Abstract
The sterile alpha motif (SAM) domain is one of the most common protein modules found in eukaryotic genomes. Many SAM domains have been shown to form helical polymer structures suggesting that SAM modules can be used to create large protein complexes in the cell. Because many polymeric SAM domains form heterogenous and insoluble aggregates that are experimentally intractable when isolated, it is likely that many polymeric SAM domains have gone uncharacterized. We, therefore, developed a method to maintain polymeric SAM domains in a soluble form that allowed rapid screening for potential SAM polymers. SAM domains were expressed as fusions to a super-negatively charged green fluorescent protein (negGFP). The negGFP imparts three useful properties to the SAM domains: (1) the charge helps to maintain solubility; (2) the charge leads to reliable migration toward the cathode on native gels; and (3) the fluorescence emission allows visualization in crude extracts. Using the negGFP-SAM fusions, we screened a large library of human SAM domains for polymerization using a native gel screen. A selected set of hSAM domains were then purified and examined for true polymer formation by electron microscopy. In this manner, we identified a set of new potential SAM polymers: ANKS3, Atherin, BicaudalC1, Caskin1, Caskin2, Kazrin, L3MBTL3, L3MBTL4, LBP, LiprinB1, LiprinB2, SAMD8, SAMD9, and STIM2. While further characterization will be necessary to verify that the SAM domains identified here truly form polymers, our results provide a much stronger working hypothesis for a large number of proteins that was possible from sequence analysis alone.
Keywords: SAM domain, polymer, green fluorescent protein, ANKS3, Atherin, BicaudalC1, Caskin, Kazrin, L3MBTL, Liprin, polycystic kidney disease
Introduction
Many eukaryotic proteins are constructed in a modular fashion in which distinct functionalities are provided by linking together protein domains.1 As a result, it is often possible to develop general functional hypotheses for a particular protein by knowing its domain structure. Consequently, considerable effort has been devoted to understanding the functions of common protein modules.
Sterile alpha motif (SAM) domains are among the most common protein modules in eukaryotic genomes2 and are somewhat unusual because they exhibit a wide range of different functions. They display a variety of oligomeric forms including both polymers3–8 and closed oligomers,5,9–12 thereby playing important roles in building cellular complexes. SAM domains also bind to other domains such as MAP kinases13 to RNA,14 and one SAM domain has been suggested to bind to lipids.15 Because of the large number of known functions, the presence of a SAM domain does not necessarily imply a particular function, but a set of possible functions. It is, therefore, necessary to characterize each SAM domain individually.
Polymerization is perhaps the most unusual and striking function of many SAM domains. SAM polymers have been found in transcriptional repressors where they may mediate spreading along chromatin3,6 as well as a wide variety of scaffolding proteins where SAM polymers are used to generate large protein complexes.4,16 SAM domains known to form polymers assembled by a head-to-tail association of monomeric subunits. These interactions are mediated by two common binding surfaces termed the mid-loop (ML) and end-helix (EH) surfaces3 [Fig. 1(A)]. Another form of SAM polymer utilizing different interaction surfaces has been proposed, but there is still no compelling evidence that this is a biologically relevant structure.17,18 The SAM domain polymer is quite versatile as the N- and C-termini project away from the axis of the polymer such that the SAM domain can be placed anywhere in a protein and still form a polymer, possibly explaining its wide exploitation in the eukaryotic genome.
Figure 1.
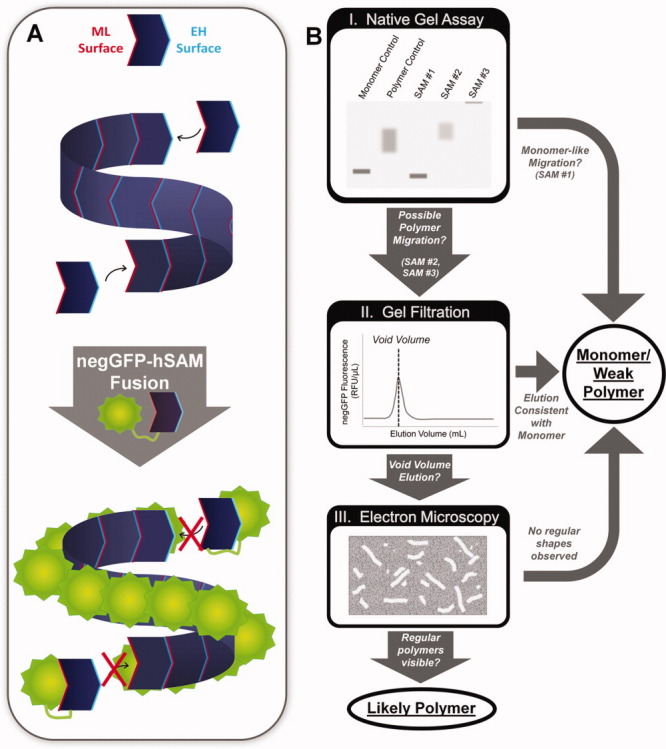
Polymerizome screening overview. (A) SAMs can form helical polymers by binding head to tail via mid-loop (ML) and end-helix (EH) interaction surfaces. Fusion to highly charged negGFP solubilizes the polymers for more facile biochemical characterization. (B) I. negGFP-hSAM fusion lysates were first analyzed by native gel electrophoresis, using the strong negative charge of negGFP to drive migration to the cathode. SAM domains that migrated in a manner similar to the monomer control were classified as monomers/weak polymers. negGFP-hSAM domains that migrated in a manner consistent with the polymer control or remained in the well were classified as possible polymers and subjected to further analysis. II. Possible polymer negGFP-hSAMs were purified and analyzed via gel filtration. SAMs that eluted in the void volume were visualized using Electron Microscopy. III. Only SAMs that form regular rod shapes consistent with a polymer were classified as likely polymers. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Polymerization is a particularly problematic function for biochemical examination because polymers are often insoluble and heterogeneous. As a result, predictions indicate that many polymeric SAM domains remain uncharacterized.19 We, therefore, sought to fill this gap in the functional annotation of this important protein module. Here, we describe the development of a high throughput screen for polymerization and its application to nearly all identified SAM domains in the human genome. We have identified a total of 17 potentially new polymer-forming SAM domains and structurally characterized 14 of them by electron microscopy (EM).
