Abstract
c-Rel is a member of the nuclear factor κB (NF-κB) transcription factor family. Unlike other NF-κB proteins that are expressed in a variety of cell types, high levels of c-Rel expression are found primarily in B and T cells, with many c-Rel target genes involved in lymphoid cell growth and survival. In addition to c-Rel playing a major role in mammalian B and T cell function, the human c-rel gene (REL) is a susceptibility locus for certain autoimmune diseases such as arthritis, psoriasis, and celiac disease. The REL locus is also frequently altered (amplified, mutated, rearranged), and expression of REL is increased in a variety of B and T cell malignancies and, to a lesser extent, in other cancer types. Thus, agents that modulate REL activity may have therapeutic benefits for certain human cancers and chronic inflammatory diseases.
Keywords: c-Rel, REL, NF-κB, signal transduction, arthritis, cancer, B cells, T cells
The c-Rel transcription factor is a unique member of the vertebrate nuclear factor κB (NF-κB) family, primarily because of its pervasive and specific role in mammalian B and T cell differentiation and function, as well as in human disease. As such, c-Rel is of interest to many researchers and clinicians studying mammalian immunology and human immunological disorders, especially autoimmune diseases and hematopoietic cancers. This broad-based review describes the attributes of c-Rel structure, function, and regulation that confer it with its normal and pathological biological activities. In this review, c-Rel is used to refer generically to c-Rel proteins, whereas REL refers specifically to human c-Rel.
Structure of c-Rel and DNA-Binding Activity
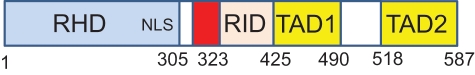
Human REL is a 587–amino acid protein (Fig. 1). Like other mammalian NF-κB proteins (i.e., NFκB1/p50, NFκB2/p52, RelA/p65, RelB), REL contains a highly conserved N-terminal DNA-binding/dimerization domain called the Rel homology domain (RHD).1 The C-terminal half of REL is composed of sequences that affect its ability to activate transcription: That is, 2 C-terminal transactivation subdomains (TAD1 and 2)2-4 are separated from the RHD by a transactivation inhibitory domain (RID).5
Figure 1.
Generalized structure of the human REL protein. The numbers below the figure indicate the limits of each domain. RHD = Rel homology domain; NLS = nuclear localization signal; RID = REL inhibitory domain; TAD1 = transactivation domain 1; TAD2 = transactivation domain 2. The red box indicates the approximate position of residues 308-330, encoded by exon 9, that are deleted in an alternatively spliced form of REL, which is overrepresented in many B lymphoma cell lines.5
REL has an optimal DNA-binding site preference for a target sequence that is slightly different from other NF-κB subunit homodimers. PCR-based site selection indicates that c-Rel has a broader target sequence recognition ability than p65 and p50, with 5′-NGGRN(A/T)TTCC-3′ identified as the optimal c-Rel binding sequence.6 The X-ray crystal structure of REL bound to a κB site (5′AGAAATTCC3′) from the IL-2 enhancer shows that c-Rel uses residues to bind DNA that are distinct from other NF-κB proteins, a property that may endow REL with its different target sequence preference. Interestingly, certain amino acids in these c-Rel-specific DNA-binding sequences are mutated in v-Rel,7 the viral oncogenic version of avian c-Rel. Mutagenesis studies also indicate that c-Rel has DNA-binding residues that are distinct from other NF-κB proteins.8 In most cells, REL exists either as a homodimer or a heterodimer with p50, but c-Rel can also form dimers with p65 and NFκB2.
Protein Modifications and Protein-Protein Interactions of c-Rel
A number of posttranslational modifications of c-Rel have been described. Within the RHD, c-Rel has been shown to undergo phosphorylation, acetylation, and, within the C-terminal sequences, phosphorylation and ubiquitination. In most cases, the functional relevance of these modifications is unclear. For example, PKA can phosphorylate a conserved site (R-R-X-S) in the c-Rel RHD domain,9,10 but whether and when such phosphorylation occurs in vivo is not known. Similarly, residues in the C-terminal half of c-Rel can be phosphorylated in vitro by IKKβ11-13 but have not been shown to be phosphorylated in vivo.13 Nevertheless, one of these IKK phosphorylation sites is mutated in some human B cell lymphomas.12 c-Rel has also been reported to be a substrate for NF-κB-inducing kinase (NIK)14 and a tyrosine kinase in myeloid cells following G-CSF stimulation.15 Although chicken c-Rel undergoes ubiquitin-mediated degradation due to modification of C-terminal lysines,16 the absence of lysine residues in the C-terminal half of REL may indicate that this is a species-specific mode of c-Rel regulation. Finally, a conserved Cys residue in a DNA-binding sequence of c-Rel allows for redox-sensitive DNA binding, where reduced REL binds DNA better than oxidized REL.17 The redox state of c-Rel also appears to affect its ability to be phosphorylated.17
c-Rel is also subject to posttranscriptional processing. For example, alternative splicing can remove part of the central transactivation inhibitory domain RID, which increases both the DNA-binding and transactivating activity of REL; of note, this alternatively spliced form of REL mRNA is overexpressed in B lymphoma cell lines.5 c-Rel can be a substrate for caspase-mediated cleavage, which renders it inactive,18 a process that may modulate its role as an anti-apoptotic regulator.
c-Rel has been shown to interact with several different types of proteins, in addition to those of the IκB and NF-κB families (Table 1). c-Rel-interacting proteins include transcription factors and nuclear shuttling proteins, as well as signaling proteins such as protein kinases (e.g., PKA, NIK, IKKβ). In addition, c-Rel has been reported to be important for the transport of certain proteins to the nucleus. For example, REL transports an alternative form of tumor suppressor p53 (ΔNP63α) into the nucleus in head and neck cancers to control proliferation19 and can transport the cell-surface protein CD40 to the nucleus in some B lymphoma cells.20 Finally, one study suggested that c-Rel may have a direct role in regulating DNA replication.21
Table 1.
