Abstract
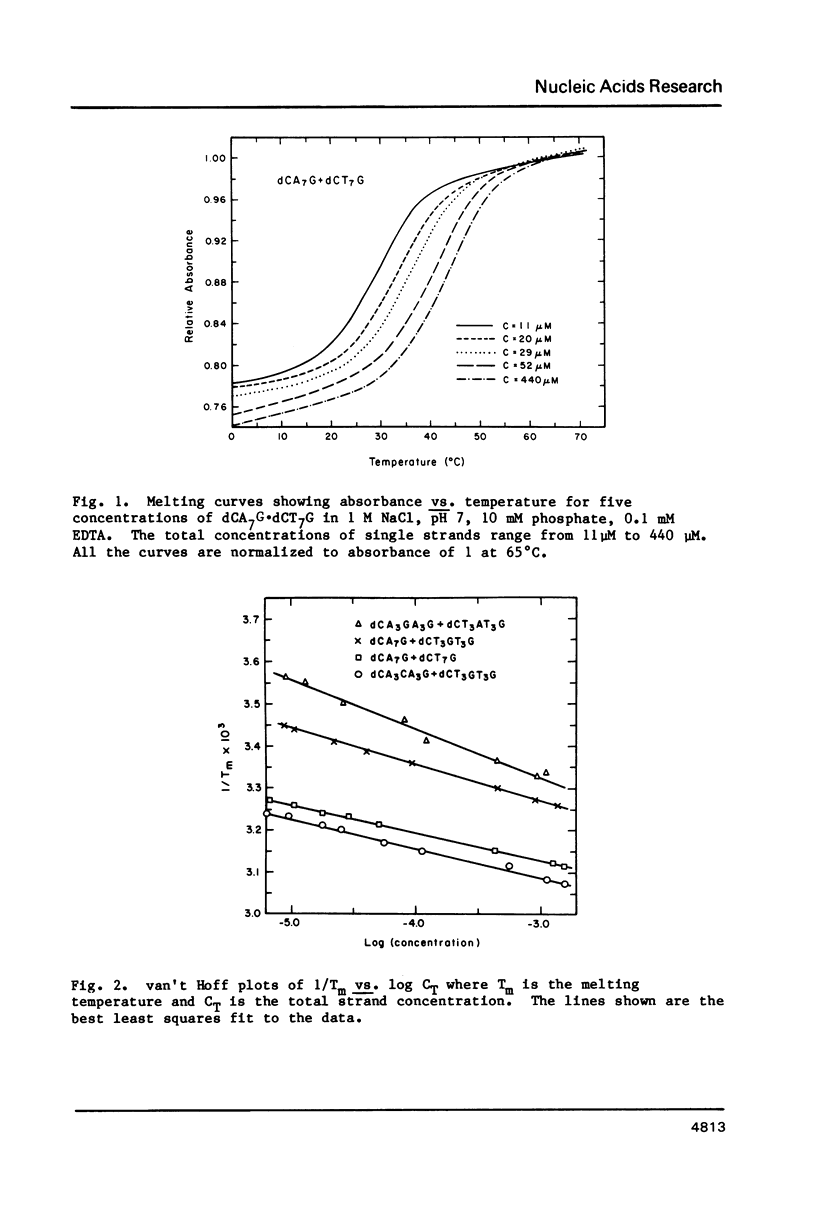
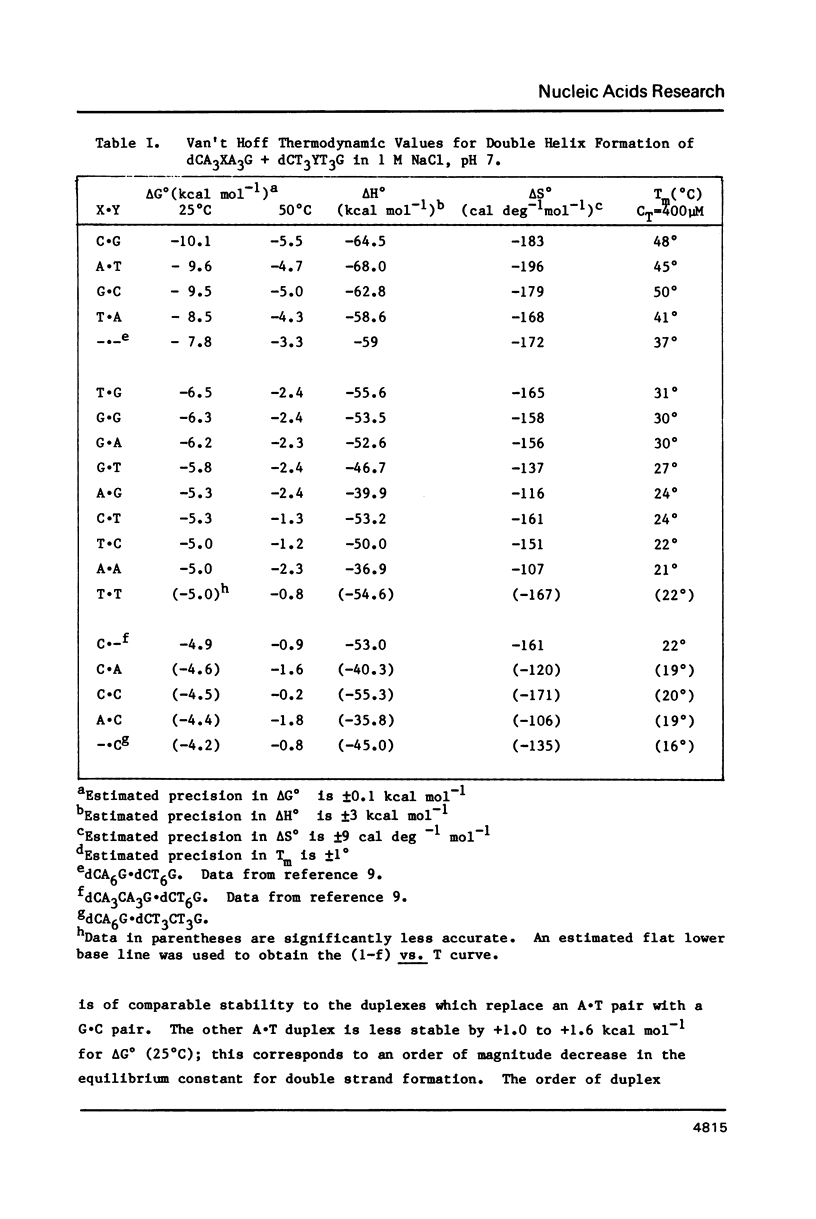
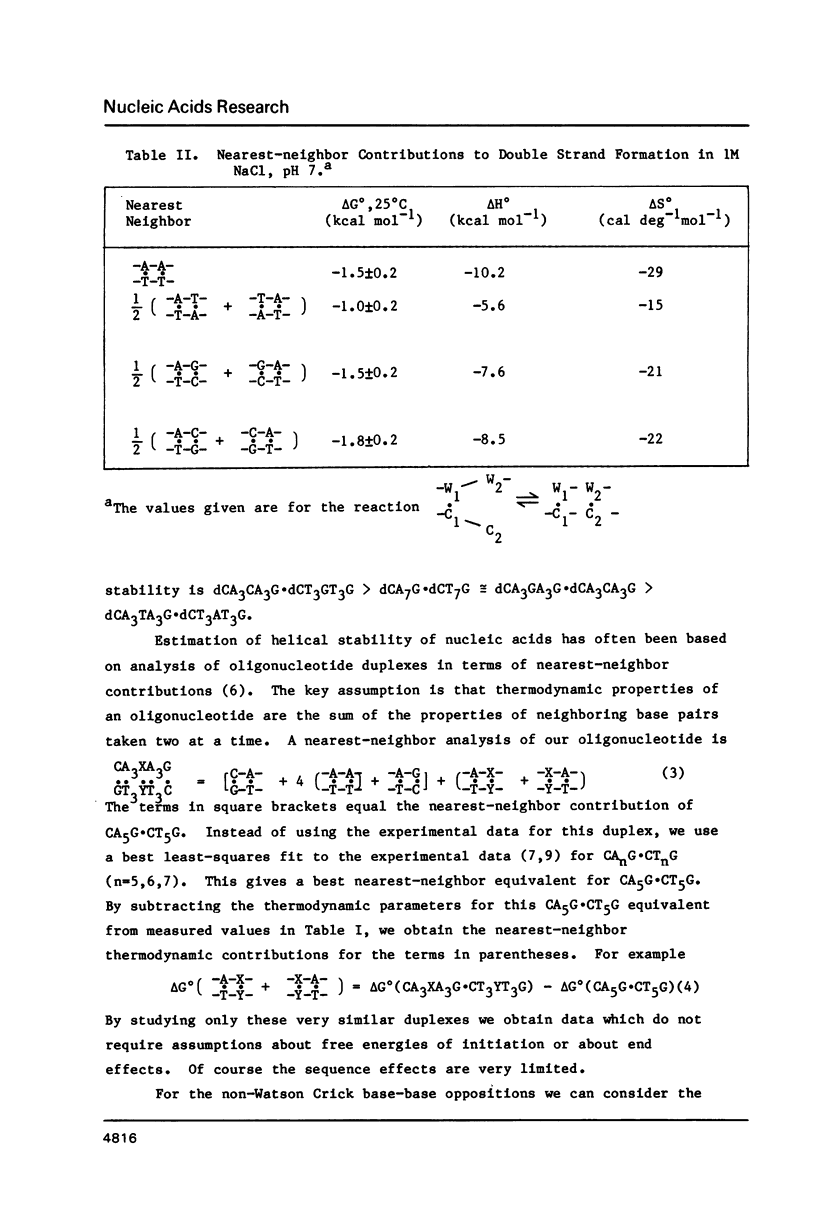
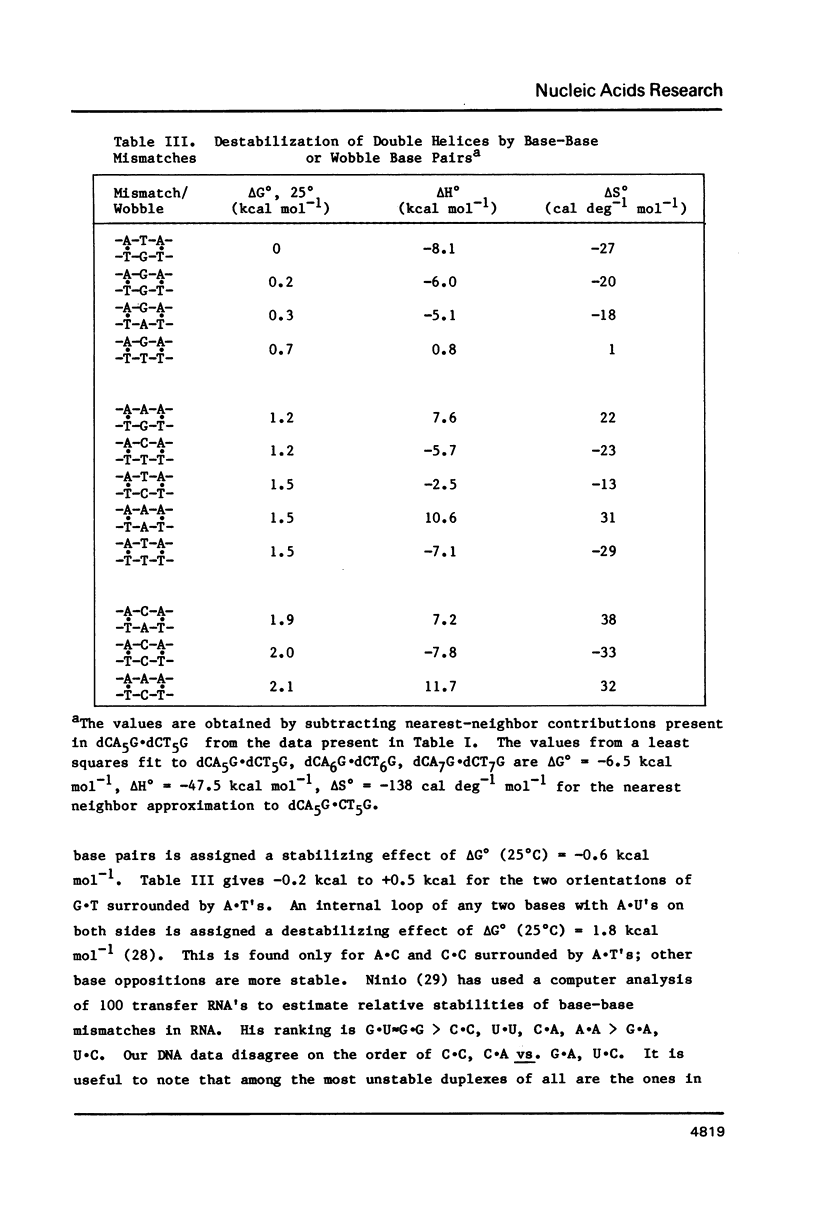
Thermodynamic parameters for double strand formation have been measured for the sixteen double helices of the sequence dCA3XA3G.dCT3YT3G, with each of the bases A, C, G and T at the positions labelled X and Y. The results are analyzed in terms of nearest-neighbors and are compared with thermodynamic parameters for RNA secondary structure. At room temperature the sequence (Formula: see text) is more stable than (Formula: see text) and is similar in stability to (Formula: see text) and (Formula: see text) are least stable. At higher temperatures the sequences containing a G.C base pair become more stable than those containing only A.T. All molecules containing mismatches are destabilized with respect to those with only Watson-Crick pairing, but