Abstract
The genomic material of hepatitis B virus (HBV) is confined within a fenestrated nucleocapsid consisting of 240 identical copies of the capsid protein, which has a rigid core and a positively charged and highly flexible C-terminal domain (CTD). Although previous mutagenesis studies have demonstrated the importance of the CTD in viral RNA packaging and reverse transcription, the microscopic structure of the CTD and its interaction with encapsidated nucleic acids at various stages of viral maturation remain poorly understood. Here, we present a theoretical analysis of the radial distributions of the CTD chains and nucleic acids in the hepatitis B virus nucleocapsid at the beginning and final stages of viral reverse transcription based on classical density functional theory and a coarse-gained model for the pertinent biomolecules. We find that a significant portion of the CTD is exposed at the surface of the RNA-containing immature nucleocapsid and that the CTD is mostly confined within the DNA-containing mature nucleocapsid. Large accumulation of cations is predicted inside both immature and mature nucleocapsids. The theoretical results provide new insights into the molecular mechanism of CTD regulation of viral reverse transcription and nucleocapsid trafficking during various stages of the viral replication processes.
Introduction
Hepatitis B virus (HBV) is a major human pathogen that causes chronic and acute hepatitis infections. A central step during HBV replication involves encapsidation of a pregenomic RNA (pgRNA) along with a virally encoded polymerase to form an immature nucleocapsid (NC) in the cytoplasm of the host cell (1, 2). The encapsulated pgRNA provides a template for the synthesis of a complete (−)-DNA strand and subsequently a complementary, incomplete (+)-DNA strand (3). The viral reverse transcription is concomitant with stepwise degradation and removal of the RNA template. In a mature HBV virion, the genome is a partially double-stranded (ds) DNA with a relaxed circular (rcDNA) conformation.
The majority of HBV capsids have an icosahedral structure with T = 4 symmetry. Each T4 capsid consists of 120 dimers of the capsid protein or the core antigen (HBcAg) that contains 183 amino acid (aa) residues. HBcAg is conventionally divided into an N-terminal assembly domain (residues 1–140), a linker (residues 141–149), and a highly charged C-terminal domain (CTD) (residues 150–183). Arginines comprise 16 of 34 residues in the CTD, making this domain highly flexible and strongly binding to the encapsidated nucleic acids. The N-terminal assembly domains of HBcAg form a rigid capsid fenestrated with pores (4). These pores, varying from 1.2 nm to 1.7 nm in diameter, allow access to the capsid lumen by monomeric nucleotides and other small molecules in the cytosol. The linkers and the CTD tails of HBcAg are positioned near the capsid pores (5, 6). Because the pore size is much larger than the diameters of typical aa residues, aa residues from CTD can be distributed either inside or outside the HBV NC.
It has been well recognized that the CTD may be involved in generating the maturation signal of the NC and could have significant effects on NC trafficking (7, 8). Although its exposure would drastically alter the surface properties of NCs, the structure of the CTD is not directly detectable through regular imaging methods such as cryo-electron microscopy (cryo-EM) (9, 10). Previous studies have given inconclusive and occasionally contradictory hypotheses as to the structural differences between immature and mature capsids, i.e., for capsids containing pgRNA and rcDNA, respectively. Early articles in the literature suggested that in mature NCs, the CTD is normally located inside the capsid shell (5, 6). Interior localization is intuitively appealing, because the CTD plays an important role in pgRNA packaging and DNA synthesis (11, 12). It was suggested that the CTD tails may become surface-exposed only when the capsid binding of the nucleic acids is weakened or prevented (13). For example, chimeric HBcAg proteins, in which the CTD was replaced with certain epitopes, were found to assemble into capsidlike structures in which these epitopes were surface-accessible. More recently, Kann et al. showed that the CTDs of RNA-containing recombinant capsids were exposed at the particle surface once they were phosphorylated in vitro by protein kinase C (14). In addition, heterologous sequences fused to the CTD have been found to localize at the capsid exterior (15, 16). The sensitivity of the CTD to proteolysis and antibody binding also suggests that its location is exterior (17, 18, 19), but perhaps this is selective at the mature NC stage (20). Rabe et al. conjectured that ∼50% and 100% of CTDs were exposed in the immature and mature NCs, respectively (21).
Previous studies indicate that the CTD is phosphorylated at multiple serine and threonine sites (22, 23, 24). Phosphorylation is required to facilitate pgRNA packaging and DNA synthesis (25). Furthermore, at least for the duck HBV (DHBV), the CTD has to be dephosphorylated at a later stage of reverse transcription to allow completion of the second-strand DNA synthesis and accumulation of mature NCs (26). Since CTD dephosphorylation has to occur after NC assembly, and since there is no indication that NCs encapsidate a phosphatase, the CTD is most likely at least partially exposed to the exterior of immature capsids to be accessible to an extracapsid phosphatase. Consistent with external exposure of the CTD, in vitro phosphatase treatment can remove the CTD phosphates from intact capsids, albeit after an extended period of time (overnight) (27), whereas after NC disruption, much more rapid CTD dephosphorylation occurred with phosphatase treatment (19). In fact, translocation of a protein domain through pores of intact capsid shells has been demonstrated in polio- (28) and parvoviruses (29). In particular, insertion of its DNA genome into parvovirus capsids induces externalization of an internally located domain of one of its capsid proteins through a channel on the capsid shell (30, 31, 32, 33).
