, and inflammatory stresses28 while mice with disrupted Keap1 exhibited high levels of Nrf2, high constitutive expression of cytoprotective enzymes, and greater resistance to environmental stresses.29,30
The Keap1-Nrf2-ARE system has been identified as a key signaling pathway in cellular defense mechanisms protecting cells from carcinogenesis process.30,31 As one of the pivotal interactions for the activation of ARE genes, the binding of Neh2 domain of Nrf2 to the Kelch domain of Keap1 could serve as a potential target for cancer chemoprevention. The interaction was first characterized using surface plasmon resonance (SPR) by Yamamoto’s group.14 In their study, GST-Nrf2 Neh2 domain protein was immobilized on a sensor chip surface coated with anti-GST antibody and the Kelch domain of Keap1 in solution was found to specifically interact with GST-Nrf2 Neh2 domain in a concentration-dependent manner with a calculated KDsurface of 580 nM.14 More recently, binding affinities between Nrf2 and Keap1 in solution were primarily characterized by isothermal titration calorimetry (ITC).32–34 The KD in solution obtained using ITC was in the single digit nanomolar range.34 Furthermore, distinct binding affinities of two evolutionary conserved binding motifs within the Nrf2 Neh2 domain, ETGE motif and DLG motif, have also been revealed by ITC analysis.32 In a “hinge and latch” model, the ETGE motif with a much higher binding affinity (KD = 5×10−9 M) is proposed to function as the “hinge” for Keap1 to recruit any newly synthesized Nrf2, while the DLG motif with a lower binding affinity (KD = 1.0×10−6 M) provides the “latch” that locks the Nrf2 to optimal positioning of its central domain for conjugation with ubiquitin.35–37 Several ETGE-containing Nrf2 peptides have been reported to interrupt the interaction between Nrf2 and Keap1. A 16mer Nrf2 peptide AFFAQLQLDEETGEFL (residues 69–84) and a 14mer Nrf2 peptide LQLDEETGEFLPIQ (residues 74–87) were able to displace Nrf2 from Keap1-Nrf2 complex effectively with comparable KD’s of 20 nM as obtained using ITC.38
The shorter 10mer Nrf2 peptide LDEETGEFLP (amino acids 76–85), however, was shown to be less effective for the displacement.38 To further investigate the interaction between Keap1 and Nrf2, we synthesized and evaluated a series of Nrf2 peptides and determined the minimal Nrf2 peptide sequence required for binding to Keap1. ITC and SPR are two of the most common methods used to measure the binding affinity of biomolecular interactions in solution or on a surface. As a biophysical technique, ITC directly measures the enthalpy and binding constants of the protein-protein interactions in solution and allows the direct examination of stoichiometry of the interactions.39 Since ITC is a pure solution phase measurement, there are no questions about the structural integrity of the proteins and the specificity of the noncovalent complex. The major drawback of ITC measurement is the large amount of sample usually required for a successful measurement. In contrast, as a real-time, label-free technique widely used in characterizing protein-protein interactions, SPR requires much less sample and still provides accurate measurements of binding events. However, the immobilization of one of the two interacting partners on a sensor surface is required for the SPR binding experiments and such immobilization may affect the conformation or activity of macromolecule immobilized and introduce artifacts into the measurements.40 The binding constants, KD, obtained using SPR represent the binding affinity on a sensor surface which might not reflect its true binding affinity in solution. To minimize sample consumption while still accurately measure the true solution binding constant between Keap1 and Nrf2 protein or peptides, we developed a SPR-based solution competition assay to evaluate the binding affinity of Nrf2 protein or peptide to Keap1 Kelch domain and employed this method to determine the minimal Nrf2 peptide sequence required for binding to Keap1.