Non-NF-κB/IκB Proteins That Can Interact with c-Rel
| Protein | Reference |
|---|---|
| Transcription factors | |
| C/EBP/β | 133 |
| IRF-8 | 134 |
| NFATc | 23 |
| Sp1 | 135 |
| Foxp3 | 136 |
| ΔNP63α | 19 |
| Androgen receptor | 137 |
| Estrogen receptor | 138, 139 |
| Transcription-mediating proteins | |
| TBP | 140 |
| TFIIB | 140 |
| p300/CBP | 108 |
| CAPERα | 141 |
| Cdk2/cyclinE | 142 |
| Aof1 | 143 |
| Nuclear import | |
| Importin alpha | 144 |
| Protein modifying/signaling | |
| BAFF-R | 145 |
| CD40 | 20 |
| Calmodulin | 146, 147 |
| Myotrophin/V1 | 148 |
| Cot/Tpl2 | 149 |
| ABIN2 | 149 |
| PAPOLA | 149 |
| Erbin | 150 |
| JNK1 | 151 |
| NIK | 152 |
| PKA-beta | 153 |
| Protein phosphatase X | 154 |
| Pin-1 | 155 |
| Viral protein | |
| HTLV-1 Tax | 156 |
Direct c-Rel Target Genes
Although there is considerable overlap in the DNA sequences and genes bound by the various NF-κB dimers, some genes do appear to be preferred targets for c-Rel, whether bound by c-Rel homodimers or by c-Rel heterodimers. Not surprisingly, based on the immune cell-specific functions of c-Rel (see below), many of these genes are involved in immune cell proliferation, survival (anti-apoptotic), or function (Table 2).
Table 2.
Partial List of c-Rel Target Genes
| Gene/Protein | Protein Function | References |
|---|---|---|
| Cell proliferation/cell growth | ||
| c-Rel | Transcription factor | 157 |
| c-Myc | Transcription factor | 44, 158 |
| IRF-4 | Transcription factor | 159 |
| E2F3a | Transcription factor | 57 |
| EP300 | Histone acetyltransferase | 160 |
| CD21 | Complement receptor | 161 |
| CD40 | Cell surface receptor | 22 |
| SFN | 14-3-3 protein | 160 |
| GM-CSF | Hematopoietic growth factor | 22, 162, 163 |
| TGFβ | Growth factor | 160 |
| IL-2 | Cytokine | 22, 41, 163 |
| IL-4 | T cell cytokine | 22, 164 |
| Apoptosis/cell survival | ||
| Bcl-2 | Anti-apoptotic | 54, 165 |
| Bfl-1/A1 | Anti-apoptotic | 60, 61 |
| Bcl-XL | Anti-apoptotic | 62 |
| miR-21 | Pro-apoptotic for β cells | 228 |
| Adhesion/cell architecture | ||
| ICAM-1 | Cell adhesion | 166, 167 |
| Selectin | Cell adhesion; binds sugars | 160 |
| MMP-1 | Metalloproteinase | 160 |
| EPHB2 | Receptor tyrosine kinase (repressed) | 168 |
| Immune cell function | ||
| Gamma1 | Ig heavy chain | 63 |
| Gamma4 | Ig heavy chain | 64 |
| TNF-α | Cytokine | 22 |
| IL-12 | p35 cytokine | 169 |
| IL-13 | Cytokine | 22 |
| IL-21 | Cytokine | 52 |
| IL-23 | Cytokine | 170 |
| CD40L | CD40 ligand | 23 |
| BlyS/BAFF | TNF-like cytokine | 171 |
| IP-10 | Chemokine | 22 |
| Ligp1 | GTPase | 22 |
| MIG | Macrophage cytokine | 22 |
| Foxp3 | Transcription factor | 172 |
| DNA repair/damage | ||
| ATM | Protein kinase | 160 |
| Claspin | Cell cycle kinase | 173 |
| Skp2 | S-phase kinase- associated factor | 174 |
Computational modeling of sequences upstream of c-Rel target genes suggests that the c-Rel regulatory module consists of multiple closely clustered c-Rel binding sites.22 Furthermore, c-Rel has been shown to bind cooperatively at certain promoters with other transcription factors; for example, c-Rel interacts with NFATc1 on the CD154 gene promoter.23
The C-terminal transactivation domain of c-Rel may have a specific role in regulating genes involved in B cell proliferation. For example, the addition of the v-Rel transactivation domain onto the RelA RHD creates a transforming protein in avian lymphoid cells, but the addition of the RelA transactivation domain onto the c-Rel RHD does not.24 Target genes of c-Rel that are involved in various developmental or disease processes are described in relevant sections of the text below.
Role of c-Rel in Normal Immune Cell Function
c-Rel is expressed at the highest levels in a wide variety of hemopoietic cells.25-28 Nevertheless, studies using knockout mice have shown that c-Rel is generally not essential for normal hematopoiesis and lymphopoiesis.29 Instead, c-Rel is required for a number of specialized functions in mature T and B cells. A summary of the phenotypes reported in studies using c-rel knockout mice is presented in Table 3.
Table 3.