The importance of the CTD throughout the life cycle of HBV has been well documented (4). For example, it has been shown that only mature NCs, i.e., those containing rcDNA, can be imported into the nucleoplasm of the host cell for genome amplification or enveloped with surface proteins for extracellular secretion (34, 35, 36). Despite numerous experiments to reveal the molecular mechanism behind the exclusion of immature NCs from envelopment and nuclear import (37, 38), little direct evidence is available at the present time on the variation of the NC structure during reverse transcription. Such microscopic details will be important for understanding HBV replication including NC interactions with the nuclear pore complex and with the viral envelope proteins. Because CTD tails are highly flexible, variation of the NC structure most likely is reflected in the CTD distribution relative to the capsid surface.
In this study we aim to address theoretically the question regarding the dynamic structure of the CTD within HBV NCs at the beginning and final stages of viral reverse transcription. Although there have been numerous theoretical studies on the internal structures of viral capsids (e.g., Devkota et al. (39), Toropova et al. (40), and Arkhipov et al. (41)), we are unaware of published theoretical work that analyzes the exposure of cationic protein tails at capsid surfaces. Because viral replication in vivo is not amenable to direct atomic simulations, we use a coarse-grained model for the pertinent biomolecules so that the structural and thermodynamic properties of the nucleocapsid can be described by classical density functional theory (42). We examine the distributions of the CTD tails, nucleic acids, and small ions for both the RNA-containing, immature NC and the dsDNA-containing, mature NC under physiological conditions. Our theoretical results indicate that for immature NCs, a remarkable portion of the CTD is exposed at the capsid surface. By contrast, for mature NCs, virtually all CTD tails are located within the lumen. Furthermore, our theoretical model predicts that cations are highly concentrated within the capsid lumen and the ion concentration in mature NCs is roughly three times higher than that in immature NCs. The theoretical results provide new insights into CTD regulation of reverse transcription and nucleocapsid trafficking during viral replication.
Molecular Model and Theory
As explained in a previous study (43), we use a coarse-grained model to describe the key components of both immature and mature HBV nucleocapsids. Fig. 1 shows schematically the molecular model studied in this work. Specifically, the nucleic acids and the CTD tails are represented by tangentially connected chains of hard spheres, and the capsid is represented by a semipermeable spherical shell. The surrounding electrolyte solution is described by the primitive model, where small ions are represented by charged spheres and the solvent is a dielectric continuum. Similar coarse-grained models have been used extensively to describe structural and thermodynamic properties of biomacromolecular systems, including packaging of DNA/RNA, protein folding, and assembly of viral capsids (44, 45, 46, 47, 48).
Figure 1.
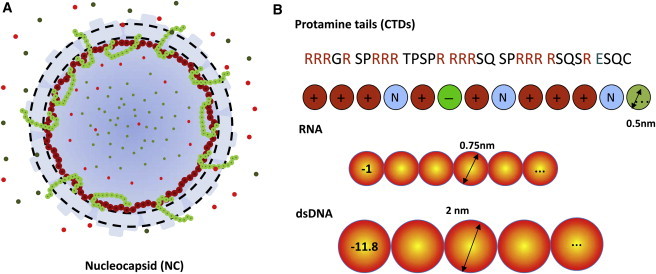
(A) Schematic representation of the HBV nucleocapsid. The capsid shell is represented by a spherical shell (dashed lines) permeable to small ions (small dots) and the coarse-grained segments of the C-terminal domain but not to the segments of pgRNA or dsDNA. (B) Coarse-grained representation of pertinent biomolecules. Shown here are only portions of the coarse-grained chains of biomacromolecules.
The encapsidated RNA is represented by a homochain with identical segments, i.e., the coarse-grained model ignores the RNA sequence and the secondary structure. Each RNA segment has a diameter of σR = 0.75 nm and valence of ZR = −1. These parameters reflect the average size and the electrical charge of individual nucleotides (47). The RNA chain is assumed to be fully flexible such that the bonding potential, Vb(R), between RNA segments can be described by the Dirac δ-function,
| (1) |
where , is the Boltzmann constant, K, and R = (r1,r2, …, rM) specifies the RNA configuration or the positions M = 3400 coarse-grained segments for pgRNA. Whereas biologists have long recognized the importance of the secondary structure of RNA on initialization of viral encapsidation, its role on the genome packaging inside the virus remains unknown. Because viral packaging, HBV included, is dominated by electrostatic interactions insensitive to the RNA sequence, we conjecture that the coarse-grained model is sufficient for capturing the essential features of the NC structure, including the distribution of CTD chains (43).
Similar to pgRNA, the CTD tails are represented by tangentially connected chains, with each segment designating one amino acid residue. For simplicity, we assume that all amino acid residues have the same diameter of σT = 0.5 nm, derived from the average van der Waals diameters of amino acids given in the Brookhaven Protein Data Bank (49). The CTD segments can be neutral, positively, or negatively charged, depending on the characteristic of the amino acid residues (see Fig. 1 B). To assign the valence of each CTD segment, we assume that under physiological conditions, each arginine residue carries one positive charge, each glutamate residue carries one negative charge, and all other amino acid residues in the CTD are free of electrostatic charge. In the immature NC, a negative charge is also assigned to each phosphoserine residue (155, 162, and 170) (26). After dephosphoration, these electrostatic charges are removed in DNA-containing mature NCs.