RESULTS AND DISCUSSION
Peptide design and synthesis
The X-ray crystal structure of human Keap1 Kelch domain in complex with a 16mer Nrf2 peptide (residues 69–84, H-AFFAQLQLDEETGEFL-OH) indicated that the 16mer Nrf2 peptide has two anti-parallel strands connected by two overlapping type I β-turns (residues 77–80 and 78–81) which are stabilized by multiple hydrogen bonds involving the peptide backbone and the side chains of D77 and T80.38 The Nrf2 peptide binds in the shallow pocket defined by loops between the D–A and B–C strands of the β-propellers on the top face of the Kelch domain. Glutamate residues E79 and E82 are the only peptide residues whose side chains make multiple specific interactions with the residues in Kelch domain including R415, R483, S508, S363, N382 and R380. The Nrf2 peptide backbone makes five contacts with the Kelch domain involving E78, E79, T80 and F83.38 The crystal structures of Kelch domain of mouse Keap1 in complex with a 9mer Nrf2 peptide (residues 76–84) and a 16mer Nrf2 peptide (residues 74–89) were shown to be similar.41 As for the 9mer Nrf2 peptide, a tight β-hairpin conformation was observed comprising the residues D77, E78, E79, T80, G81 and E82. The β-hairpin conformation is stabilized by three intramolecular hydrogen bonds between D77 and G81, D77 and E79, T80 and E82, and such conformation allows the Nrf2 peptide to strongly interact with the Kelch domain of Keap1.41
Several ETGE-containing peptides have been reported to bind Keap1 Kelch domain with varying affinity, and these include the 16mer Nrf2 peptide (residues 69–84), 14mer Nrf2 peptide (residues 74–87) and 10mer Nrf2 peptide (residues 76–85).38 All these peptides overlapped with each other on the peptide sequence from residues 76–84. To find out the minimal peptide sequence required to maintain the Keap1 binding affinity, we designed and synthesized a series of Nrf2 peptides based on the 16mer Nrf2 peptide sequence (residues 69–84) as shown in Figure 1 and determined their binding affinity to Keap1 Kelch domain. After we determined that the minimal Nrf2 sequence is the 9mer between residues 76 and 84, we also explored the effect of end-capping on the binding affinity of the 9mer Nrf2 peptide to Keap1 Kelch domain by acetylation of the N-terminus and amidation of the C-terminus. In order to ensure accuracy of data obtained in our SPR experiments, the concentrations of Nrf2 peptides used were determined by amino acid analyses through acid hydrolysis and o-phthaldialdehyde (OPA)/N-Boc-L-Cysteine (NBC) derivatization as described previously.42
Figure 1.
Sequences of the 16mer Nrf2 peptide (residues 69–84) and its N-terminal deletion peptides designed and synthesized, all containing the key binding motif ETGE (A) and the 9mer Nrf2 peptide and its analogues with modification on the N-terminus and the C-terminus (B).
SPR assay development
In surface plasmon resonance (SPR), binding of molecules in solution to the sensor chip surface alters the refractive index of the medium close to the sensor chip surface which can be monitored in real time as changes in SPR signal expressed in response units. The resulting plots of the SPR signal (proportional to the mass bound) against time are referred to as sensorgrams and provide real time information of the interactions. In SPR experiments, one of the interacting components is immobilized on a sensor chip surface and is known as the ligand, while the other interacting component continuously flows over the sensor chip surface and is known as the analyte. The SPR measurement of the interactions between Keap1 Kelch domain and Nrf2 peptides could be performed on Biacore in two possible experimental setups depending on which protein is immobilized on the chip surface. It was reported by Yamamoto’s group that Keap1 Kelch domain has a surface dissociation equilibrium constant (KDsurface) of 5.8 × 10−7 M when the fusion protein of GST and the Neh2 domain of Nrf2 was immobilized on sensor chip surface through anti-GST antibody.14
We first explored the direct immobilization of either a GST-Nrf2 fusion protein or the Keap1 Kelch domain protein to a CM5 chip surface through the standard amine coupling procedure and measured for the surface binding affinity between Keap1 Kelch domain protein and Nrf2 protein or peptide. When the Keap1 Kelch domain protein was immobilized as the ligand, we did obtain concentration dependent SPR sensorgrams of both GST-Nrf2 fusion protein and 16mer Nrf2 peptide (data not shown). Although a relatively large SPR response of 80 RU could be achieved for GST-Nrf2 protein at 100 nM concentration, a limited response of only about 15 RU was achieved for the 16mer Nrf2 peptide at 10 μM concentration. Such low levels of SPR response were not sufficient to differentiate the binding affinity of low molecular weight Nrf2 peptides or small molecule analytes. Therefore, the low SPR response was a major concern when Keap1 Kelch domain was immobilized as the ligand for direct binding analysis. The instability of Keap1 Kelch domain on the sensor chip surface was another problem; during the cycles of sensor surface regeneration, decreases as much as 15% in SPR response were observed from one run to the next (data not shown). Therefore, immobilization of Keap1 Kelch domain on CM5 chip surface was not a viable option for reliable, sensitive measurement of the binding affinity between Keap1 Kelch domain protein and Nrf2 protein or peptide.