Phenotypes of c-rel Mouse Knockouts
| Disrupted Genes | Phenotype | References |
|---|---|---|
| c-rel | Reduced B cell proliferation, survival, and antibody expression | 40 |
| Reduction in induced arthritis | 75 | |
| Sensitivity to Toxoplasma gondii | 175 | |
| Increased sepsis | 176 | |
| T cell defects | 49, 177 | |
| Dendritic cell defects | 178, 179 | |
| Reduction in induced diabetes | 180 | |
| Reduction in induced colitis | 181 | |
| Liver regeneration defect | 182 | |
| Reduced synaptic plasticity and memory | 183, 184 | |
| c-relΔCOOH | Increased B cell numbers in lymph nodes | 185 |
| c-rel/nfkb1 | B cell developmental defects, TLR signals impaired | 186 |
| Impaired CD4+ T cell responses | 50, 56, 179 | |
| c-rel/relA | Embryonic lethal, multiple hematopoietic defects | 44, 187 |
| c-rel/relA/tnf | Neonatal lethal, multiple epidermal defects | 76 |
c-Rel plays a key role in the development of regulatory T cells
Several recent studies have shown that c-Rel is required for the development in the thymus of CD4 regulatory T cells (Tregs).30-37 Tregs, which consist of a subset of T lymphocytes that express the lineage-specific transcription factor Foxp3, are required to suppress the activity of autoreactive T cells that escape negative selection and limit the duration and strength of normal T cell responses. In c-rel knockout mice (c-rel–/– mice), only ~15% of normal Treg numbers develop in the thymus.31 The reduced Treg population in c-rel–/– mice is due to a thymocyte-intrinsic defect that reflects the high level of c-Rel expression in Tregs.31 Although the total number of thymic Tregs is substantially reduced in c-rel–/– mice, the Tregs that do develop appear to possess normal T cell suppressive activity in culture and in vivo.31 This is consistent with the residual c-Rel-deficient Tregs expressing normal levels of Foxp3, which is essential for maintaining the Treg lineage-specific pattern of gene expression required for the immune-modulating functions of these cells.38 Importantly, the absence of autoimmune disease in c-rel–/– mice31 indicates that these remaining Tregs are sufficient to keep autoreactive T cells in check. Collectively, these findings point to c-Rel activity as being important, although not essential, for the development of Treg cells in the thymus. Once Treg development is complete, c-Rel appears to be dispensable for the function of mature Treg cells, which instead is orchestrated by Foxp3. The localization of c-Rel to the cytoplasm of mature Foxp3-positive Tregs is consistent with such a model.31
Treg development proceeds via a 2-step process.39 The initial step involves the T cell receptor (TCR) plus CD28-dependent generation of Treg precursors (CD25+GITR+Foxp3– CD4 thymocytes) from CD4+CD8+ double-positive (DP) thymocytes. These cells are then converted into functional Foxp3-positive Tregs by the action of the common γ-chain cytokines IL-2 and IL-15. The role of c-Rel in the generation of Treg precursors is almost certainly linked with the process of T cell selection. Normally, DP thymocytes, the precursors of almost all T cells (including Tregs), receive a TCR signal, the strength of which dictates their developmental fate. DP thymocytes receiving antigen-dependent TCR signals that are deemed to be either too weak or too strong to be compatible with normal T cell function are eliminated during positive and negative selection, respectively, by apoptosis. Those cells expressing a TCR that delivers an antigen signal to developing thymocytes within a tolerated range are able to complete the differentiation process. Although c-Rel is activated by TCR and CD28 signals in mature conventional CD4 T cells,40 it remains to be determined whether the reduction in Treg precursor numbers in c-rel–/– mice is due to impaired signaling downstream of the TCR, CD28, or both receptors during selection. The reduction in Treg precursor numbers in c-rel–/– mice suggests that either the positive or negative selection of these cells is impaired. Although Bcl-2 transgene expression can override cell death in wild-type mice arising from both modes of selection, it cannot rescue the loss of Treg cell numbers in c-rel–/– mice.31 That result indicates that instead of functioning in an anti-apoptotic capacity, c-Rel serves an instructive role promoting differentiation in response to antigen signals encountered during positive selection. Given that thymocytes directed toward Treg development tolerate stronger TCR signals during selection than conventional T cells, the Tregs that develop in the absence of c-Rel are likely to represent cells receiving a TCR signal at either the stronger or weaker end of the spectrum compatible with Treg differentiation.
The second step in Treg development, the induction of foxp3 transcription, is dependent on common γ-chain cytokine signals.39 Emerging evidence indicates that c-Rel directly controls the induction of foxp3 transcription during Treg differentiation. Within the foxp3 locus, there are c-Rel binding sites in the promoter,32,33,36 in a conserved element (CNS2) essential for stable foxp3 transcription,36 and in the CNS3 element, which like c-Rel is required for the development of the majority of Foxp3-positive Tregs in the thymus.36 The finding that Foxp3 expression in the residual c-rel–/– Tregs is normal31 suggests that c-Rel does not play a direct role in controlling the maintenance of foxp3 transcription but instead that c-Rel is required to establish a chromatin structure within the foxp3 gene that allows other transcription factors access to the locus. Given that c-Rel is not activated by the common γ-chain cytokine signals required for the induction of Foxp3 expression, any remodeling of the foxp3 gene by c-Rel must precede the formation of the Treg precursors. Many questions as to how c-Rel dictates Treg development remain to be answered, with the emerging data offering exciting insight into a previously unknown developmental role for c-Rel in mammalian cells that may provide mechanistic clues as to how c-Rel regulates other normal as well as pathological processes.
c-Rel has multiple roles in mature T cell growth, proliferation, and survival
Naive T cells exiting the thymus are sustained in the periphery through a combination of cytokine and weak TCR signals induced by self-antigens,40 the latter at levels below the threshold required to promote cell cycle entry. Typically, the activation of naive T cells involves antigen-presenting cells (APCs) such as dendritic cells delivering dual antigen/MHC and B7 co-stimulatory signals via the TCR and CD28, respectively, with CD28 optimizing TCR signals to reduce the threshold for antigen-dependent T cell activation. Depending on the type of cytokines that activated T cells receive from an APC, naive CD4 T cells can differentiate into Th1, Th2, Th17, and follicular helper T cells, each of which represents a different class of effector T cell.
Nuclear NF-κB activity in naive T cells is minimal, and c-Rel is only present at low levels in the cytoplasm of resting T cells.41 With the homeostatic maintenance of naive T cells unaffected by the absence of c-Rel,42 the preexisting c-Rel does not appear to serve a critical “housekeeping” function. Although TCR signals rapidly activate preexisting RelA in resting T cells, the cytoplasmic stores of c-Rel are not readily mobilized. This difference reflects the type of IκB proteins that bind to RelA versus c-Rel. RelA associates with IκBα, which is degraded in response to TCR signals, whereas c-Rel is bound to IκBβ, which is relatively resistant to degradation following TCR stimulation.43 Instead, TCR signals promote a delayed induction of c-Rel expression, resulting from NFAT-mediated activation of c-rel transcription.44,45 The different kinetics of c-Rel and RelA recruitment to the nucleus is indicative of a need to activate distinct NF-κB transcriptional targets in a temporally ordered manner during T cell activation. Why RelA, like NFAT, is in the first wave of transcription factors mobilized by TCR signals, whereas c-Rel is a secondary transcriptional effector, is still not known, and this temporal order contrasts with B cells, where c-Rel is among the first transcription factors activated by B cell antigen receptor (BCR) signaling.