Results
Supercharged GFP fusion approach to solubilizing and screening SAM polymers
Our goal was to comprehensively screen human SAM (hSAM) domains for polymerization. We, therefore, attempted to develop a method that would reliably solubilize polymeric SAM domains so that they could be examined. In addition, we sought a way to rapidly screen for their ability to oligomerize. The approach we took is outlined in Figure 1. We expressed all SAM domains as fusions to a super-negativley charged green fluorescent protein (negGFP)20 with a net charge of −30. As described below, the highly charged fusion effectively solubilizes SAM domains. We also exploited the extreme negative charge of the fusion proteins to develop a rapid initial screen for oligomerization. In particular, the negatively charged fusions reliably run toward the cathode in a native gel so that much of the uncertainty about migration in a native gel is eliminated. We also exploited the negGFP fluorescence so that we could screen for migration in native gels and in gel filtration columns using crude extracts, obviating the need for laborious purification. Those SAM domains that appeared to form hetergenous aggregates in native gels were then examined further by gel filtration and, in selected cases, electron microscropy (EM).
Supercharged GFP fusions solubilize SAM polymers
To test whether the negGFP could solubilize SAM polymers, we chose the SAM domain from TEL (TEL-SAM) because it forms long, very stable polymers that are largely insoluble.3 We also used as controls two monomeric mutants of TEL-SAM: TEL-SAM-A61D, which bears the mutation on the ML surface, and TEL-SAM-V80E, which bears the mutation on the EH surface. The single mutants are soluble because they cannot self associate, but they bind to each other to form a dimer with a Kd of ∼2 nM via the lone intact ML and EH surfaces.21,3 We expressed negGFP-TEL fusions in E. coli and observed soluble protein in the cell lysate for all fusions.
To examine whether the soluble wild-type TEL-SAM fusion remained polymeric when fused to the highly charged negGFP, we analyzed the fusion protein by gel filtration chromatography and EM. By monitoring fluorescence, it was possible to observe the elution position of negGFP fusions on a gel filtration column run using crude extract. As shown in Figure 2(A), negGFP-TEL-WT eluted in the void volume of a gel filtration column, while the negGFP-TEL-V80E fusion eluted as a monomer as expected. To verify that the high molecular weight material was polymer and not amorphous aggregates, negGFP-TEL-WT was purified and examined by EM [Fig. 2(B)]. The negGFP-TEL-WT polymers were readily apparent across the entire grid. The diameter of a polymer is roughly 12 nm compared to the 8 nm diameter of the TEL-SAM polymers visualized previously,3 with the extra width presumably supplied by the negGFP fusion protein.
Figure 2.
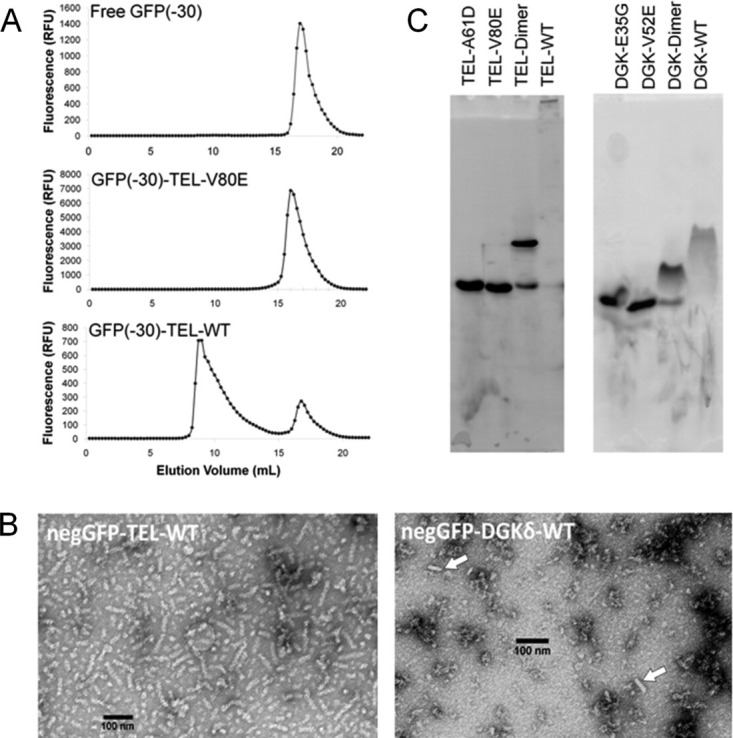
Testing assays for polymerization. (A) Gel filtration analysis of the soluble fraction from cell lysates containing free negGFP, a monomeric SAM negGFP-TEL-V80E, and a high-affinity polymer negGFP-TEL-WT. (C) Electron micrographs of negGFP-TEL-SAM, a high-affinity polymer, and negGFP-DGKδ-SAM, a low-affinity polymer. The high-affinity polymer shows almost entirely rod-shaped structures, while the low-affinity polymer shows occasional rod-shaped forms. (C) Lysates containing expressed negGFP-hSAM were run on a native gel and the bands visualized using by fluorescence.