We then explored the opposite experimental setup by immobilizing the GST-Nrf2 fusion protein on a CM5 chip surface as the ligand, which could allow solution competition assays to be performed for the determination of solution binding affinity between Keap1 Kelch domain as the analyte and a Nrf2 protein or peptide as the competitor. GST-Nrf2 fusion protein was immobilized as the ligand at a surface density around 1000 RU and was used to explore the kinetics of the binding of Keap1 Kelch domain to surface-immobilized GST-Nrf2 protein. The KDsurface was calculated to be 74 nM by fitting the sensorgrams, though poorly, to a 1:1 Langmuir model (data not shown). Although the SPR response can be increased with the increasing surface density of GST-Nrf2 protein, the biggest problem associated with this experimental setup was the instability of GST-Nrf2 protein immobilized on the sensor chip surface; the GST-Nrf2 protein quickly loses binding affinity to Keap1 Kelch domain from run to run (data not shown).
While peptides are much more stable in solution and on the sensor chip surface than proteins, the binding motif in an immobilized peptide cannot be too close to the chip surface to avoid adversely affecting the binding affinity of the immobilized peptide to its receptors. Thus, the selection of linker length and site of attachment is very important. When we examined the sequence of the 16mer Nrf2 peptide (residues 69–84), we noticed that the N-terminus of the 16mer Nrf2 peptide is ten residues away from the critical ETGE motif, whereas the C-terminus is only two residues away. Furthermore, X-ray crystal structure of the Keap1 Kelch domain protein in complex with the 16mer Nrf2 peptide reveals that six of the N-terminal amino acid residues are neither involved in the stabilization of the β-turn nor involved in the interaction with the binding pocket. In addition, it has been shown that the shorter 14mer Nrf2 peptide (residues 74–87) maintains similar binding affinity as 16mer Nrf2 peptide, indicating some of the N-terminal amino acids are not important for the interaction with Keap1 Kelch domain and can serve as part of the linker for immobilization to sensor chip surface.
Since there is only one free amino group (i.e., the N-terminal amino) in the 16mer Nrf2 peptide, immobilization via standard amine coupling method can be used to attach the peptide onto sensor chip surface. Alternatively, the 16mer Nrf2 peptide can be labeled with biotin at its N-terminus and captured onto a SA sensor chip surface via high affinity biotin/streptavidin interaction. To perform a kinetic binding analysis, it is important that a low ligand density surface is used to avoid avidity and to simplify data analysis.43 For immobilization through streptavidin capture, immobilization levels can be accurately controlled to a few RUs; however, immobilization via standard amine coupling procedure required injections of high refractive index solutions, such as EDC/NHS and ethanolamine solutions which make it difficult to control the immobilization level for the low molecular weight peptide. To compare these two immobilization methods, a low density surface was first prepared and tested in the kinetic analysis of the interaction between the immobilized 16mer Nrf2 peptide as the ligand and Keap1 Kelch domain protein as the analyte.