IL-2, which is required for autocrine-dependent T cell proliferation, is induced by c-Rel in response to TCR and CD28 signaling.41,46 c-Rel controls il2 gene transcription by remodeling the chromatin structure of the locus to make it permissive for transcription,41 which is similar to the way that c-Rel is thought to control foxp3 transcription in Treg cells,36 suggesting that chromatin remodeling is a general feature of c-Rel-dependent transcriptional regulation. Although c-rel–/– T cells display a TCR plus CD28- dependent proliferative defect in culture due to a failure to express IL-2,46 the relatively normal IL-2-dependent T cell functions found in c-rel–/– mice in vivo suggest that IL-2 can be effectively regulated by c-Rel-independent pathways. Aside from its restricted role in controlling IL-2 transcription, c-Rel does not appear to serve a general, nonredundant function during T cell proliferation. Rather, the role of c-Rel in T cell expansion appears to be stimulus dependent. For example, c-Rel promotes the clonal expansion of Th1 effectors in a T cell–intrinsic manner during the immune response to Toxoplasma gondii 47 but is dispensable for influenza-specific CD8 T cell division in infected mice.48 This differential need for c-Rel during T cell activation presumably reflects the type of stimulatory signals invoked by specific pathogens. Notwithstanding the different stimulus-specific requirements for c-Rel in T cell proliferation, T cells (like B cells) use c-Rel in an overlapping manner with another NF-κB transcription factor, in this case RelA, to induce the c-Myc-dependent growth of mitogen-activated T cells.44 Although c-Myc rescues the c-rel–/–/rela–/– T cell growth defect, the failure of these cells to proliferate in response to TCR signals indicates that c-Rel and RelA play additional roles in the cell cycle that are independent of cell growth.
TCR-activated CD4 and CD8 T cells display a differential dependence on c-Rel for survival. Whereas TCR activation of PKCθ employs c-Rel to promote the survival of CD8 T cells, the PKCθ/c-Rel pathway is dispensable for CD4 T cell survival.49 However, c-Rel in combination with NFκB1 is critical for the survival of activated CD4 T cells.50 This emphasizes how c-Rel is differentially employed by CD4 and CD8 T cells to provide TCR-induced survival functions.40
c-Rel also controls the differentiation of CD4 Th cells during diverse immune responses that include central nervous system inflammation, islet allograft rejection, and microbial challenge by using both T cell autonomous and nonautonomous mechanisms.29 For example, Th1 cell development in experimental autoimmune encephalopathy involves c-Rel-induced expression of IL-12 by APC, yet in T. gondii infection, this aspect of Th1 differentiation is dispensable, with c-Rel instead serving a T cell–intrinsic role. Follicular helper (FH) T cells are required for the formation and maintenance of germinal centers and play a key role in plasma cell and memory B cell differentiation.51 IL-21, a cytokine produced by activated CD4 T cells that is important for the expansion and differentiation of FH T cells, requires c-Rel for the transcription of the il21 gene.52 Consistent with this finding, c-rel–/– mice exhibit defective FH T cell development and germinal center formation.52 Although IL-21 administration rescues the defect in development of c-rel –/– FH T cells, germinal center formation in c-rel–/– mice remains impaired,52 indicating an IL-21-independent role for c-Rel in the formation of these crucial secondary lymphoid structures.
Finally, c-Rel is important in the regulation of cytokine expression by activated T cells. In addition to promoting the T cell–specific expression of IL-2, GM-CSF, IL-3, IFN-γ, and IL-21 (Table 2), in naive CD4 T cells exposed to inflammatory cytokines, an exchange of the IκB factor that tethers c-Rel in the cytoplasm from IκBβ to IκBα creates a T cell that expresses cytokines at a faster rate and in larger amounts during subsequent TCR activation.53 This c-Rel priming mechanism that permits a T cell to respond rapidly and efficiently in an antigen-specific manner may serve as a temporary stop gap role during an immune response while a more robust T effector response has time to evolve.
c-Rel plays a role in activated B cell growth, proliferation, and survival
In keeping with c-Rel being dispensable for the antigen-independent B cell development that occurs in the bone marrow,54 c-Rel levels are low in B cell precursors, and p50/RelA is the major NF-κB dimer found in these cells.55 However, during the developmental transition from a pre–B cell to a naive mature B lymphocyte, an up-regulation of c-Rel and p50 expression causes p50/c-Rel to become the predominant NF-κB dimer in peripheral IgM-positive B lymphocytes. This shift to c-Rel-dominated NF-κB heterodimers in mature B cells highlights the importance of c-Rel in the events linked to the immune activation and differentiation of mature peripheral B cells.
B cell activation in response to antigens, Toll-like receptor (TLR) ligands, or T cell (CD40 ligand) signals typically promotes cell cycle entry and cell division, which are accompanied by isotype switching and antibody secretion. Entry of a mature B lymphocyte into the cell cycle requires a quiescent cell to initiate and coordinate multiple biochemical processes. If a B cell is not primed to enter the cell cycle when it receives an activation signal, then programmed cell death appears to be initiated as a default option to eliminate any inappropriately stimulated cells. Indeed, upwards of 30% of normal B cells undergo apoptosis following BCR engagement,56 suggesting that, at any given time, a significant proportion of quiescent B cells are not correctly poised to respond to mitogenic signals. To counteract this situation, it has been proposed that activated B cells engage a survival pathway that provides a window of protection from apoptosis to those cells that are able to be biochemically or metabolically reconfigured to successfully enter the cell cycle.