Not all polymers are as stable as TEL-SAM and may be more susceptible to steric and charge repulsion imparted by the negGFP fusion. To test the applicability of the negGFP fusion to weak SAM polymers, we tested the approach on the human DGKδ-SAM polymer, which has an intersubunit Kd of roughly 6 μM,7 more than 1000-fold weaker than TEL-SAM. EM images of purified negGFP-DGKδ-WT are shown in Figure 2(B). The negGFP-DGKδ-WT polymers are not as easily identified as they are shorter than the TEL polymers and infrequently spread across the grid. Many round particles of the same diameter as the polymers can be seen in the background of the grid and may be single turns or very short polymers laying on the grid perpendicular to the polymer axis. Nevertheless, scattered rods are clearly visible on the grid. Thus, negGFP can be used to solubilize SAM polymers and it does not completely eliminate the ability of even weak polymers to self-associate.
Testing a rapid native gel screen for SAM polymers
To rapidly screen for potential SAM polymers, we used native gel analysis. The approach makes use of two key features of negGFP fusions: (1) the extreme negative charge ensures that virtually all proteins migrate toward the cathode and (2) the green fluorescence can be used to monitor the migration of the fusion protein in crude lysates. Figure 2(C) shows the migration of negGFP fusions containing TEL-WT, the monomeric mutants TEL-V80E and TEL-A61D, as well as the mixture of TEL-V80E and TEL-A61D that should form a dimer. These controls demonstrate that monomeric and dimeric populations can be easily distinguished. The polymeric negGFP-TEL-WT exhibited a dramatic shift in migration and largely remains in the gel well. These results indicate that native gels can readily identify highly stable polymers like TEL-SAM in crude cell lysates.
Can weaker polymers still be identified by native gel analysis? Native gel analysis of negGFP fusions containing the weakly associating DGKδ-WT and two DGKδ monomeric mutants, E35G (ML surface) and V52E (EH surface), is shown in Figure 2(C). Like TEL, the DGKδ monomer fusions could easily be distinguished from the DGK dimer created by mixing E35G and V52E mutants, indicating that the negGFP fusion does not completely disrupt subunit association. negGFP-DGKδ-WT, unlike negGFP-TEL-WT, entered the gel but displayed a retarded migration relative to the monomer and dimer controls. In addition, both the dimer and polymer bands of DGKδ-SAM are more diffuse than seen for TEL-SAM, which may reflect subunit dissociation during electrophoresis or heterogeneity in the starting population. Nevertheless, the results indicate that native gels can clearly identify possible polymers, even one as weakly assembled as DGKδ-SAM.
Screening human SAM domains for possible polymer formation
To screen for new SAM polymers in the human proteome, we attempted to generate negGFP fusions from as many hSAM domains as possible. hSAM domains were identified through the SMART database22,23 and annotations in the NCBI gene database. Most of the identified hSAM domains were obtained by PCR amplification from the Stratagene MegaMan Human Transcriptome Library. For some hSAM domains that could not be obtained from the cDNA library, we were able to obtain clones from a variety of sources. Several proteins contain more than one SAM domain placed in tandem within the protein. For these proteins, we attempted to clone the tandem SAMs both in isolation and as a single, combined construct. We initially targeted 114 unique hSAM domains from 92 proteins in the human genome. Of the 114 targets, we were able to prepare negGFP-hSAM fusions for 96 unique hSAM domains from 77 proteins.
All the negGFP-hSAM fusions were expressed in E. coli and the lysates analyzed by native gel electrophoresis. negGFP-DGKδ-V52E and negGFP-DGKδ-WT were used in each gel as monomer and polymer controls. As DGKδ-SAM is the weakest hSAM polymer interaction characterized thus far, most other polymeric SAMs should migrate more slowly through the gel. Concentrations were adjusted to approximately the same level by measuring fluorescence intensity. Some SAM domains that were expressed at low levels were partially purified, so they could be concentrated before loading on a gel. (Supporting Information Fig. S1, boxed section). We were unable to analyze the hSAM domains from five proteins because they were either proteolyzed or poorly expressed. Indeed, most constructs containing a single SAM derived from a tandem SAM protein could not be stably expressed in isolation, suggesting that most tandem SAMs form a structural unit. We were able to screen the hSAMs (and tandem hSAMs) from 72 separate proteins for their ability to polymerize. The results, summarized in Figure 3, indicate that close to half of the members of the hSAM domain family may form polymers or homo-oligomers.