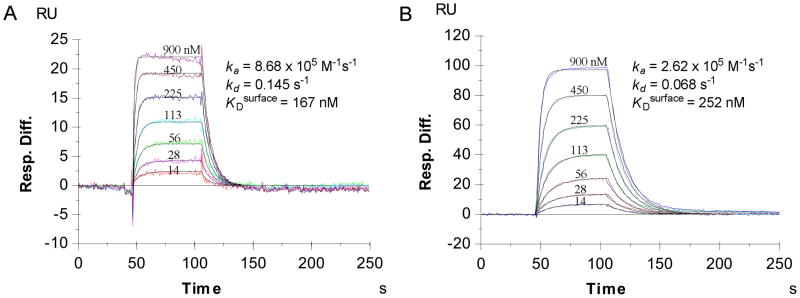
As a result, we were able to achieve 9 RU of biotin-labeled 16mer Nrf2 peptide on the SA chip through biotin-streptavidin complex and 200 RU of 16mer Nrf2 peptide on CM5 chip through direct amine coupling. Figure 2 shows the sensorgrams of the binding of Keap1 Kelch domain to the 16mer Nrf2 peptide immobilized on the sensor surfaces via the two different immobilization methods. Clearly, the smaller amount of biotin-labeled 16mer Nrf2 peptide immobilized on the low density SA chip (9 RU) binds more effectively to Keap1 Kelch domain and achieved about 5-fold higher SPR response than the larger amount of 16mer Nrf2 peptide immobilized on the high density CM5 chip (200 RU) when the same concentrations of Keap1 Kelch domain flowed through the chip surfaces. Apparently, most of the peptide immobilized on the CM5 chip through direct amine coupling was not able to bind to Keap1 Kelch domain. Analysis of the concentration-dependent sensorgrams shown in Figure 2 indicated that the binding between Keap1 Kelch domain and the immobilized 16mer Nrf2 peptides was stoichiometrically 1:1 on both surfaces with similar KDsurface (167 nM on CM5 chip vs 252 nM on SA chip). The affinity of the immobilized 16mer Nrf2 peptide to Keap1 Kelch domain was significantly lower than that obtained by ITC (KDsolution = 20 nM).14 This is not surprising if we consider the effect of surface immobilization on the binding affinity. It should also be noted that the quality of sensorgrams and their fitting to the 1:1 Langmuir binding model shown in Figure 2 are greatly improved as compared to those for the immobilization on CM5 chip surfaces of Keap1 Kelch domain protein and Nrf2 protein.
Figure 2.
Binding kinetics of Kelch domain of Keap1 to 16mer Nrf2 peptide immobilized on CM5 chip using standard amine coupling protocol with a ligand density of 200 RU (A) and biotin-labeled 16mer Nrf2 peptide captured on SA chip with a ligand density of 9 RU (B).
Preparation of high density sensor chip surface with biotin-labeled 16mer Nrf2 peptide and measurement of the solution binding affinity between Keap1 Kelch domain and 16mer Nrf2 peptide. Because the biotin-16mer Nrf2 peptide surface obtained via streptavidin capture binds Keap1 Kelch domain protein more effectively and the amount of biotin-labeled 16mer Nrf2 peptide on the SA chip surface can be accurately controlled and easily elevated by adjusting the ligand concentration or the immobilization time, two high density surfaces of biotin-16mer Nrf2 peptide on a SA chip (30 RU and 300 RU) were prepared and used to measure the concentrations of free Keap1 Kelch domain in solution competition assays. In such solution competition assays, Keap1 Kelch domain protein at a fixed concentration was incubated with varying concentrations of inhibitory Nrf2 peptides and the concentrations of the free (i.e., unbound) Keap1 Kelch domain protein at equilibrium were measured to calculate the binding affinity between Keap1 Kelch domain and the inhibitory Nrf2 peptides in solution.
The chip surface with a ligand density of 30 RU was shown to have 3-fold higher SPR sensitivity than the chip surface with a ligand density of 9 RU for detecting the free Keap1 Kelch domain protein, while the SA chip with a higher ligand density of 300 RU was shown to have around 20× higher SPR sensitivity than the chip surface with a ligand density of 9 RU as shown in Figure 3. No equilibrium could be reached during the 1 min association period on either sensor chip surfaces with higher ligand density; however, sensorgrams obtained from the 30 RU surface can still be fitted properly to a simple 1:1 Langmuir model and the KDsurface calculated was in good agreement with that obtained on the chip surface with a ligand density of 9 RU. Two separate standard curves were obtained for the 30 and 300 RU surfaces by plotting the initial slopes of the association phase against the concentration of Keap1 Kelch domain and both could be used to calculate the free Keap1 Kelch domain concentration in the presence of varying concentrations of inhibitory Nrf2 peptides in solution competition assays. To achieve the highest sensitivity for the detection of free Keap1 Kelch domain protein, the SA chip with the high ligand density of 300 RU was used in solution competition assays to determine the binding affinity of inhibitory Nrf2 peptides to Keap1 Kelch domain protein in solution.