The rcDNA in a mature nucleocapsid is also represented by a coarse-grained model. Specifically, we assume that each DNA segment has a diameter the same as the cross-section diameter of a DNA double helix (σD = 2 nm) (50). Because the separation between two neighboring DNA basepairs in the DNA helix is 0.34 nm, each segment corresponds to 5.9 basepairs and carries a negative charge of ZD = −11.8. In contrast to that for pgRNA, the coarse-grained model for rcDNA includes a bending energy to account for both the chain connectivity and the backbone rigidity. The DNA bonding potential has the form
| (2) |
with
| (3) |
to account for chain connectivity and
| (4) |
to account for the bending potential. In (2), (3), (4), N = 508 corresponds to the number of coarse-grained segments for the partially double-strained rcDNA (∼3200 basepairs), v1 and v2 represent the Boltzmann factors of the backbone potential due to bond stretching and bond bending, respectively, θ is the angle between two neighboring bond vectors, and εD characterizes the stiffness of the rcDNA chain. If εD = 0, the DNA bonding potential becomes identical to that for the tangent-chain model. Conversely, εD = ∞ corresponds to a rigid-rod chain.
Using the bonding potential described by (2), (3), (4), Woodward and Forsman (51) showed that without the self-avoiding effects, the polymer persistence length, λp, can be approximated by λp ≈ ɛDσ, where σ is the diameter of the polymer segment. Accordingly, we estimate the stiffness constant (ɛD ≈ 25) of the coarse-grained model for rcDNA based on the intrinsic persistence length (∼50 nm). In this study, we find from calculations over a range of ɛD values that the final results are not sensitive to a small variation of the stiffness constant.
The HBV capsid is fenestrated with >120 holes, ∼1.5 nm in diameter on average, uniformly distributed on the surface. These holes occupy ∼30% of the surface area. As a result, the HBV capsid is permeable to small ions but not to nucleic acids. Because small ions and the CTD segments are not restricted to the viral lumen, we assume that the HBV capsid can be depicted as a rigid, semipermeable spherical shell of finite thickness ( = 2 nm, approximately the same as the diameter of the core domain of the capsid protein) (6). Based on the NC structure reported by Wynne et al. (52), the inner radius of the capsid is RC = 13 nm. The capsid inner surface is tethered with 240 copies of the CTD tails, one from each capsid protein. In addition, the capsid surface carries uniform charge density QC = 0.7 nm−2 (53). These charges are uniformly distributed over a spherical surface located at from the capsid center. Because the capsid pores (1.2–1.7 nm in diameter) are much larger than individual amino acid residues (∼0.5 nm), the CTD tails are able to distribute on both sides of the capsid wall. On the other hand, protection of the viral genome from nuclease digestion requires that the encapsulated RNA/DNA segments are fully contained within the capsid.
Viral replication occurs on a timescale of days, but given a viral configuration, the structure is relaxed at microscopic timescales. As a result, the nucleocapsids are virtually in thermodynamic equilibrium with their surroundings at each stage of replication. In this work, the viral environment is represented by a bulk NaCl solution at physiological concentration (i.e., CS =140 mM). Because viral packaging entails strong electrostatic interactions, we expect that the intracellular osmotic pressure arising from macromolecular crowding has a relatively insignificant effect on the viral packaging (43). As in the primitive model of aqueous electrolyte solutions, the cations (Na+) and anions (Cl−) are represented by spherical particles with diameters of σNa = 0.39 nm and σCl = 0.36 nm, respectively. Water is represented by a continuous dielectric medium with a dielectric constant of 78.4.
According to our coarse-grained representations of biomolecules (DNA/RNA/CTD), the pair potential between nonbonded segments is identical to that between a pair of small ions in the primitive model of aqueous electrolyte solutions. This potential includes a term accounting for the hard-sphere repulsion and the direct Coulomb energy:
| (5) |
where and = 0.714 nm stands for the Bjerrum length of pure water at 298 K. The CTD chains are tethered at the inner surface of the capsid with the ends freely distributed both inside and outside the capsid lumen. Because of the strong electric charge, we assume that the high affinity of CTD for nucleic acids mainly arises from electrostatic interactions.
In addition to interacting with CTD, the DNA/RNA segments experience a confining potential due to the capsid shell:
| (6) |
Because the total pore volume is negligible in comparison with the total volume of the capsid shell, we assume that the small ions are distributed either inside or outside the nucleocapsid, but not within the shell.
A classical density functional theory (DFT) is used to calculate the radial distributions of nucleic acids, CTD segments, and small ions in both immature and mature nucleocapsids (54). Since macroscopic units are not appropriate for measuring the local distributions of biomacromolecules within a capsid of no more than 30 nm in diameter, in this work all the radial distributions are reported in the units of the number densities (nm−3). In the DFT calculations, the total numbers of nucleic acids and CTD segments are fixed to prescribed values, whereas chemical potentials of small ions are kept constant to the values of bulk solutions. The numerical performance of the nonmean-field method has been well documented for inhomogeneous electrolyte solutions and for polymers (42, 55, 56, 57). As detailed in the Supporting Material, the DFT equations are essentially identical to those for confined polyelectrolytes, and their application to viral packaging has been reported before (43, 50).