Figure 3.
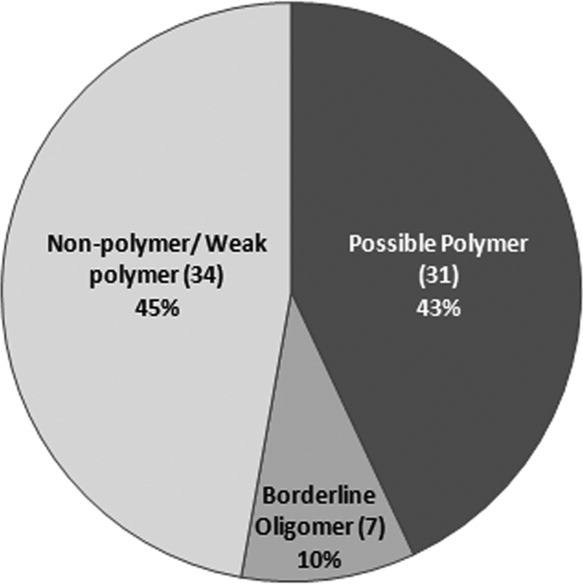
Summary of the behavior of the negGFP-hSAM fusions on native gels. SAMs and tandem-SAM constructs with unique sequences were categorized by their oligomerization ability in the Native gel assay. Single hSAM constructs derived from a tandem SAM context were not included in this analysis. Samples that were either proteolyzed or poorly expressed in lysate but classifiable in Native gel analysis of purified protein were categorized according to their behavior in the latter assay.
All the hSAMs that have been previously characterized to form polymers or have homologues that form polymers behaved as aggregates in the native gel analysis. This group includes diacylglycerol kinase δ (DGKδ),7 diacylglycerol kinase η (DGKη), translocation-ETS-leukemia (TEL/ETV6),3 translocation-ETS-leukemia 2 (TEL2), all polyhomeotic homologues,6 sex comb on midleg homologue 1 (SCMH1),6 Shank-2, Shank-3,8 Tankyrase-1,24 Tankyrase-2, and SCM-like with Four MBT domains 1 (SFMBT1) (Supporting Information Table 1). Ephrin receptor B2 (EPHB2) behaved as a monomer, but this is expected as experimentally observed oligomer forms were only found to occur at very high concentration.18 Likewise, hSAMs that have been previously shown to be monomers, such as V-ETS Erythroblastosis virus E26 oncogene homolog 1 (ETS1),25 behaved as monomers (Supporting Information Table 1). These hSAMs, therefore, serve as additional controls for the technique and indicate that the vast majority of bone fide SAM polymers can be identified if they express at sufficient levels, without false positives.
Overall, hSAMs from 30 proteins displayed a migration pattern in the gel consistent with a polymer, 17 of which had not been previously identified as polymer-forming hSAMs. This group included Ankyrin repeat and sterile alpha motif domain containing 3 (ANKS3), Atherin, BicaudalC1, CASK interacting protein 1 (Caskin1), CASK interacting protein 2 (Caskin2), Kazrin, L(3)mbt-like 3 (L3MBTL3), L(3)mbt-like 4 (L3MBTL4), lipopolysaccharide binding protein (LBP), Liprinβ1, Liprinβ2, sterile alpha motif domain containing 3 (SAMD3), sterile alpha motif domain containing 8 (SAMD8), sterile alpha motif domain containing 9 (SAMD9), sterile alpha motif domain containing 13 (SAMD13), sterile alpha and TIR motif containing 1 (SARM1), and stromal interaction molecule 2 (STIM2).
Identification of polymeric hSAM domains by EM
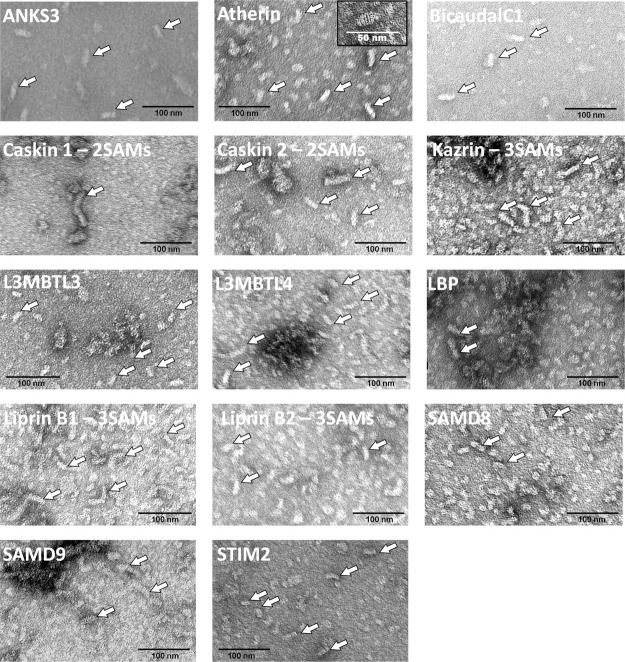
To distinguish polymers from amorphous aggregates, we examined selected negGFP-hSAM fusions by negative-stain EM. As we were interested in new polymers, we did not further examine any hSAM domains for which a close homolog had been previously shown to be polymeric (see Supporting Information Table 1), leaving 17 hSAM domains to be characterized. Of these, we found 14 hSAMs that appear able to form polymers rather than amorphous aggregates as judged by the presence of elongated rods. The SAM domains that may be polymer forming are ANKS3, Atherin, BicaudalC1, Caskin1, Caskin2, Kazrin, L3MBTL3, L3MBTL4, LBP, Liprinβ1, Liprinβ2, SAMD8, SAMD9, and STIM2. (Fig. 4) The hSAM domains that formed aggregates by native gel analysis but did not form obvious polymers when examined by EM were SAMD3, SAMD13, and SARM1 (data not shown).