Figure 3.
SPR sensorgrams of the binding of varying concentrations of Kelch domain of Keap1 to biotin-labeled 16mer Nrf2 peptide captured on SA chip with a ligand density of 300 RU (A) and plot of the initial slopes highlighted in red from 5 s to 15 s in the association phase of sensorgrams in A as a function of free Kelch domain of Keap1 concentrations as calibration curve (B).
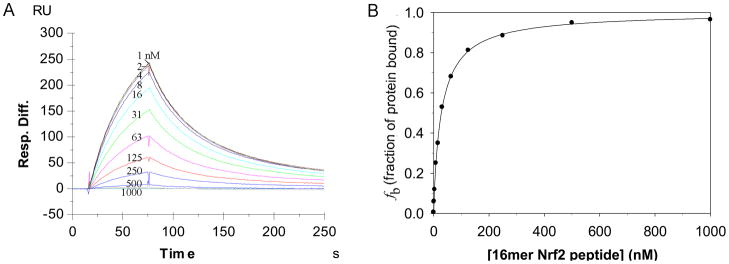
Competition experiments were first performed with a fixed concentration of Keap1 Kelch domain (40 nM) in the presence of varying concentrations of the inhibitory 16mer Nrf2 peptide (1 nM – 1 μM). The KDsolution of the 16mer Nrf2 peptide calculated from the competition study as shown in Figure 4 was 23.9 nM. This KD in solution is in good agreement with the 20 nM KD reported using ITC.38 It should be pointed out that the KDsolution between Keap1 Kelch domain and Nrf2 peptides in SPR competition experiments were calculated based on the assumption of 1:1 binding stoichiometry. Although other scenarios could not be excluded from our SPR competition experiments alone, our assumption of 1:1 binding stoichiometry is reasonable considering that the co-crystal structures of Keap1 Kelch domain with an Nrf2 peptide show only one binding site for Nrf2 peptides38,41 and that previous ITC experiments demonstrated 1:1 binding stoichiometry between Keap1 Kelch domain and Nrf2 peptides.32–34
Figure 4.
SPR sensorgrams of the binding of Keap1 Kelch domain (40 nM) to biotin-labeled 16mer Nrf2 peptide captured on a SA chip with ligand density (~300 RU) in the presence of varying concentrations (1 nM to 1 μM) of 16mer Nrf2 peptide (A) and the binding curve as a plot of the fraction of Keap1 Kelch domain protein bound against the concentration of the inhibitory 16mer Nrf2 peptide was fitted to the quadratic equation to derive the equilibrium dissociation constant between the Keap1 Kelch domain and the inhibitory 16mer Nrf2 peptide (B).
Ranking of inhibitory Nrf2 peptides
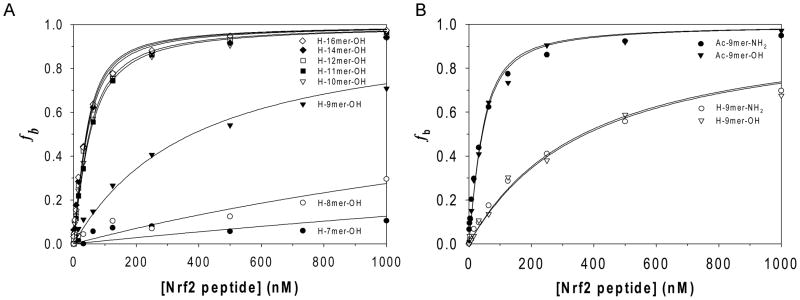
To determine the minimal sequence of Nrf2 peptides required for binding to Keap1 Kelch domain, a series of Nrf2 peptides containing the binding motif ETGE were synthesized and evaluated using the solution competition assay developed. As shown in Figure 5A, 7mer and 8mer Nrf2 peptides showed very low binding affinities to Keap1 Kelch domain with a KDsolution much greater than 1 μM, while 9mer Nrf2 peptide exhibited a moderate affinity to Keap1 Kelch domain with a KDsolution of 352 nM. Nrf2 peptides greater than 9 amino acid residues were shown to have comparable binding affinities with KDsolution ranging from 22 nM to 31 nM considering the errors from the concentration determination using amino acid analyses. These results indicate that the minimal Nrf2 peptide sequence required for binding to Keap1 Kelch domain is the 9mer Nrf2 peptide L76DEETGFEL84 and that at least three amino acid residues (L76, D77 and E78) close to the N-terminal end of ETGE motif are necessary to maintain their high Keap1 binding affinity.