Intuitively, the DFT used in this work can be understood as an extension of the polyelectrolyte Poisson-Boltzmann equation (PBE). Different from conventional applications of PBE, however, the DFT incorporates the packaging effects due to molecular volume, and the electrostatic and chain correlations that are ignored by the mean-field method. It has been shown before that the DFT is accurate for both the electrostatic interactions and the excluded-volume effects important for RNA/DNA packaging (43, 50).
Results and Discussion
Structure of immature HBV nucleocapsid
We first examine the radial distributions of CTD segments, nucleic acids, and small ions in an immature NC. In a wide-type HBV capsid, the pgRNA consists of 3400 nucleotides and each CTD chain includes 34 amino acid residues. Accordingly, the number of coarse-grained segments for the encapsidated pgRNA and that for CTD are M = 3400 and 34, respectively. Phosphorylation of S155, S162, and S170 at the CTD are considered by assigning a negative charge to each of the phosphoserine residues.
Fig. 2 shows the local density profiles for the coarse-grained segments of the RNA and CTD in an immature NC. As observed in the cryo-EM map of an RNA-containing HBV capsid (10), RNA is mainly located within a thin layer near the inner surface of the NC. The theory predicts a strong shell (8–12.5 nm) of the packaged RNA and parts of the C-terminal basic tails in very good agreement with the cryo-EM images. The thickness of the RNA-CTD complex is approximately the same as that for the CTD on the inner surface of the capsid, which is best described as a brush tethered to the inner capsid wall. Beyond the brush region of the tethered CTD, the density of RNA segments reaches a low plateau and remains approximately the same through the NC center. The central region is mainly occupied by a small number of RNA segments and cations that help to maintain the charge neutrality (see also Figure 4, Figure 8). Because the shell-CTD interaction was not considered other than the CTD tethering at the inner surface, the segment density within the capsid shell represents only a mean-field average. Nevertheless, it is clear that the capsid pores allow a significant number of the CTD tails to extend outside the NC surface (Fig. 2).
Figure 2.
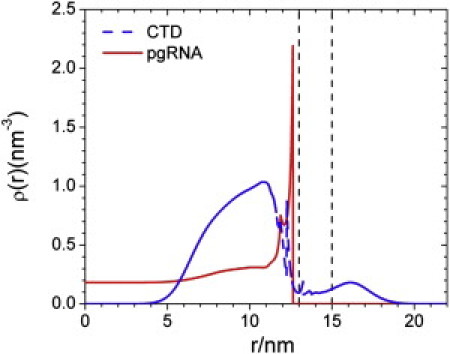
Density profiles for the coarse-grained segments of pgRNA and CTDs in an immature HBV nucleocapsid. In this and other figures, the position of the capsid wall is indicated by the perpendicular dashed lines.
Figure 4.
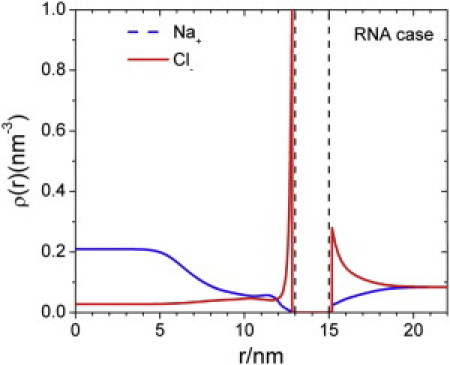
Density profiles of cations and anions inside and outside of an immature HBV nucleocapsid.
Figure 8.
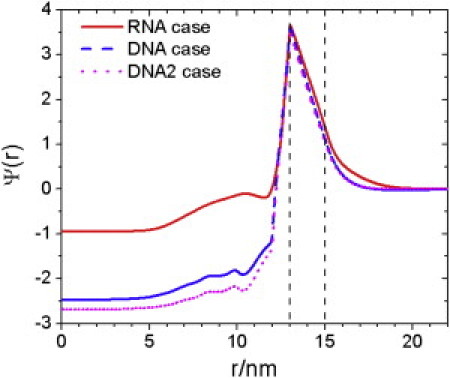
Reduced electrostatic potential, Ψ(r), inside and outside a HBV nucleocapsid containing pgRNA (RNA case), rcDNA (DNA case), and rcDNA without dephosphorylation of CTD (DNA2 case).
The DFT calculation also allows us to examine in detail the distributions of individual CTD segments both inside and outside the capsid. As shown in Fig. 3, a great number of positively charged amino acids are associated with the RNA at the inner surface of the NC (Fig. 2). Because of the chain connectivity, the density profiles of neutral and negative segments of CTD also show strong accumulation near the capsid inner surface. The weak CTD brush at the NC external surface closely resembles the electronic radial density profile of HBV capsid produced in Escherichia coli. (Fig. 4 A d of Wingfield et al. (6)). Exposure of the CTD tails increases the dimension of HBV capsid by up to 2 nm, which is also in good agreement with the experiment.
Figure 3.
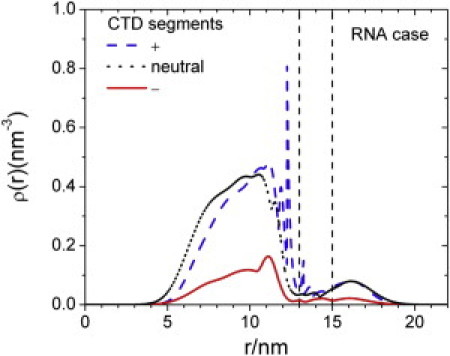
Radial distributions of positively charged (+), negatively charged (−), and neutral (0) amino acid residues from the CTDs of an immature HBV nucleocapsid.