Figure 4.
Novel polymer-forming hSAMs identified by electron microscopy. EM images representative of each population of purified negGFP-hSAM proteins are shown. Examples of polymers are indicated by white arrows. The inset of the Atherin image shows a single polymer at higher resolution in which striations are present that may be indicative of helical repeats.
We were intrigued to find that a number of proteins that contain two or three tandem SAM domains formed likely polymeric structures. The tandem hSAM proteins were Caskin 1, Caskin 2, Kazrin, Liprin β1, and Liprin β2. So far, all previously characterized SAM domain polymers are made from single SAM domain units. We, therefore, chose one of these, Caskin1, which contains two tandem SAM domains to characterize further. We were able to obtain a crystal structure of the Caskin1 tandem SAM domains, revealing a novel helical polymer structure, containing four tandem SAM domains per turn (submitted for publication).
Discussion
We have identified a number of potential SAM polymers which provides insight into how these proteins might function. We previously developed a prediction algorithm for possible polymer-forming SAM domains.19 Of the 51 human, SAM domains predicted previously that were also examined here, 46 are consistent with our experimental findings (the SAM domains used for algorithm training excluded). The few discrepancies included several SAMs predicted to be nonpolymers: SAMHD1 and SEC23IP, which behaved as borderline oligomers, and SAMD8, which we identified as a polymeric SAM. Additional discrepancies included were two SAMs predicted to be polymers: c14orf174, which behaved as a borderline oligomer, and INPPL1, which behaved as a nonpolymer/weak polymer. While it is not possible to rigorously validate polymer formation for any protein without considerably more work, our results provide a much stronger working hypothesis than can be had from sequence analysis alone. Some interesting possibilities emerged.
Atherin is a protein found in atherosclerotic lesions in vessels where it potentially acts to anchor low-density lipoproteins (LDL) to the arterial wall.26 We found the Atherin hSAM to form polymers suggesting that SAM-mediated polymers could act as a physical scaffold for atherosclerotic plaques.
Bicaudal C1, which contains another SAM from our list of likely novel polymers, is a conserved RNA-binding protein that has been linked to polycystic kidney disease (PKD). Mutation of the Bicaudal C1 gene which leads to truncation of the protein short of the SAM domain gives rise to a mouse model of PKD.27 Functional Bicaudal C1 containing the SAM domain is required for the alignment of renal primary cilia such that directional fluid flow can be achieved. Bicaudal C1-hSAM concentrates the protein in cytoplasmic RNA-processing bodies, called P-bodies, where it is thought to potentially play a role in downregulation of canonical WNT signaling and thereby facilitate the formation of the planar cell polarity, required for coordination of cilia orientation.28 Localization of the Bicaudal C1 protein at P-bodies could conceivably be organized by SAM-mediated polymers such as those observed in this study. Interestingly, Bicaudal C1 has been shown to physically associate with another SAM-containing protein with a role in PKD, ANKS6. ANKS6 has been shown to self-associate in a manner dependent on both its SAM and K homology domains.29 While the ANKS6 hSAM was classified as a borderline oligomer in this study due to its weak association in the native gel assay, formation of strong polymers might ensue with additional interactions mediated by the K homology domain. As the Bicaudal C1–ANKS6 association is dependent on their respective ankyrin repeats but not their SAMs, the ankyrin repeat binding sites could conceivably join these proteins in a polymer network of Bicaudal C1 and ANKS6 hSAM homopolymers. This topology would be distinct from previously proposed SAM copolymers that are formed by polycomb group proteins.6
STIM1 and 2 are type-I transmembrane proteins that play an essential role in calcium homeostasis and signaling. STIM1 provides a critical role in store-operated calcium entry (SOCE) by sensing calcium depletion in the lumen of the ER and transducing that signal to factors at the plasma membrane to allow influx of extracellular calcium.30,31 STIM2 is thought to play roles in maintaining basal calcium levels as well as potentially regulating SOCE.32 The ER-signal peptide, calcium-sensing EF domain, and SAM domain reside within the ER lumen while the remainder of the protein projects into the cytosol.33 Previous work showed STIM1 and STIM2 aggregate in a SAM-dependant manner in response to calcium depletion, a step required for relocalization of STIM1 into puncta in close proximity to the plasma membrane.34,35 This puncta formation is required for signal transduction and the eventual opening of Ca++ channels.36 A crystal structure of a construct containing the STIM1 EF and SAM domains shows that the two domains pack together when the EF hand is in the Ca++-loaded state.37 Oligomerization of both STIM1 and STIM2 appears to be linked to partial unfolding35,38 suggesting the possibility that the loss of Ca++ binding might potentially destabilize all or part of the EF domain exposing a surface of the SAM domain required for oligomer formation. The rod-shaped structures we observed suggest that STIM2 assembly may be well organized rather than just simple aggregation of unfolded polypeptide chains.