Figure 5.
Binding curves as plots of the fraction of Keap1 Kelch domain protein bound against the concentration of inhibitory Nrf2 peptides. The data were fitted to the quadratic equation to derive the equilibrium dissociation constants between Keap1 Kelch domain and the inhibitory 7mer to 16mer Nrf2 peptides (A) and between Keap1 Kelch domain and the C-terminal/N-terminal modified 9mer Nrf2 peptides (B).
As 9mer Nrf2 peptide showed significantly lower binding affinity than 10mer to 16mer Nrf2 peptide, we postulated that the positive charge of its free amino group at the N-terminus and the negative charge of the carboxylate at the C-terminus could adversely affect its binding to Keap1 Kelch domain. To eliminate these electric charges and better mimic the peptide backbone of Nrf2 protein, we further synthesized the 9mer Nrf2 peptide amide and their corresponding N-acetylated 9mer Nrf2 peptides as shown in Figure 1B. N-Acetylated 9mer Nrf2 peptide acid and amide were found to have a KDsolution of 21 to 23 nM, which is 15× lower than the corresponding non-acetylated 9mer Nrf2 peptides, while C-terminal amidation did not result in any improvement in binding to Keap1 Kelch domain as shown in Figure 5B.
In conclusion, we developed a reliable SPR assay to accurately measure the binding affinity of Nrf2 protein or Nrf2 peptides to Keap1 Kelch domain and we investigated different SPR experimental setups and methods of surface immobilization, and compared surface binding assays to solution competition assays. The instability of Nrf2 and Keap1 Kelch domain proteins immobilized on sensor chip surface was found to be the major problem with protein immobilization. When the 16mer Nrf2 peptide with high binding affinity to Keap1 Kelch domain was immobilized on sensor chip surface as the ligand, the surface was stable over a long period of time and could be used repeatedly to obtain reliable surface as well as solution binding affinity for months without loss of activity. We also demonstrated that immobilization of a biotin-labeled 16mer Nrf2 peptide as the ligand on a SA chip is the optimal immobilization method which provided sensitive and stable sensor chip surfaces for either kinetic analysis of Keap1-Nrf2 peptide interaction or detection of the free Keap1 Kelch domain protein concentration in solution competition assays. Using a SPR competition assay, the unlabeled 16mer Nrf2 peptide was shown to have a KDsolution of 23.9 nM, which is in good agreement with the KD obtained using ITC. Analysis of shorter Nrf2 peptides using the SPR competition assay revealed that 10mer to 14mer Nrf2 peptides had similar binding affinity to Keap1 Kelch domain ranging from 22 nM to 31 nM while the 9mer Nrf2 peptide had moderate binding affinity of 352 nM and Nrf2 peptides less than 9 amino acid residues failed to bind to Keap1 Kelch domain. These results suggest that 9mer Nrf2 peptide containing residues 76–84 is the minimal sequence required for binding to Keap1. In order to improve the binding affinity of 9mer Nrf2 peptide, we then investigated the effect of N-terminal acetylation and C-terminal amidation of 9mer Nrf2 peptide on its binding affinity to Keap1 Kelch domain. As compared to the unmodified 9mer Nrf2 peptide, N-acetylated 9mer Nrf2 peptide acid and amide were shown to have about 15× increase in the binding affinity to Keap1 Kelch domain while no effect was observed from the C-terminus amidation of 9mer Nrf2 peptide. These data demonstrated the adverse effect of the positive charge at the N-terminus of 9mer Nrf2 peptide on its binding affinity to Keap1, suggesting that there exist repulsive interactions between positively charged N-terminus of 9mer Nrf2 peptide and the positively charged bottom surface of the Kelch domain.