In mature B cells, c-Rel serves a dual role of promoting cell division and survival when these cells are activated.29 Although BCR, CD40, TLR4, and TLR9 signals all rapidly mobilize c-Rel dimers in resting B cells,40 studies with c-rel–/– mice have established that the requirement of c-Rel for mitogen-induced B cell division and survival is stimulus dependent.56 Whereas c-Rel is largely dispensable for promoting these functions in TLR9-stimulated B lymphocytes (S. Gerondakis, unpublished results), c-Rel is essential for these roles in antigen receptor–activated cells.56 c-Rel has two distinct cell cycle functions in B cells. In BCR-activated cells, c-Rel promotes the transition from G1- to S-phase.56 The genes that c-Rel directly regulates to control this phase of the cycle remain to be determined, although c-Rel induction of transcription factor E2F3a and cyclin E have been proposed to play a role in G1-to-S phase progression.57,58 c-Rel also promotes cell growth during G1 by up-regulating c-myc transcription,59 a step associated with ribosome biosynthesis that is essential for subsequent cell division. Once BCR-activated B cells enter S-phase, c-Rel appears to be dispensable for progression through the remainder of the cell cycle.56 Although the combined roles of c-Rel and p50 in B cell growth appear to be a common requirement for all mitogenic signals,59 the need for c-Rel to promote S-phase entry is more selective, with its importance in the hierarchy of mitogenic stimuli headed by BCR signaling.56
In the absence of c-Rel, mitogen-activated B cells display elevated levels of apoptosis.56,60 c-Rel promotes the survival of these cells by inhibiting a cell intrinsic death pathway through the direct transcriptional induction of genes encoding the Bcl-2 family pro-survival proteins A1/Blf-160,61 and Bcl-xL.60-62 With Bcl-2 transgene expression able to block the apoptosis of activated c-rel–/– B cells,56 coexpression of the closely related pro-survival proteins A1 and Bcl-XL in activated B cells could represent a fail-safe mechanism. However, enforced expression of A1 alone only confers partial protection to BCR-activated c-rel–/– cells,60 indicating that A1 and Bcl-xL serve distinct survival roles. That A1 and Bcl-xL have distinct roles in BCR-activated cell survival is supported by their kinetics of induction in these cells. A1 expression is rapidly induced with kinetics that coincide with the initial nuclear induction of c-Rel, whereas Bcl-xL expression is delayed, instead tracking with the second wave of nuclear c-Rel expression observed in mitogen-activated B cells.55 Different survival roles for A1 and Bcl-xL in B cell activation could involve A1 protecting cells from apoptosis during cell cycle entry (G0-to-G1 transition), whereas Bcl-xL may protect B cells later in the cell cycle or during DNA rearrangement events linked to isotype switching.
Mitogenic activation, in conjunction with cytokine signals such as IL-4 or IFN-γ, promotes isotype switching in a process whereby the assembled VH gene is expressed in conjunction with Cµ and different CH regions. This process uses nonhomologous DNA rearrangement between regions of repetitive sequence (switch or S-regions) that flank Cµ and downstream CH genes. TLR4 and cytokine signals specifically target different CH genes for rearrangement by using transcription to allow the switch recombination machinery access to the S-regions upstream of targeted CH loci. A 3′IgH enhancer located downstream of the CH locus also controls isotype switching, albeit via mechanisms that remain to be determined. c-Rel is necessary for efficient switching to IgG1 and IgE,46 with c-Rel promoting S-region transcription prior to DNA rearrangement.63,64 c-Rel interaction with the 3′IgH enhancer65 may also contribute to the efficiency of the switching process. Although DNA rearrangement during switching depends on B cell proliferation, a general role for c-Rel in B cell division is difficult to reconcile with the differential impact that the loss of c-Rel has on switching to specific isotypes. However, a genome-wide scan has shown that c-Rel is enriched on DNA at sites of recombination.66
Role of c-Rel in Human Disease
REL as a susceptibility locus in human immune diseases
Multiple reports have recently linked SNPs (single-nucleotide polymorphisms) within or near the REL locus with an increased susceptibility to certain human autoimmune diseases and cancer. These REL-associated diseases include rheumatoid arthritis,67-69 celiac disease,70 psoriasis,71 ulcerative colitis,72 primary sclerosing cholangitis,73 and B cell lymphoma74 (Table 4).
Table 4.
Association of REL Alterations with Human Disease
| Disease | Changes |
|---|---|
| Celiac disease | SNPa |
| Inflammatory bowel disease | SNP |
| Psoriasis | SNP |
| Rheumatoid arthritis | SNP |
| Sclerlosing cholangitis | SNP |
| Leukemia/lymphoma | Gene amplification, chromosomal rearrangement, mutation, SNP |
| Oral carcinoma | Gene deletion, gene amplification |
| Metastatic lung cancer | Gene amplification |
SNP = single-nucleotide polymorphism associated with the disease.
The 2 REL polymorphisms associated with increased susceptibility to autoimmune disease are located within REL introns: one within intron 4 has been linked to rheumatoid arthritis and Crohn’s disease, whereas the SNP in intron 2 is associated with celiac disease. If these SNPs are relevant to these human diseases, their presence within REL introns implies that altered REL transcription or mRNA maturation increases susceptibility to autoimmune disorders. In one study of potential celiac disease patients, the REL polymorphism did correlate with increased REL protein expression.227 However, the ability to find a change in REL expression that is associated with these SNPs is complicated by not knowing the cell type or the stage in disease development (initiation or maintenance) for which a change in REL expression is important. For example, although c-Rel has been shown to be required in mouse models of arthritis,75 the relative contribution of c-Rel expression in immune infiltrates (T cells, neutrophils, and eosinophils) versus stromal cells is unclear. Even though c-Rel expression is perceived as being strictly linked with hematopoietic cells, it is also normally expressed at low levels in endothelial and epithelial cells. In the case of skin epithelium, the loss of NF-κB activity of which c-Rel is one component leads to the development of potent inflammatory skin lesions,76 highlighting the possibility that a loss or reduction of c-Rel expression in certain cell types, including epithelial cells, increases susceptibility to autoimmune disease.
If these REL polymorphisms do alter REL protein expression, one might guess that their impact on autoimmune disease is due to a change in the expression of REL target genes or to the altered expression of non-REL targets arising from an overall imbalance in the NF-κB complexes caused by a change in REL levels. Furthermore, the penetrance of REL SNPs in the susceptibility to autoimmune disease will almost certainly be influenced by multiple loci. The finding that tnfaip3, which encodes the de-ubiquitinase A20, a key terminator of NF-κB signaling, is another rheumatoid arthritis and celiac disease–associated locus77 indicates that polymorphisms in the NF-κB pathway are likely to become increasingly important in human disease susceptibility.