A number of proteins containing our newly identified polymeric hSAMs are known scaffolding proteins such as Liprin β1 and β2, Kazrin, Caskin1 and Caskin2. Like other characterized polymeric SAM domains in scaffolding proteins, polymerization provides a way to organize large multiprotein complexes. Unlike previously characterized SAMs found within scaffolding proteins, however, all of these proteins contain multiple, adjacent SAMs. Thus for the first time, we have discovered likely polymeric forms of tandem SAM domains. In one case, Caskin1, we followed up on our findings from this screen and obtained a new polymeric SAM structure made from two tandem SAM domains (submitted for publication). Like the Caskin1 example, we hope that our screen will provide a starting point for further investigations of the structural and functional implications of SAM polymerization.
Materials and Methods
Cloning
A clone for negGFP including an N-terminal 6xHis tag20 was kindly provided by the Liu lab (Harvard University). The negGFP sequence including the His tag was cloned into the pBAD-HisA vector (Invitrogen) using NcoI and KpnI sites. Next, an oligomer cassette was inserted downstream of the negGFP sequence using KpnI and HindIII restriction sites. The cassette included the sequence for a Gly-Ser linker (GGSGGS) followed by the sequences for the following combination of restriction sites: NotI–AsiSI-PISceI–Asc-FseI.
All hSAMs were identified by either the SMART database or through annotations included in the NCBI database. The SAM domains were then PCR amplified from the MegaMan Human Transcriptome Library (Stratagene) and the correct clones validated by sequencing. Platinum PCR Mix (Invitrogen) was used initially to amplify constructs from the cDNA library. Clones that were not amplified with Platinum PCR mix could often be amplified using Phusion polymerase (New England Biolabs). Several hSAMs were cloned from alternative sources of DNA. Constructs containing the Caskin 2, Liprin α2, Liprin α3, Liprin α4, Liprin β1, and Liprin β2 sequences were obtained from Open Biosystems. A Liprin α1 construct was kindly provided by Michael Streuli. The Caskin 1 (KIAA1306) construct was provided by the Kazusa DNA Research Institute. KazrinE was obtained from ATCC (9891927). Finally, SAM domains were inserted in-frame into a pBAD-HisA vector containing the negGFP sequences using the NotI and FseI sites.
negGFP-hSAM Lysate preparation
The negGFP-hSAM constructs were transformed into ARI814 cells.39 LB media (50 mL) containing 100 μg/mL ampicillin was inoculated with 0.5 mL of a saturated overnight culture grown in the same media and incubated with shaking at 37°C until the cell density reached an OD of 0.8. Cells were then transferred to 16°C, induced with 0.2% arabinose, and incubated for an additional 12 h. After harvesting by centrifugation, cells were resuspended in 0.5 mL of 20 mM Tris (pH 7.5), 1M NaCl, 1 mM TCEP, 5 mM MgCl2 buffer containing lysozyme (5 mg/mL), DnaseI (20 μg/mL), 0.5 mM PMSF, and half a tablet of complete mini protease inhibitor (Roche). Cells were subjected to three rounds of freeze-thaw followed by two 10-s rounds of sonication. The cell lysate was centrifuged at 13,000g for 20 min and the pellet discarded. Lysate fluorescence was measured by transferring 200 μL of lysate into a 96-well clear bottom, black-sided plate (Nunc). negGFP fluorescence was monitored on a Molecular Devices Spectramax M5 plate reader using an excitation wavelength of 488 nm, an emission wavelength of 510 nm, and a cutoff filter of 515 nm.
Gel filtration-fluorescence assay
Cell lysate or purified protein (0.5 mL) was loaded onto a Superdex200-10/300GL gel filtration column (Amersham Biosciences) at 0.5 mL/min in 20 mM Tris (pH 7.5), 500 mL NaCl. 0.25 mL fractions were collected, and 0.2 mL of each fraction was transferred to 96-well clear bottom, black-sided plates (Nunc). Fluorescence was monitored on a Molecular Devices Spectramax M5 plate reader using an excitation wavelength of 488 nm, an emission wavelength of 510 nm, and a cutoff filter of 515 nm.
Native gels
Cell lysate (22.5 μL) prepared as described above was mixed with 7.5 μL 4X RunBlue Native Sample Buffer (Expedeon) and loaded onto a 20% RunBlue 12-well Native gel (Expedeon). The amounts of lysate samples run in the control gel were adjusted such that each sample had an equal fluorescence for comparison purposes. Gels were run in 40 mM Tricine, 60 mM Tris buffer for 20 h at 4°C and visualized on a Bio-Rad Molecular Imager FX Pro-Plus using an excitation wavelength of 488 nm and an emission wavelength of 510 nm. Possibly polymeric SAMs were defined as those that displayed a diffuse and retarded migration compared to the monomeric DGK control. Borderline oligomers were defined as those that had either a sharply defined band at a position in the gel that was significantly higher than the monomeric control or a sharp band with a slight diffuse trail. Monomeric/weak polymer SAMs were defined as those with a sharp band roughly consistent with the monomer control (some variation is to be expected as not all constructs are the same molecular weight.