That some of the CTD is exposed at the surface of an immature NC is consistent with previous experiments by Kann (14, 58). It was shown that CTD in purified recombinant capsids is phosphorylated by protein kinase C in vitro. Because the CTD of the immature NC needs to be dephosphorylated as it matures, and because serine/threonine phosphatases are not allowed to enter into the NC due to their size (59, 60), the phosphorylated serines in the CTD are most likely exposed outside and dephosphorylated. In the presence of phosphoserines, the exposed CTD could be transported to the nuclear pore complex (NPC) via importin α/β complex.
Fig. 4 shows the distributions of cations and anions both inside and outside the NC. Qualitatively, the ionic density profiles at the external surface of the NC are virtually identical to those in a conventional electric double layer. The capsid surface is positive primarily because of the surface charge and the exposure of the arginine segments of the CTD. As a result, the anions are concentrated near the NC outer surface, declining to the bulk value within 5 nm from the NC outer wall. Despite the lining of RNA segments at the inner surface, there is also a significant accumulation of anions in this region. The accumulation of anions in this region occurs because most CTD segments are positively charged near the tethering surface. The drastic difference in the densities of cations and anions at the center of the capsid arises from the small but finite density of RNA segments.
Structure of mature HBV nucleocapsid
Next we examine the radial distributions of DNA, CTD, and small ions in the mature NC. As mentioned before, the partially double-strained DNA is represented by a semiflexible chain with N = 508 coarse-grained segments, approximately corresponding to the DNA length in a wide-type virus. A value of εD = 25 is used for the stiffness parameter so that the dsDNA has an intrinsic persistence length of ∼50 nm. Based on the results of Perlman et al. (26) and Basagoudanavar et al. (25), we assume that the three phosphoserine residues at CTD are dephosphorylated after the reverse transcription, i.e., the electrical charges assigned to these residues are removed accordingly.
Fig. 5 shows the density profiles for the coarse-grained segments of rcDNA and the CTD tails in a mature capsid. In comparison to the RNA distribution in the immature NC, rcDNA segments are even more strongly accumulated near the inner surface of the NC. The narrower shell (9–12 nm) of the nucleic-acid-CTD complex in the mature NC is consistent with the cryo-EM images (10). Because rcDNA is extremely condensed near the NC inner surface, there is virtually no DNA density at the capsid center. The strong localization of DNA segments at the inner surface makes the density profile for CTD segments significantly contracted, giving rise to strong density oscillations in the CTD brush. In dramatic contrast to the presence of a CTD brush outside the immature NC, in the mature NC virtually all CTD segments are confined within the lumen. The containment of CTD segments can be attributed to the stronger electrostatic interaction with the localized DNA segments. Although the CTD becomes less exposed in mature capsids, our theoretical model does not preclude their instantaneous accessibility outside the capsid due to thermal fluctuations.
Figure 5.
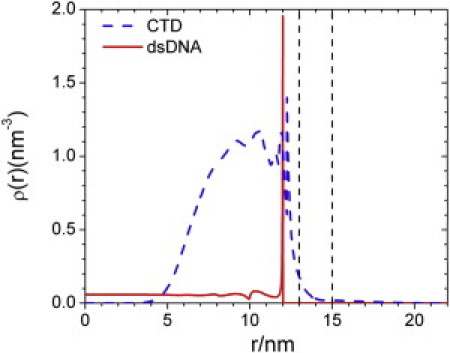
Density profiles for the coarse-grained segments of dsDNA and CTDs in a mature HBV nucleocapsid.
Fig. 6 presents the density profiles of amino acid residues from CTD according to their electrostatic charges. Similar to the distributions in the immature capsid, positively charged residues are strongly accumulated at the inner surface of the NC, accompanied by slight depletion of negatively charged amino acid residues. Beyond the dense layer of nucleic acid, the distribution of positively charged residues approximately coincides with that for the neutral segments. Different from that in an immature capsid, the density profile for the positively charged residues displays a significant layering structure.
Figure 6.
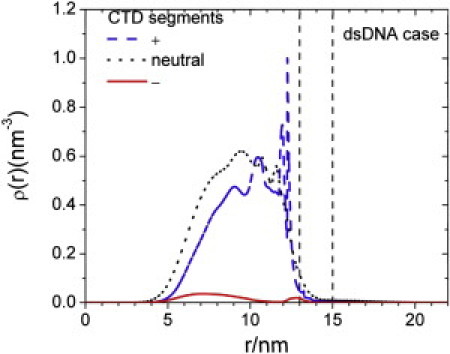
TRadial distributions of positively charged (+), negatively charged (−), and neutral (0) amino acid residues from the CTDs of a mature HBV nucleocapsid.