Role of c-Rel in Lymphoid Cell Cancer
In vitro transformation of lymphoid cells
Based on the ability of the viral oncoprotein v-Rel to transform a variety of avian hematopoietic cells in culture,78 it is not surprising that chicken, mouse, and human c-Rel also have this transforming activity in primary chicken spleen cell cultures.79,80 Although full-length versions of chicken and human c-Rel can transform chicken lymphoid cells in vitro, their transforming activity is enhanced when either of the 2 C-terminal transactivation domains is removed.4,81 In contrast, removal of the entire transactivation domain of c-Rel or mutations that abolish DNA binding inactivates c-Rel’s transforming activity.4 Taken together, such studies indicate that c-Rel must bind to DNA and activate target genes to effect transformation and that chronic low-level induction of target gene expression—such as occurs with certain transactivation domain deletions—is optimal for transformation.4,82
In contrast to c-Rel, overexpression of p50, p52, RelB, RelA, and a constitutively active version of IKKβ is not transforming in avian lymphoid cells.82 However, substitution of the RelA transactivation domain with that of v-Rel can confer transforming activity onto the RelA-v-Rel hybrid, even though the reciprocal hybrid protein (v-Rel-RelA) cannot transform chicken lymphoid cells.82 This result suggests that the unique ability of c-Rel to transform avian lymphoid cells in culture resides in its C-terminal transactivation domain. Nevertheless, there is clearly flexibility in the type of transactivation domain that can induce avian lymphoid cell transformation when fused to an appropriate RHD, given that there is only about 10% sequence identity between the transactivation domains of chicken and human c-Rel and that the herpes virus activator protein VP16 can also substitute for the REL transactivation domain in transforming assays.4
There have been no reports of v-Rel or c-Rel being able to transform mouse lymphoid cells in culture. Overexpression of a human REL mutant (RELΔTAD1) missing one transactivation domain can, however, enhance the oncogenic properties of the human B lymphoma cell line BJAB.83 Furthermore, RELΔTAD1 changes the GCB-like mRNA expression profile of BJAB cells into one more similar to the ABC-like DLBCLs, including increasing the expression of REL targets such as BCL2, IRF4, and miR-155,83 as well as reducing the expression of the GCB-like marker CD10.84 These results provide strong support for REL contributing to oncogenesis in human B cells.
REL gene amplifications in human B and T cell malignancies
The REL gene is located at human chromosomal position 2p16.1-15, which is a common site of gene amplification in a variety of B and T cell malignancies (Table 5). Among B cell lymphomas, amplifications of REL have been found at relatively high frequency in Hodgkin’s lymphoma (~46%) and non-Hodgkin’s B cell lymphomas, such as diffuse large B cell lymphoma (DLBCL) (~15%), Burkitt’s lymphoma (~7%), and follicular (~17%) and mediastinal (~21%) lymphoma. At least for DLBCL, REL appears to be the only gene in the minimally amplified region.85 In one follicular lymphoma, double minute chromosome amplification of REL has also been reported.86 Overall, REL gene amplifications occur primarily in lymphomas with a mature B cell phenotype, which corresponds to the stage in development where c-Rel plays a major role in normal B cell function. Surprisingly, only one B lymphoma cell line has been reported to have REL gene amplification.87 REL gene amplifications have also been found in some leukemias (chronic lymphocytic leukemia) and T cell lymphomas (peripheral T cell, anaplastic large cell, natural killer).
Table 5.
REL Gene Amplification in Human Lymphoma/Leukemia
| Lymphoma/Leukemia Type | % Amplified REL (Total Cases Analyzed) | References |
|---|---|---|
| B cell malignancies | ||
| Hodgkin’s | 46 (175) | 101, 188-192 |
| DLBCL | 15 (1379) | 90, 91, 94, 118, 193-207 |
| Follicular | 17 (310) | 99, 193, 197, 208-212 |
| Primary mediastinal | 21 (136) | 91, 118, 199, 202, 213, 214 |
| Cutaneous B cell | 63 (31) | 215 |
| CLL | 5 (525) | 193, 211, 216-219 |
| Lymphoplasmacytic | 10 (10) | 211 |
| Burkitt’s | 7 (45) | 193, 220, 221 |
| Marginal zone | 7 (42) | 195, 211 |
| Pythorax associated | 28 (7) | 198 |
| T cell malignancies | ||
| Peripheral T cell | 11 (47) | 222 |
| Cutaneous CD30+ anaplastic large cell | 75 (8) | 223 |
| Natural killer T cell | 40 (5) | 224 |
CLL = chronic lymphocytic leukemia; DLBCL = diffuse large B cell lymphoma.
Gene expression profiling studies indicate that increased NF-κB target gene expression and sensitivity to NF-κB pathway inhibitors can be used to classify one class of aggressive DLBCL—namely, the so-called activated B cell–like (ABC) DLBCL.88,89 Curiously, REL gene amplifications have been reported in one study to be confined to the germinal center B (GCB) subtype of DLBCL, which does not have particularly high NF-κB target gene expression and is generally resistant to NF-κB inhibitors.90,91 The restriction of REL gene amplifications to GCB-like DLBCL led Shaffer et al.92 to suggest that increased REL activity, as provided by REL gene amplification, is required at an early initiation stage of GCB DLBCL development but not for late-stage maintenance of tumor cell growth. However, independent analyses of DLBCL samples have found REL gene amplifications equally distributed among both the ABC and GCB molecular subtypes93,94 and have suggested that REL amplifications are more strongly associated with MYC gene rearrangements than with either the ABC or GCB subtype.94 Alternatively, REL gene amplification may be required for GCB-like DLBCL cell growth in vivo but not in vitro. Of note, there are no human DLBCL cell lines with high-level REL gene amplification, indicating that high-level REL gene/protein expression may be incompatible with in vitro DLBCL cell growth. Indeed, in some situations, high-level expression of REL can induce cell cycle arrest.95 Overall, the frequency of REL gene amplification in human lymphoma is striking; nevertheless, the role that REL gene amplification plays in DLBCL is still not entirely clear.