Protein purification
Cultures (4 L) of ARI814 cells expressing each construct were grown and lysed in 35 mL of lysis buffer as described above, except that 10 mM imidazole was included in the lysis buffer. Cell lysate was incubated with 2 mL of Ni-NTA Superflow resin (Qiagen) for 1 h at 4°C and washed with 100 mL of 20 mM Tris (pH 7.5), 1M NaCl, 1 mM TCEP, and 10 mM imidazole followed by 150 mL of 20 mM Tris (pH 7.5), 1M NaCl, 1 mM TCEP, and 20 mM imidazole. Protein was eluted with 15 mL of 20 mM Tris (pH 7.5), 1M NaCl, 1 mM TCEP, 200 mM imidazole and then dialyzed by two successive transfers into 2 L of 20 mM Tris (pH 7.5), 300 mM NaCl, 1 mM TCEP. Full-length negGFP-hSAM protein samples contaminated with free, proteolyzed negGFP were subjected to a second, gel-filtration purification step. Protein samples were concentrated to 500 μL using an Amicon Ultra(-10) centrifugal filter concentrator unit (Millipore) and were loaded onto a Superdex200-10/300GL gel filtration column (Amersham Biosciences) at 0.5 mL/min in 20 mM Tris pH 7.5/300 mM NaCl/1 mM TCEP. 0.25 mL fractions were collected and assayed for GFP fluorescence as described above. Samples containing GFP fluorescence at an elution volume consistent with the molecular weight for a full-length monomer or higher were pooled. The final pure protein samples were concentrated as described until protein began to fall out of solution or a final volume of 200 μL was achieved.
Electron microscopy
Carbon-coated parlodion support films mounted on copper grids were made hydrophilic immediately before use. Approximately 3 μL of each protein sample was applied to separate grids and allowed to adhere for several minutes. Grids were rinsed with distilled water and negatively stained with 1% uranyl acetate. Samples were examined in a Hitachi H-7000 electron microscope at an accelerating voltage of 75 kV.
Acknowledgments
The authors thank Heedeok Hong, Robert Jefferson, Tyler Korman, and Ryan Stafford for helpful comments on the manuscript.
Supplementary material
References
- 1.Pawson T, Raina M, Nash P. Interaction domains: from simple binding events to complex cellular behavior. FEBS Lett. 2002;513:2–10. doi: 10.1016/s0014-5793(01)03292-6. [DOI] [PubMed] [Google Scholar]
- 2.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 3.Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, Faham S, Bowie JU. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 2001;20:4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311:531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- 5.Qiao F, Song H, Kim CA, Sawaya MR, Hunter JB, Gingery M, Rebay I, Courey AJ, Bowie JU. Derepression by depolymerization; structural insights into the regulation of Yan by Mae. Cell. 2004;118:163–173. doi: 10.1016/j.cell.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Kim CA, Sawaya MR, Cascio D, Kim W, Bowie JU. Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J Biol Chem. 2005;280:27769–27775. doi: 10.1074/jbc.M503055200. [DOI] [PubMed] [Google Scholar]
- 7.Harada BT, Knight MJ, Imai S, Qiao F, Ramachander R, Sawaya MR, Gingery M, Sakane F, Bowie JU. Regulation of enzyme localization by polymerization: polymer formation by the SAM domain of diacylglycerol kinase delta1. Structure. 2008;16:380–387. doi: 10.1016/j.str.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 8.Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311:531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- 9.Douziech M, Sahmi M, Laberge G, Therrien M. A KSR/CNK complex mediated by HYP, a novel SAM domain-containing protein, regulates RAS-dependent RAF activation in Drosophila. Genes Dev. 2006;20:807–819. doi: 10.1101/gad.1390406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leone M, Cellitti J, Pellecchia M. NMR studies of a heterotypic Sam-Sam domain association: the interaction between the lipid phosphatase Ship2 and the EphA2 receptor. Biochemistry. 2008;47:12721–12728. doi: 10.1021/bi801713f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajakulendran T, Sahmi M, Kurinov I, Tyers M, Therrien M, Sicheri F. CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling. Proc Natl Acad Sci USA. 2008;105:2836–2841. doi: 10.1073/pnas.0709705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachander R, Kim CA, Phillips ML, Mackereth CD, Thanos CD, McIntosh LP, Bowie JU. Oligomerization-dependent association of the SAM domains from Schizosaccharomyces pombe Byr2 and Ste4. J Biol Chem. 2002;277:39585–39593. doi: 10.1074/jbc.M207273200. [DOI] [PubMed] [Google Scholar]