In summary, our SPR-based solution competition assay developed can reliably derive solution equilibrium dissociation constants of the interaction between Keap1 Kelch domain and Nrf2 protein or peptides. The 9mer Nrf2 peptide (LDEETGFEL) has been shown to be the minimal peptide sequence required for binding to Keap1 Kelch domain protein with a KDsolution of 352 nM. Its binding affinity to Keap1 Kelch domain can be increased by 15× by removing the positive charge at the N-terminus with acetylation. The present study provides more insights into the interaction between Nrf2 and Keap1 and the SPR-based solution competition assay can be used for the screening of small molecules as inhibitors of Keap1-Nrf2 interaction.
Table 1.
Solution equilibrium dissociation constants (KDsolution) of Nrf2 peptides as measured by a SPR-based solution competition assay
| Nrf2 peptides | Peptide sequence | KDsolution (nM) |
|---|---|---|
| H-7mer-OH | H-EETGEFL-OH | ≫ 1,000 |
| H-8mer-OH | H-DEETGEFL-OH | ≫ 1,000 |
| H-9mer-OH | H-LDEETGEFL-OH | 352 |
| H-10mer-OH | H-QLDEETGEFL-OH | 27.3 |
| H-11mer-OH | H-LQLDEETGEFL-OH | 31.3 |
| H-12mer-OH | H-QLQLDEETGEFL-OH | 23.8 |
| H-14mer-OH | H-FAQLQLDEETGEFL-OH | 22.5 |
| H-16mer-OH | H-AFFAQLQLDEETGEFL-OH | 23.9 |
Table 2.
Solution equilibrium dissociation constants (KDsolution) of 9mer Nrf2 peptides as measured by a SPR-based solution competition assay
| Nrf2 peptides | Peptide sequence | KDsolution (nM) |
|---|---|---|
| Ac-9mer-NH2 | Ac-LDEETGEFL-NH2 | 21.4 |
| H-9mer-NH2 | H-LDEETGEFL-NH2 | 355 |
| Ac-9mer-OH | Ac-LDEETGEFL-OH | 23.1 |
| H-9mer-OH | H-LDEETGEFL-OH | 352 |
Acknowledgments
We gratefully acknowledge the financial support of grants CA133791 and CA125868 from the National Institutes of Health.
ABBREVIATIONS
- ARE
antioxidant response element
- BTB domain
bric-a-brac domain
- bZIP CNC protein
basic leucine zipper-type Cap n′ Collar protein
- CM5 chip
carboxymethyl dextran chip
- EDC
N-ethyl-N′-(dimethylaminopropyl)carbodiimide
- Fc1–4
flow cell 1–4
- GSH
glutathione
- GST
glutathione S-transferase
- HO1
heme oxygenase 1
- ITC
isothermal titration calorimetry
- Keap1
Kelch ECH-associated protein 1
- NBC
N-Boc-L-cysteine
- Neh1–6
Nrf2-ECH homologous domains 1–6
- NQO1
NAD(P)H:quinone oxidoreductase 1
- NHS
N-hydroxysuccinimide
- Nrf2
Nuclear factor erythroid 2 (Nf-E2)-related factor 2
- OPA
o-phthaldialdehyde
- RU
response unit
- SPR
surface plasmon resonance
References
- 1.Wilkinson J, Clapper ML. Detoxication enzymes and chemoprevention. Proc Soc Exp Biol Med. 1997;216:192–200. doi: 10.3181/00379727-216-44169. [DOI] [PubMed] [Google Scholar]
- 2.Kensler TW. Chemoprevention by inducers of carcinogen detoxication enzymes. Environ Health Persp. 1997;105:965–970. doi: 10.1289/ehp.97105s4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prestera T, Talalay P. Electrophile and Antioxidant Regulation of Enzymes That Detoxify Carcinogens. Proc Natl Acad Sci USA. 1995;92:8965–8969. doi: 10.1073/pnas.92.19.8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takagi Y, Mitsui A, Nishiyama A, Nozaki K, Sono H, Gon Y, Hashimoto N, Yodoi J. Overexpression of thioredoxin in transgenic mice attenuates focal ischemic brain damage. Proc Natl Acad Sci USA. 1999;96:4131–4136. doi: 10.1073/pnas.96.7.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yueh MF, Tukey RH. Nrf2-Keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. J Biol Chem. 2007;282:8749–8758. doi: 10.1074/jbc.M610790200. [DOI] [PubMed] [Google Scholar]
- 6.Talalay P. Chemoprotection against cancer by induction of Phase 2 enzymes. Biofactors. 2000;12:5–11. doi: 10.1002/biof.5520120102. [DOI] [PubMed] [Google Scholar]