there is a wide range of destabilization. At room temperature the most stable mismatches are those containing guanine (G.T, G.G, G.A); the least stable contain cytosine (C.A, C.C). At higher temperatures pyrimidine-pyrimidine mismatches become the least stable.
Full text
PDF




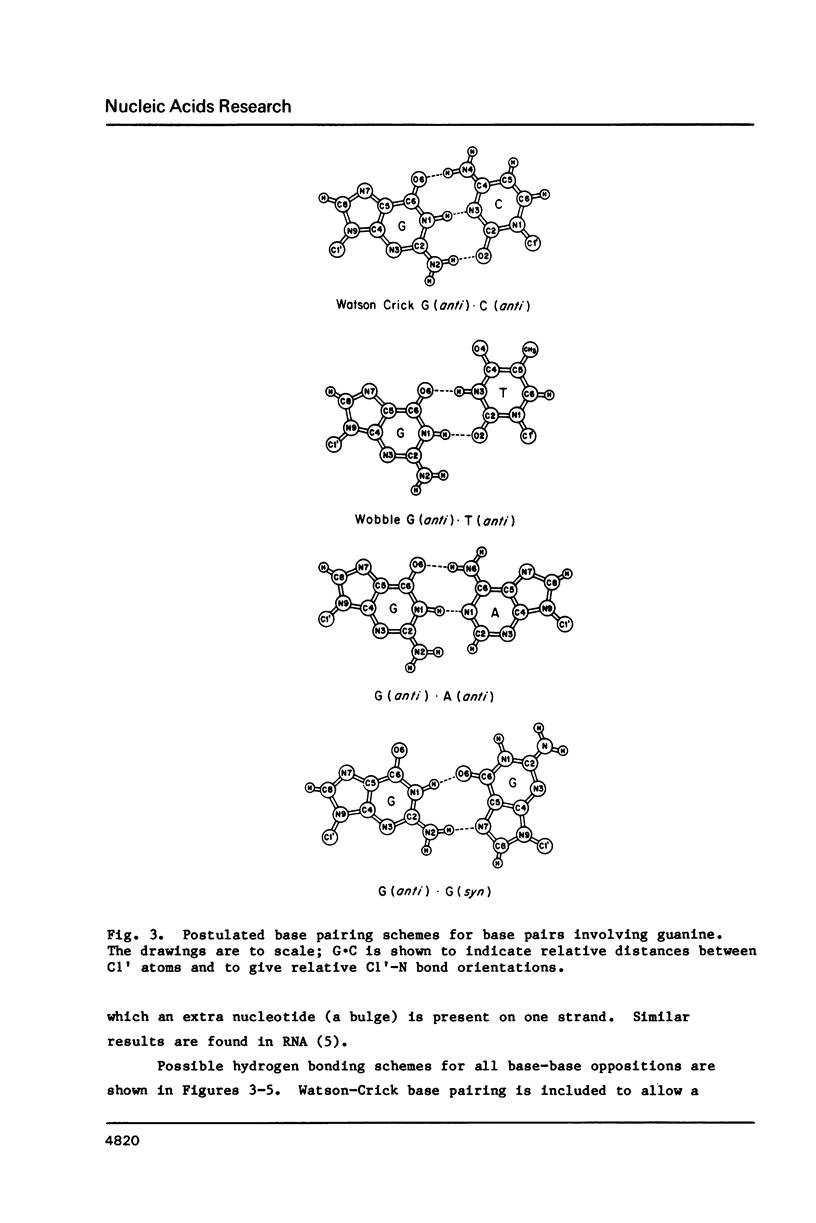
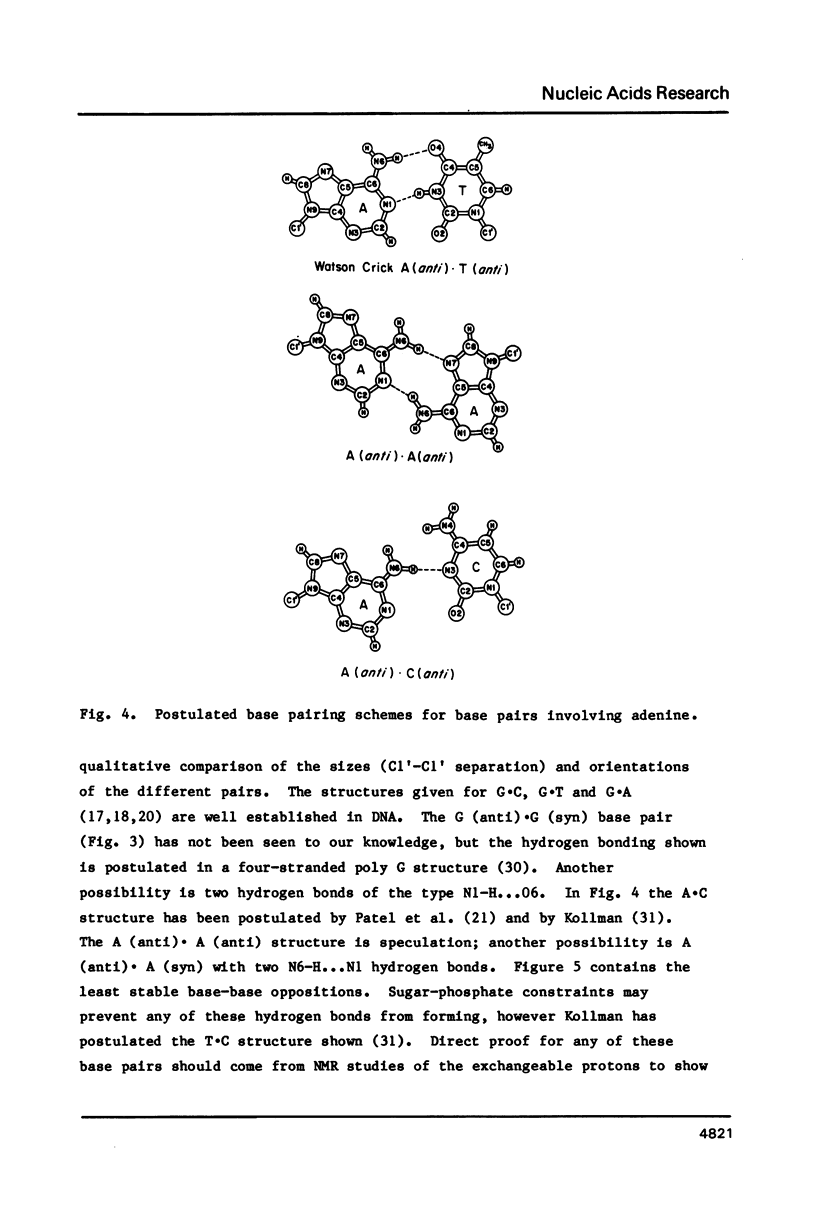
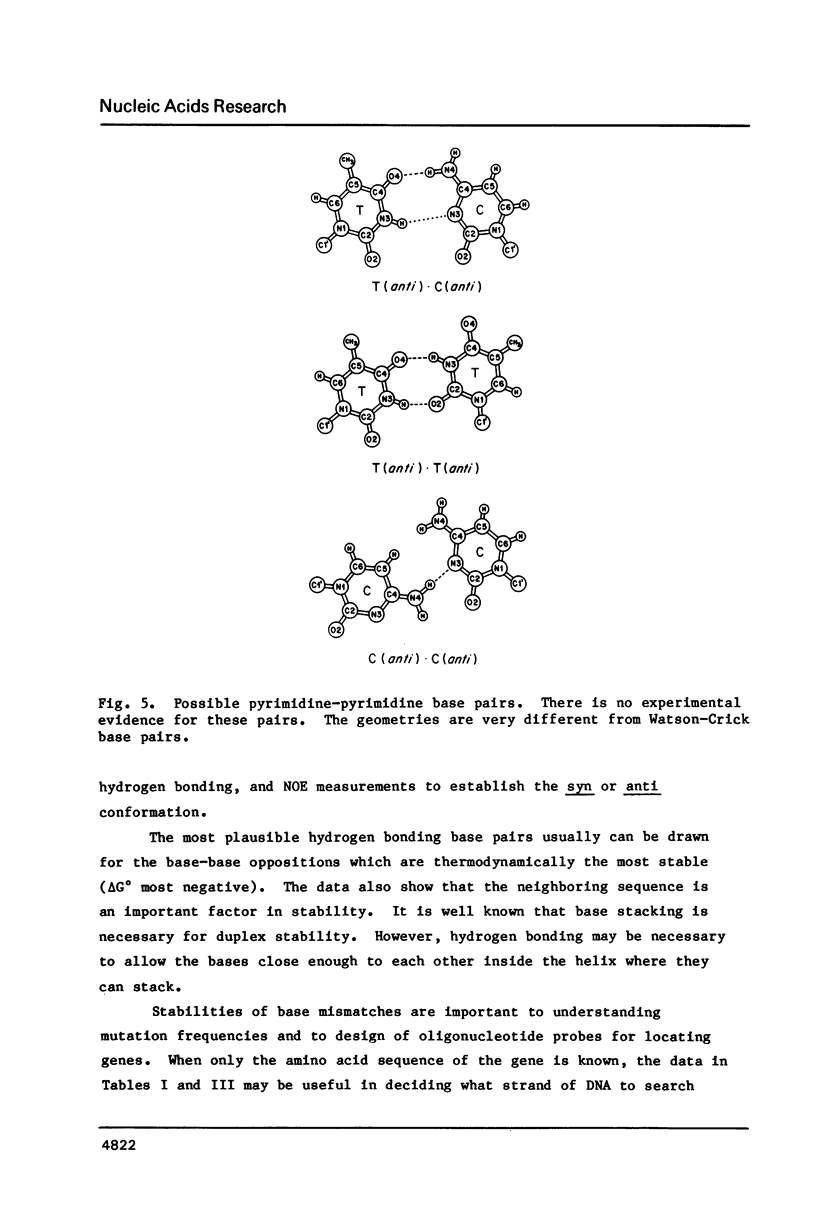


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Cornelis A. G., Haasnoot J. H., den Hartog J. F., de Rooij M., van Boom J. H., Cornelis A. Local destabilisation of a DNA double helix by a T--T wobble pair. Nature. 1979 Sep 20;281(5728):235–236. doi: 10.1038/281235a0. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Dodgson J. B., Wells R. D. Synthesis and thermal melting behavior of oligomer-polymer complexes containing defined lengths of mismatched dA-dG and dG-dG nucleotides. Biochemistry. 1977 May 31;16(11):2367–2374. doi: 10.1021/bi00630a009. [DOI] [PubMed] [Google Scholar]