- 13.Qiao F, Harada B, Song H, Whitelegge J, Courey AJ, Bowie JU. Mae inhibits pointed-P2 transcriptional activity by blocking its MAPK docking site. EMBO J. 2006;25:70–79. doi: 10.1038/sj.emboj.7600924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson PE, Donaldson LW. RNA recognition by the Vts1p SAM domain. Nat Struct Mol Biol. 2006;13:177–178. doi: 10.1038/nsmb1039. [DOI] [PubMed] [Google Scholar]
- 15.Bhunia A, Domadia PN, Mohanram H, Bhattacharjya S. NMR structural studies of the Ste11 SAM domain in the dodecyl phosphocholine micelle. Proteins. 2009;74:328–343. doi: 10.1002/prot.22166. [DOI] [PubMed] [Google Scholar]
- 16.De Rycker M, Venkatesan RN, Wei C, Price CM. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem J. 2003;372:87–96. doi: 10.1042/BJ20021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smalla M, Schmieder P, Kelly M, Ter Laak A, Krause G, Ball L, Wahl M, Bork P, Oschkinat H. Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites. Protein Sci. 1999;8:1954–1961. doi: 10.1110/ps.8.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thanos CD, Goodwill KE, Bowie JU. Oligomeric structure of the human EphB2 receptor SAM domain. Science. 1999;283:833–836. doi: 10.1126/science.283.5403.833. [DOI] [PubMed] [Google Scholar]
- 19.Meruelo AD, Bowie JU. Identifying polymer-forming SAM domains. Proteins. 2009;74:1–5. doi: 10.1002/prot.22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran HH, Kim CA, Faham S, Siddall M, Bowie JU. Native interface of the SAM domain polymer of TEL. BMC Struct Biol. 2:5–5. doi: 10.1186/1472-6807-2-5. (YEAR) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Rycker M, Price CM. Tankyrase polymerization is controlled by its sterile alpha motif and poly (ADP-ribose) polymerase domains. Mol Cell Biol. 2004;24:9802–9812. doi: 10.1128/MCB.24.22.9802-9812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson ML, Kang H, Lee GM, Blaszczak AG, Lau DW, McIntosh LP, Graves BJ. Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP. Proc Natl Acad Sci USA. 2010;107:10026–10031. doi: 10.1073/pnas.0915137107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lees AM, Deconinck AE, Campbell BD, Lees RS. Athera newly identified, lesion-specific, LDL-binding protein in human atherosclerosis. Atherosclerosis. 2005;182:219–230. doi: 10.1016/j.atherosclerosis.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 27.Cogswell C, Price SJ, Hou X, Guay-Woodford LM, Flaherty L, Bryda EC. Positional cloning of jcpk/bpk locus of the mouse. Mamm Genome. 2003;14:242–249. doi: 10.1007/s00335-002-2241-0. [DOI] [PubMed] [Google Scholar]
- 28.Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, Beyer T, Blum M, Constam DB. Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development. 2009;136:3019–3030. doi: 10.1242/dev.038174. [DOI] [PubMed] [Google Scholar]
- 29.Stagner EE, Bouvrette DJ, Cheng J, Bryda EC. The polycystic kidney disease-related proteins Bicc1 and SamCystin interact. Biochem Biophys Res Commun. 2009;383:16–21. doi: 10.1016/j.bbrc.2009.03.113. [DOI] [PubMed] [Google Scholar]
- 30.Roos J, DiGregorio PJ, Yeromin AV, Ohlsen K, Lioudyno M, Zhang S, Safrina O, Kozak JA, Wagner SL, Cahalan MD, Veliçelebi G, Stauderman KA. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnstone LS, Graham SL, Dziadek MA. STIM proteins: integrators of signalling pathways in development, differentiation and disease. J Cell Mol Med. 2010;14:1890–1903. doi: 10.1111/j.1582-4934.2010.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams RT, Manji SS, Parker NJ, Hancock MS, Van Stekelenburg L, Eid JP, Senior PV, Kazenwadel JS, Shandala T, Saint R, Smith PJ, Dziadek MA. Identification and characterization of the STIM (stromal interaction molecule) gene family: coding for a novel class of transmembrane proteins. Biochem J. 2001;357:673–685. doi: 10.1042/0264-6021:3570673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stathopulos PB, Zheng L, Ikura M. Stromal interaction molecule (STIM) 1 and STIM2 calcium sensing regions exhibit distinct unfolding and oligomerization kinetics. J Biol Chem. 2009;284:728–732. doi: 10.1074/jbc.C800178200. [DOI] [PubMed] [Google Scholar]
- 36.Luik RM, Wu MM, Buchanan J, Lewis RS. The elementary unit of store-operated Ca2+ entry: local activation of CRAC channels by STIM1 at ER–plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stathopulos PB, Zheng L, Li G, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Stathopulos PB, Li G, Plevin MJ, Ames JB, Ikura M. Stored Ca2+ depletion-induced oligomerization of stromal interaction molecule 1 (STIM1) via the EF-SAM region. J Biol Chem. 2006;281:35855–35862. doi: 10.1074/jbc.M608247200. [DOI] [PubMed] [Google Scholar]
- 39.Schatz PJ, Cull MG, Martin EL, Gates CM. Screening of peptide libraries linked to lac repressor. Meth Enzymol. 1996;267:171–191. doi: 10.1016/s0076-6879(96)67012-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.