REL gene rearrangements and point mutations in B cell lymphoma
Experiments in chickens and mice established that the c-rel gene could be activated by retroviral insertional mutagenesis in B cell lymphoma.96-98 REL gene rearrangements and mutations have also been detected in human B lymphoma samples and cell lines. The RC-K8 DLBCL cell line has a large deletion on chromosome 2, which results in the expression of a chimeric REL protein (REL-NRG) in which the REL DNA-binding/dimerization domain is fused to sequences of unknown function (Non-Rel-Gene).99,100 Similarly, in one Hodgkin’s lymphoma sample, a genomic alteration near the 3′ end of REL results in the expression of a C terminally truncated REL protein that is constitutively nuclear.101 In another Hodgkin’s lymphoma sample, REL is translocated to a position near the light chain enhancer,102 and in a third Hodgkin’s lymphoma cell line, an EBV genome has integrated near to REL, resulting in increased REL mRNA expression.103 Lastly, a point mutation that converts Ser-to-Pro at amino acid 525 within the transactivation domain of REL has been detected in 2 human B cell lymphomas.12 In one lymphoma patient, the S525P mutation is a germ-line mutation, suggesting that it is a predisposing mutation for lymphoma. The REL-S525P protein shows increased transforming ability in chicken spleen cells and altered transactivation properties.12 It would not be surprising if other human B cell lymphomas, with or without REL gene amplification, have activating mutations in REL, especially given that a variety of point mutations and deletions within the REL transactivation domain can enhance the in vitro transforming activity of REL.4,104
Of particular interest, the RC-K8 DCLBL cell line has (at least) 4 mutational events that affect the REL/NF-κB pathway: (1) a deletion that results in the expression of the chimeric REL-NRG protein99,100; (2) inactivating mutations in the gene encoding IκBα105; (3) inactivating mutations in A20, an upstream negative regulator of NF-κB signaling106,107; and (4) a disabling mutation in the REL transcriptional co-activator p300.108,109 As a consequence of these mutations, RC-K8 cells contain high levels of nuclear κB site DNA-binding complexes consisting of wild-type REL and REL-NRG, and several REL target genes, including BCL-x, TRAF1, Bfl-1/A1, and ICAM-1, are expressed at high levels.105 Reexpression of IκBα blocks the proliferation of RC-K8 cells.105 On the basis of these findings, it has been proposed that constitutive REL-directed gene expression is required for the growth and survival of RC-K8 cells and that this chronic gene induction is tuned to an optimal oncogenic level by cooperation with nonactivating REL-NRG dimers and defective p300 protein.108
REL mRNA and protein expression in B cell lymphoma
The findings described above suggest that the aberrant and chronic REL-induced gene expression seen in B cell lymphoma leads to enhanced mature B cell proliferation and survival. In some cases, this simple hypothesis is supported by experimental data. For example, many REL target genes are overexpressed in de novo DLBCLs,110 and increased REL mRNA expression has been correlated with a poorer prognosis in splenic marginal B cell lymphoma.111 Moreover, down-regulation of REL expression by siRNA or chemical inhibitors has been shown to block B cell lymphoma growth.112 However, Kluiver et al.113 did not find a correlation between REL amplification and high-level REL mRNA expression in classical Hodgkin’s lymphoma.
REL protein expression or nuclear activity in primary human B cell lymphomas has been less thoroughly analyzed than REL mRNA expression. Curry et al.114 analyzed 68 newly diagnosed DLBCL samples and found that 65% showed nuclear REL expression. Moreover, those patients with nuclear REL and the GCB-like gene expression profile had a worse overall survival. Houldsworth et al.93 did not find a correlation between nuclear REL expression and REL gene amplification—that is, some DLBCL samples with high-level REL gene amplification had less nuclear REL staining than some DLBCL samples with no REL gene amplification. Enhanced nuclear REL staining has, however, been reported to be a marker for certain types and stages of B cell lymphoma, as nuclear REL staining was reported in ~85% of Reed-Sternberg cells of classic Hodgkin’s lymphoma,115 in a majority of Hodgkin’s-like large cell lymphoma,116 and in mediastinal large B cell lymphoma.117,118 Furthermore, Rodig et al.119,120 presented data indicating that nuclear REL staining and high-level expression of the adaptor protein TRAF1, a possible REL target gene product, can be used to distinguish classical Hodgkin’s lymphoma cells from other types of B cell lymphomas, such as anaplastic large cell lymphoma, lymphocyte-predominant Hodgkin’s lymphoma, and nonmediastinal DLBCL. Nevertheless, given that c-Rel complexes are normally in the nucleus of mature B cells, one cannot determine by immunolocalization whether nuclear REL staining in specific human B cell lymphomas is driving malignancy or is simply a marker for the developmental stage of the given tumor.
REL in nonlymphoid cancers
Alterations in the REL gene or REL protein activity have been detected in nonlymphoid human cancers. Amplification of the gene encoding IKKϵ occurs in many breast cancers, and the resultant increased IKKϵ activity leads to enhanced nuclear accumulation of REL.121 Similarly, transgenic mice in which c-Rel is expressed from a mammary cell–specific promoter develop mammary tumors.122 In one study, REL gene deletions were detected in 7 of 7 oral carcinomas,123 but REL amplifications were found in these cancers in another study.124 REL gene amplifications have also been found in about 10% of metastatic lung cancers.125 In cell-based studies, inhibition of c-Rel by shRNA slowed lung cancer cell growth126 and decreased REL protein expression correlated with reduced pancreatic cancer stem cell growth in a combination drug treatment regimen.127 Finally, nuclear REL staining was seen in 50% of endometrial cancer samples128 and is enhanced in retinoblastomas129 and a mouse squamous cell epithelial cancer model.130
Modulation of REL Activity for Therapeutic Purposes
Given the role of REL in human cancer and autoimmune disease, modulation of REL activity or its transcriptional output could provide a therapeutic target. That inhibition of REL activity might prove a relevant strategy is bolstered by the facts that c-Rel expression is generally restricted to immune cells and that c-Rel knockout mice are fully viable (see above). Indeed, siRNA-mediated inhibition of c-Rel can block the growth of certain mouse B lymphoma cells,112 and knockdown of c-Rel can affect lung cancer cell growth in vitro.126 Moreover, c-Rel knockout mice show increased resistance to experimentally induced arthritis.75
In principle, specific inhibition of c-Rel could be achieved by down-regulation of c-Rel expression (e.g., by siRNA), by inhibition of a protein that is required for c-Rel expression or activity, or by inhibition of c-Rel transactivation activity (see Table 6). One compound with anti-inflammatory activity in clinical trials (STA-5326) has been reported to be a selective inhibitor of c-Rel nuclear translocation.131 Also, the immunosuppressant FK506 can specifically block c-Rel nuclear translocation in some cell systems.132,133 Direct inhibitors of c-Rel activity have not been identified. Given the similarity within the RHD between REL and other NF-κB family members, it is unlikely that sequences within the RHD could provide a specific therapeutic target; however, the REL transactivation domain, which is required for REL’s oncogenic and immune cell functions, might provide a specific target for inhibition. Finally, induction of REL activity could serve as a means of inducing Treg cell development to suppress autoimmune disease.
Table 6.
c-Rel Inhibitors
Concluding Remarks
c-Rel is an important component of the mammalian immune system, especially in terms of its roles in the development of regulatory T cells and in the regulation of mature T and B cell proliferation (Fig. 2). Moreover, the REL gene and its downstream pathway are altered in many T and B cell malignancies and autoimmune diseases. It seems likely that future genome-wide mutational screenings of other human lymphoid cell malignancies, with or without REL gene amplification, will uncover mutations in REL. Therefore, direct or downstream inhibitors of REL signaling may have therapeutic uses in human immune cell diseases.