Fig. 7 shows the density profiles of small ions in the mature capsid. Although encapsidation of rcDNA and dephosphorylated CTD significantly increases the total amounts of negative charges in mature capsid, the NC particle as a whole exhibits only a small amount of positive net charge at the surface. The overall charge neutrality is evident from the weak density inhomogeneity outside the capsid shell. The ionic distributions quickly decline with distance to the bulk concentration. Because of the encapsidation of a highly charged dsDNA, the anions are almost completely excluded from the NC lumen, with only a small amount of anions accumulating near the NC wall due to the tethered, positively charged CTD. On the other hand, the cation density inside the NC is significantly larger than the bulk density. As mentioned above, the structure of mature capsid is little influenced by the DNA bending energy. A calculation with the bending energy turned off shows almost indistinguishable changes in the density profiles (data not shown), indicating that the electrostatic charge is the determining factor.
Figure 7.
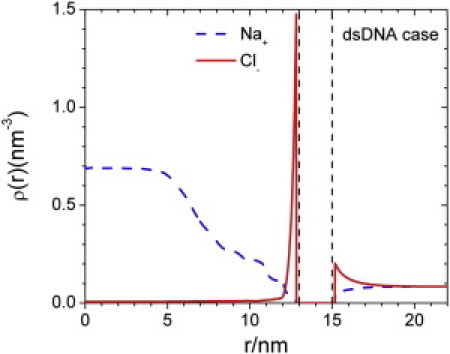
Density profiles of cations and anions inside and outside of a mature HBV nucleocapsid.
Finally, Fig. 8 shows the distributions of the electrostatic potential (in units of kBT/e) in immature and mature NC capsids. Here, DNA2 stands for the scenario if the serine residues (S155, S162, and S170) were not dephosphorylated. Consistent with strong accumulation of cations within the NC, the electrostatic potential is highly negative at the capsid center. The negative potential also explains the very low concentration of anions at the capsid center. As expected, dephosphorylation raises the electrostatic potential inside the NC, favoring DNA packaging in mature NCs.
Conclusions
We have analyzed the structure of immature and mature nucleocapsids of HBV on the basis of a coarse-grained model of the pertinent biomolecular components. Classical DFT was used to quantify the radial distributions of the CTD of capsid proteins, the nucleic acid chains, and small ions both inside and outside the NCs. The theoretical investigations indicate that whereas the majority of CTD resides in the capsid lumen to neutralize the negatively charged pgRNA or rcdsDNA, a significant portion of CTD is exposed on the surface of immature NC. In a mature capsid, however, virtually all CTDs are confined within the capsid lumen. The strong containment of CTD in the mature capsid is mainly due to the electrostatic interactions between arginine residues and rcDNA. To our knowledge, this work represents the first theoretical investigation on the exposure of the flexible domains of capsid proteins at viral surfaces. The low-resolution computational method is adequate not only because of its computational efficiency but also because of the lack of atomistic details for the entire NC and the in vivo environment associated with viral replication.
The theoretical predictions for the structure of the immature NC appear to be in good agreement with most previous experiments. As shown in cryo-EM images, pgRNA and CTD form a complex located within a thin layer near the inner surface of the nucleocapsid. The CTD tails exposed at the NC external surface closely resemble the electronic radial density profile of HBV capsid produced in Escherichia coli. Although the total amount of negative charge from rcDNA in a mature NC is significantly larger than that from pgRNA in an immature NC, the mature NC shows less net charge due to an elevated concentration of counterions inside the capsid lumen.
The predicted CTD topology as a function of capsid maturation has obvious implications on viral replication and trafficking. CTD has been shown to be dispensable for viral capsid assembly but indispensable for RNA encapsidation (61). This observation implies that negative charges derived from encapsidated RNA must draw most, if not all, of the CTD into the NC lumen. According to Perlman et al. (26) and Basagoudanavar et al. (25), phosphoserines on CTD of DHBV in the immature NC are dephosphorylated as the NC matures. Given that there is no report so far showing that the NC has the phosphatase activity and serine/threonine phosphatases are not allowed to enter into the NC (59, 60), the phosphoserines at the CTD must be dephosphorylated externally, indicating at least partial exposure of the CTD on the NC exterior, consistent with our result.
Whereas our theoretical results demonstrate that the reverse transcription of pgRNA to dsDNA makes the CTD less exposed, the experimental evidence for the distribution of the CTD in mature NCs is ambiguous. It is conceivable that the increasing negative charge conferred by dsDNA needs to be neutralized by more CTD if the charge balance hypothesis plays a dominant role in DNA replication (62). Although the strong confinement of the CTD in mature NCs predicted by the DFT appears to be in good agreement with the early literature (5, 6), there is yet no consensus on such distribution (21). Future scattering measurements may be able to help validate the theoretical predictions.
Acknowledgments
This work utilizes supercomputers from the National Energy Research Scientific Computing Center.
The authors are grateful to the National Institutes of Health (R21-AI077532) for financial support. Additional support for this work was provided by the National Science Foundation (NSF-CBET-0852353).
Editor: Nathan Baker.
Footnotes
Polyelectrolyte density functional theory and numerical detailsare available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(11)01188-X.