- Early T. A., Olmsted J., 3rd, Kearns D. R., Lezius A. G. Base pairing structure in the poly d(G-T) double helix: wobble base pairs. Nucleic Acids Res. 1978 Jun;5(6):1955–1970. doi: 10.1093/nar/5.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Petersheim M., Hickey D. R., Turner D. H. Thermodynamic studies of RNA stability. J Biomol Struct Dyn. 1984 Mar;1(5):1229–1242. doi: 10.1080/07391102.1984.10507514. [DOI] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. 3. Small internal loops resulting from mismatches. J Mol Biol. 1973 Aug 5;78(2):301–319. doi: 10.1016/0022-2836(73)90118-6. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Chandrasegaran S., Pulford S. M., Miller P. S. Detection of a guanine X adenine base pair in a decadeoxyribonucleotide by proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4263–4265. doi: 10.1073/pnas.80.14.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keepers J. W., Schmidt P., James T. L., Kollman P. A. Molecular-mechanical studies of the mismatched base analogs of d(CGCGAATTCGCG)2:d(CGTGAATTCGCG)2, d(CGAGAATTCGCG)2, d(CGCGAATTCACG)2, d(CGCGAATTCTCG)2, and d(CGCAGAATTCGCG).d(CGCGAATTCGCG). Biopolymers. 1984 Dec;23(12):2901–2929. doi: 10.1002/bip.360231214. [DOI] [PubMed] [Google Scholar]
- Lomant A. J., Fresco J. R. Structural and energetic consequences of noncomplementary base oppositions in nucleic acid helices. Prog Nucleic Acid Res Mol Biol. 1975;15(0):185–218. doi: 10.1016/s0079-6603(08)60120-8. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calorimetric determination of base-stacking enthalpies in double-helical DNA molecules. Biopolymers. 1982 Nov;21(11):2185–2194. doi: 10.1002/bip.360211107. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Uhlenbeck O. C., Doty P. Self-complementary oligoribonucleotides: adenylic acid-uridylic acid block copolymers. J Mol Biol. 1971 Apr 28;57(2):201–215. doi: 10.1016/0022-2836(71)90341-x. [DOI] [PubMed] [Google Scholar]
- Nelson J. W., Martin F. H., Tinoco I., Jr DNA and RNA oligomer thermodynamics: the effect of mismatched bases on double-helix stability. Biopolymers. 1981 Dec;20(12):2509–2531. doi: 10.1002/bip.1981.360201204. [DOI] [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of imino protons in the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix and in two similar helices that contain a G . T base pair, d(C-G-T-G-A-A-T-T-C-G-C-G), and an extra adenine, d(C-G-C-A-G-A-A-T-T-C-G-C-G). Biochemistry. 1982 Dec 7;21(25):6567–6574. doi: 10.1021/bi00268a038. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyadenosine-deoxycytidine pairing in the d(C-G-C-G-A-A-T-T-C-A-C-G) duplex: conformation and dynamics at and adjacent to the dA X dC mismatch site. Biochemistry. 1984 Jul 3;23(14):3218–3226. doi: 10.1021/bi00309a016. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Deoxyguanosine-deoxyadenosine pairing in the d(C-G-A-G-A-A-T-T-C-G-C-G) duplex: conformation and dynamics at and adjacent to the dG X dA mismatch site. Biochemistry. 1984 Jul 3;23(14):3207–3217. doi: 10.1021/bi00309a015. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Broka C., Rice J. A., Itakura K., Breslauer K. J. Premelting and melting transitions in the d(CGCGAATTCGCG) self-complementary duplex in solution. Biochemistry. 1982 Feb 2;21(3):428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Dallas J., Itakura K., Breslauer K. J. Structure, dynamics, and energetics of deoxyguanosine . thymidine wobble base pair formation in the self-complementary d(CGTGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):437–444. doi: 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):445–451. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Sasisekharan V., Zimmerman S., Davies D. R. The structure of helical 5'-guanosine monophosphate. J Mol Biol. 1975 Feb 25;92(2):171–179. doi: 10.1016/0022-2836(75)90221-1. [DOI] [PubMed] [Google Scholar]
- Strauss F., Gaillard C., Prunell A. Helical periodicity of DNA, Poly(dA) . poly(dT) and poly(dA-dT). poly(dA-dT) in solution. Eur J Biochem. 1981 Aug;118(2):215–222. doi: 10.1111/j.1432-1033.1981.tb06389.x. [DOI] [PubMed] [Google Scholar]
- Tibanyenda N., De Bruin S. H., Haasnoot C. A., van der Marel G. A., van Boom J. H., Hilbers C. W. The effect of single base-pair mismatches on the duplex stability of d(T-A-T-T-A-A-T-A-T-C-A-A-G-T-T-G) . d(C-A-A-C-T-T-G-A-T-A-T-T-A-A-T-A). Eur J Biochem. 1984 Feb 15;139(1):19–27. doi: 10.1111/j.1432-1033.1984.tb07970.x. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]