- 7.Griffith OW, Mulcahy RT. The enzymes of glutathione synthesis: gamma-glutamylcysteine synthetase. Adv Enzymol. 1999;73:209–267. doi: 10.1002/9780470123195.ch7. [DOI] [PubMed] [Google Scholar]
- 8.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 9.Immenschuh S, Ramadori G. Gene regulation of heme oxygenase-1 as a therapeutic target. Biochem Pharmacol. 2000;60:1121–1128. doi: 10.1016/s0006-2952(00)00443-3. [DOI] [PubMed] [Google Scholar]
- 10.Kwak MK, Kensler TW. Targeting NRF2 signaling for cancer chemoprevention. Toxicol Appl Pharm. 2010;244:66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kundu JK, Surh YJ. Nrf2-Keap1 Signaling as a Potential Target for Chemoprevention of Inflammation-Associated Carcinogenesis. Pharm Res. 2010;27:999–1013. doi: 10.1007/s11095-010-0096-8. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 13.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 14.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of Nf-E2-Related Factor-2 (Nrf2), a Nf-E2-Like Basic Leucine-Zipper Transcriptional Activator That Binds to the Tandem Nf-E2/Ap1 Repeat of the Beta-Globin Locus-Control Region. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 17.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 20.Nioi P, Nguyen T, Sherratt PJ, Pickett CB. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol Cell Biol. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate for proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Prod Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cullinan SB, Gordan JD, Jin JO, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, Kensler T. Nrf2 as a target for cancer chemoprevention. Mutat Res. 2005;591:93–102. doi: 10.1016/j.mrfmmm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, Harada T, Engel JD, Yamamoto M. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 30.Kensler TW, Wakabayash N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 31.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 32.Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eggler AL, Liu GW, Pezzuto JM, van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 disrupt binding to the protein is insufficient to Nrf2 domain Neh2. Proc Natl Acad Sci USA. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “Tethering” mechanism - A two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 36.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 37.Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Different electrostatic Potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. Embo J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velazquez-Campoy A, Leavitt SA, Freire E. Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol Biol. 2004;261:35–54. doi: 10.1385/1-59259-762-9:035. [DOI] [PubMed] [Google Scholar]
- 40.Panayotou G, Ladbury J. Analysis of SH2 domain--phosphopeptide interactions by isothermal titration calorimetry and surface plasmon resonance. Methods Mol Biol. 2001;124:295–311. doi: 10.1385/1-59259-059-4:295. [DOI] [PubMed] [Google Scholar]
- 41.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Wu X, Hu L. Efficient amidation from carboxylic acids and azides via selenatriazoline: application to the coupling of amino acids and peptides with azides. J Org Chem. 2007;72:765–774. doi: 10.1021/jo061703n. [DOI] [PubMed] [Google Scholar]
- 43.Murphy M, Jason-Moller L, Bruno J. Curr Protoc Protein Sci. Unit 19. Chapter 19. 2006. Using Biacore to measure the binding kinetics of an antibody-antigen interaction; p. 14. [DOI] [PubMed] [Google Scholar]