Supporting Material
References
- 1.Bartenschlager R., Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porterfield J.Z., Dhason M.S., et al. Zlotnick A. Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J. Virol. 2010;84:7174–7184. doi: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Summers J., Mason W.S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 4.Bruss V. Hepatitis B virus morphogenesis. World J. Gastroenterol. 2007;13:65–73. doi: 10.3748/wjg.v13.i1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watts N.R., Conway J.F., et al. Wingfield P.T. The morphogenic linker peptide of HBV capsid protein forms a mobile array on the interior surface. EMBO J. 2002;21:876–884. doi: 10.1093/emboj/21.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wingfield P.T., Stahl S.J., et al. Steven A.C. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 7.Schmitz A., Schwarz A., et al. Kann M. Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog. 2010;6:e1000741. doi: 10.1371/journal.ppat.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewellyn E.B., Loeb D.D. The arginine clusters of the carboxy-terminal domain of the core protein of hepatitis B virus make pleiotropic contributions to genome replication. J. Virol. 2011;85:1298–1309. doi: 10.1128/JVI.01957-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zlotnick A., Cheng N., et al. Wingfield P.T. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc. Natl. Acad. Sci. USA. 1997;94:9556–9561. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roseman A.M., Berriman J.A., et al. Crowther R.A. A structural model for maturation of the hepatitis B virus core. Proc. Natl. Acad. Sci. USA. 2005;102:15821–15826. doi: 10.1073/pnas.0504874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowther R.A., Kiselev N.A., et al. Pumpens P. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 12.Zhou S., Yang S.Q., Standring D.N. Characterization of hepatitis B virus capsid particle assembly in Xenopus oocytes. J. Virol. 1992;66:3086–3092. doi: 10.1128/jvi.66.5.3086-3092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshikawa A., Tanaka T., et al. Mayumi M. Chimeric hepatitis B virus core particles with parts or copies of the hepatitis C virus core protein. J. Virol. 1993;67:6064–6070. doi: 10.1128/jvi.67.10.6064-6070.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kann M., Sodeik B., et al. Helenius A. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 1999;145:45–55. doi: 10.1083/jcb.145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schödel F., Moriarty A.M., et al. Milich D.R. The position of heterologous epitopes inserted in hepatitis B virus core particles determines their immunogenicity. J. Virol. 1992;66:106–114. doi: 10.1128/jvi.66.1.106-114.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borisova G.P., Berzins I., et al. Siakkou H. Recombinant core particles of hepatitis B virus exposing foreign antigenic determinants on their surface. FEBS Lett. 1989;259:121–124. doi: 10.1016/0014-5793(89)81509-1. [DOI] [PubMed] [Google Scholar]
- 17.Gallina A., Bonelli F., et al. Milanesi G. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J. Virol. 1989;63:4645–4652. doi: 10.1128/jvi.63.11.4645-4652.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seifer M., Standring D.N. A protease-sensitive hinge linking the two domains of the hepatitis B virus core protein is exposed on the viral capsid surface. J. Virol. 1994;68:5548–5555. doi: 10.1128/jvi.68.9.5548-5555.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlicht H.J., Bartenschlager R., Schaller H. The duck hepatitis B virus core protein contains a highly phosphorylated C terminus that is essential for replication but not for RNA packaging. J. Virol. 1989;63:2995–3000. doi: 10.1128/jvi.63.7.2995-3000.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H., Mao R., et al. Guo J.T. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J. Virol. 2010;84:387–396. doi: 10.1128/JVI.01921-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabe B., Vlachou A., et al. Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. Proc. Natl. Acad. Sci. USA. 2003;100:9849–9854. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machida A., Ohnuma H., et al. Mayumi M. Phosphorylation in the carboxyl-terminal domain of the capsid protein of hepatitis B virus: evaluation with a monoclonal antibody. J. Virol. 1991;65:6024–6030. doi: 10.1128/jvi.65.11.6024-6030.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu M.S., Summers J. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J. Virol. 1994;68:2965–2969. doi: 10.1128/jvi.68.5.2965-2969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao W., Ou J.H. Phosphorylation and nuclear localization of the hepatitis B virus core protein: significance of serine in the three repeated SPRRR motifs. J. Virol. 1995;69:1025–1029. doi: 10.1128/jvi.69.2.1025-1029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basagoudanavar S.H., Perlman D.H., Hu J.M. Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J. Virol. 2007;81:1641–1649. doi: 10.1128/JVI.01671-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perlman D.H., Berg E.A., et al. Hu J. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc. Natl. Acad. Sci. USA. 2005;102:9020–9025. doi: 10.1073/pnas.0502138102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pugh J., Zweidler A., Summers J. Characterization of the major duck hepatitis B virus core particle protein. J. Virol. 1989;63:1371–1376. doi: 10.1128/jvi.63.3.1371-1376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q., Yafal A.G., et al. Chow M. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of these sequences at physiological temperature. J. Virol. 1994;68:3965–3970. doi: 10.1128/jvi.68.6.3965-3970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kronenberg S., Böttcher B., et al. Kleinschmidt J.A. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 2005;79:5296–5303. doi: 10.1128/JVI.79.9.5296-5303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cotmore S.F., D'Abramo A.M., Jr., et al. Tattersall P. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology. 1999;254:169–181. doi: 10.1006/viro.1998.9520. [DOI] [PubMed] [Google Scholar]
- 31.Reguera J., Carreira A., et al. Mateu M.G. Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid. Proc. Natl. Acad. Sci. USA. 2004;101:2724–2729. doi: 10.1073/pnas.0307748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maroto B., Valle N., et al. Almendral J.M. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J. Virol. 2004;78:10685–10694. doi: 10.1128/JVI.78.19.10685-10694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufmann B., Chipman P.R., et al. Rossmann M.G. Visualization of the externalized VP2 N termini of infectious human parvovirus B19. J. Virol. 2008;82:7306–7312. doi: 10.1128/JVI.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerelsaikhan T., Tavis J.E., Bruss V. Hepatitis B virus nucleocapsid envelopment does not occur without genomic DNA synthesis. J. Virol. 1996;70:4269–4274. doi: 10.1128/jvi.70.7.4269-4274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summers J., Smith P.M., et al. Yu M.S. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y., Tavis J.E., Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J. Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponsel D., Bruss V. Mapping of amino acid side chains on the surface of hepatitis B virus capsids required for envelopment and virion formation. J. Virol. 2003;77:416–422. doi: 10.1128/JVI.77.1.416-422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pairan A., Bruss V. Functional surfaces of the hepatitis B virus capsid. J. Virol. 2009;83:11616–11623. doi: 10.1128/JVI.01178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devkota B., Petrov A.S., et al. Harvey S.C. Structural and electrostatic characterization of pariacoto virus: implications for viral assembly. Biopolymers. 2009;91:530–538. doi: 10.1002/bip.21168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toropova K., Basnak G., et al. Ranson N.A. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J. Mol. Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 41.Arkhipov A., Freddolino P.L., Schulten K. Stability and dynamics of virus capsids described by coarse-grained modeling. Structure. 2006;14:1767–1777. doi: 10.1016/j.str.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 42.Wu J.Z., Li Z.D. Density-functional theory for complex fluids. Annu. Rev. Phys. Chem. 2007;58:85–112. doi: 10.1146/annurev.physchem.58.032806.104650. [DOI] [PubMed] [Google Scholar]
- 43.Jiang T., Wang Z.G., Wu J.Z. Electrostatic regulation of genome packaging in human hepatitis B virus. Biophys. J. 2009;96:3065–3073. doi: 10.1016/j.bpj.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Angelescu D.G., Linse P. Modelling of icosahedral viruses. Curr. Opin. Colloid Interface Sci. 2008;13:389–394. [Google Scholar]
- 45.Hagan M.F., Chandler D. Dynamic pathways for viral capsid assembly. Biophys. J. 2006;91:42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrov A.S., Harvey S.C. Structural and thermodynamic principles of viral packaging. Structure. 2007;15:21–27. doi: 10.1016/j.str.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 47.Zhang D.Q., Konecny R., et al. McCammon J.A. Electrostatic interaction between RNA and protein capsid in cowpea chlorotic mottle virus simulated by a coarse-grain RNA model and a Monte Carlo approach. Biopolymers. 2004;75:325–337. doi: 10.1002/bip.20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elrad O.M., Hagan M.F. Encapsulation of a polymer by an icosahedral virus. Phys. Biol. 2010;7:045003. doi: 10.1088/1478-3975/7/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolinski A., Godzik A., Skolnick J. A general method for the prediction of the 3-dimensional structure and folding pathway of globular proteins: application to designed helical proteins. J. Chem. Phys. 1993;98:7420–7433. [Google Scholar]
- 50.Li Z.D., Wu J.Z., Wang Z.G. Osmotic pressure and packaging structure of caged DNA. Biophys. J. 2008;94:737–746. doi: 10.1529/biophysj.107.112508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woodward C.E., Forsman J. Density functional theory for flexible and semiflexible polymers of infinite length. Phys. Rev. E. 2006;74:010801. doi: 10.1103/PhysRevE.74.010801. [DOI] [PubMed] [Google Scholar]
- 52.Wynne S.A., Crowther R.A., Leslie A.G.W. The crystal structure of the human hepatitis B virus capsid. Mol. Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 53.Kegel W.K., Schoot Pv P. Competing hydrophobic and screened-coulomb interactions in hepatitis B virus capsid assembly. Biophys. J. 2004;86:3905–3913. doi: 10.1529/biophysj.104.040055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang T., Li Z.D., Wu J.Z. Structure and swelling of grafted polyelectrolytes: predictions from a nonlocal density functional theory. Macromolecules. 2007;40:334–343. [Google Scholar]
- 55.Wu J.Z. Density functional theory for chemical engineering: from capillarity to soft materials. AIChE J. 2006;52:1169–1193. [Google Scholar]
- 56.Li Z.D., Wu J.Z. Density functional theory for polyelectrolytes near oppositely charged surfaces. Phys. Rev. Lett. 2006;96:048302. doi: 10.1103/PhysRevLett.96.048302. [DOI] [PubMed] [Google Scholar]
- 57.Li Z.D., Wu J.Z. Density functional theory for planar electric double layers: closing the gap between simple and polyelectrolytes. J. Phys. Chem. B. 2006;110:7473–7484. doi: 10.1021/jp060127w. [DOI] [PubMed] [Google Scholar]
- 58.Kann M., Schmitz A., Rabe B. Intracellular transport of hepatitis B virus. World J. Gastroenterol. 2007;13:39–47. doi: 10.3748/wjg.v13.i1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Y.H., Xing Y.N., et al. Shi Y. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 60.Shi Y.G. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 61.Birnbaum F., Nassal M. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J. Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chua P.K., Tang F.M., et al. Shih C. Testing the balanced electrostatic interaction hypothesis of hepatitis B virus DNA synthesis by using an in vivo charge rebalance approach. J. Virol. 2010;84:2340–2351. doi: 10.1128/JVI.01666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