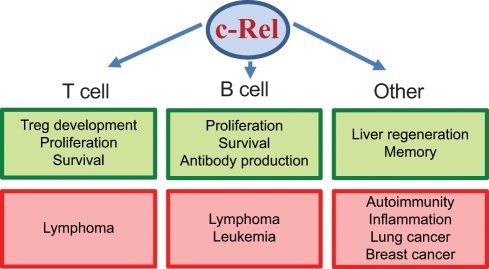
Figure 2.
c-Rel in normal development and disease. Shown are the major normal biological processes (green boxes) and pathologies (red boxes) in which c-Rel plays a role in T cells, B cells, and other cell types.
Acknowledgments
We thank Francis Wolenski for help with figures.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Research efforts in our laboratories are supported by the National Institutes of Health [grant CA047763-21S3] to T.D.G. and NHMRC [257502] and Leukemia and Lymphoma Society [SCOR grant 7015] program grants to S.G.
References
- 1. Gilmore TD. Introduction to NF-κB: players, pathways, perspectives. Oncogene. 2006;25:6680-4 [DOI] [PubMed] [Google Scholar]
- 2. Martin AG, Fresno M. Tumor necrosis factor-α activation of NF-κB requires the phosphorylation of Ser-471 in the transactivation domain of c-Rel. J Biol Chem. 2000;275:24383-91 [DOI] [PubMed] [Google Scholar]
- 3. Martin AG, San-Antonio B, Fresno M. Regulation of nuclear factor κB transactivation: implication of phosphatidylinostitol 3-kinase and protein kinase C ζ in c-Rel activation by tumor necrosis factor α. J Biol Chem. 2001;276:15840-9 [DOI] [PubMed] [Google Scholar]
- 4. Starczynowski DT, Reynolds JG, Gilmore TD. Deletion of either C-terminal transactivation subdomain enhances the in vitro transforming activity of human transcription factor REL in chicken spleen cells. Oncogene. 2003;22:6928-36 [DOI] [PubMed] [Google Scholar]
- 5. Leeman JR, Weniger MA, Barth TF, Gilmore TD. Deletion analysis and alternative splicing define a transactivation inhibitory domain in human oncoprotein REL. Oncogene. 2008;27:6770-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kunsch C, Ruben SM, Rosen CA. Selection of optimal κB/Rel DNA-binding motifs: interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang D, Chen Y, Ruetsche M, Phelps CB, Ghosh G. X-ray crystal structure of proto-oncogene product c-Rel bound to the CD28 response element of IL-2. Structure. 2001;9:669-78 [DOI] [PubMed] [Google Scholar]
- 8. Sanjabi S, Williams KJ, Saccani S, et al. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 2005;19:2138-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mosialos G, Gilmore TD. v-Rel and c-Rel are differentially affected by mutations at a consensus protein kinase recognition sequences. Oncogene. 1993;8:721-30 [PubMed] [Google Scholar]
- 10. Mosialos G, Hamer P, Capobianco AJ, Laursen R, Gilmore TD. A protein kinase-A recognition sequence is structurally linked to transformation by p59v- rel and cytoplasmic retention of p68c- rel . Mol Cell Biol. 1991;11:5867-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lawrence T, Bebien M, Liu GY, Nizet V, Karin M. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature. 2005;434:1138-43 [DOI] [PubMed] [Google Scholar]
- 12. Starczynowski DT, Trautmann H, Pott C, et al. Mutation of an IKK phosphorylation site within the transactivation domain of REL in two patients with human B-cell lymphoma enhances REL’s in vitro transforming activity. Oncogene. 2007;26:2685-94 [DOI] [PubMed] [Google Scholar]
- 13. Garbati MR, Gilmore TD. Ser484 and Ser494 in REL are the major sites of IKK phosphorylation in vitro: evidence that IKK does not directly enhance GAL4-REL transactivation. Gene Expr. 2008;14:195-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sánchez-Valdepeñas C, Martin AG, Ramakrishnan P, Wallach D, Fresno M. NF-κB-inducing kinase is involved in the activation of the CD28 responsive element through phosphorylation of c-Rel and regulation of its transactivating activity. J Immunol. 2006;176:4666-74 [DOI] [PubMed] [Google Scholar]
- 15. Neumann M, Tsapos K, Scheppler JA, Ross J, Franza BR., Jr Identification of complex formation between two intracellular tyrosine kinase substrates: human c-Rel and the p105 precursor of p50 NF-κB. Oncogene. 1992;7:2095-104 [PubMed] [Google Scholar]
- 16. Chen E, Hrdlicková R, Nehyba J, Longo DL, Bose HR, Jr, Li C-CH. Degradation of proto-oncoprotein c-Rel by the ubiquitin-proteasome pathway. J Biol Chem. 1998;273:35201-7 [DOI] [PubMed] [Google Scholar]
- 17. Glineur C, Davioud-Charvet E, Vandenbunder B. The conserved redox-sensitive cysteine residue of the DNA-binding region in the c-Rel protein is involved in the regulation of the phosphorylation of the protein. Biochem J. 2000;352:583-91 [PMC free article] [PubMed] [Google Scholar]
- 18. Barkett M, Dooher JE, Lemonnier L, et al. Three mutations in the retroviral oncoprotein v-Rel render it resistant to cleavage by caspase-3. Biochim Biophys Acta. 2001;1526:25-36 [DOI] [PubMed] [Google Scholar]
- 19. King KE, Ponnamperuma RM, Allen C, et al. The p53 homologue ΔNP63α interacts with nuclear factor–κB pathway to modulate epithelial cell growth. Cancer Res. 2008;68:5122-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou HJ, Pham LV, Tamayo AT, et al. Nuclear CD40 interacts with c-Rel and enhances proliferation in aggressive B-cell lymphoma. Blood. 2007;110:2121-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishikawa H, Asano M, Kanda T, Gélinas C, Ito Y. Two novel functions associated with the Rel oncoproteins: DNA replication and cell-specific transcriptional activation. Oncogene. 1993;8:2889-96 [PubMed] [Google Scholar]
- 22. Bunting K, Rao S, Hardy K, et al. Genome-wide analysis of gene expression in T cells to identify targets of the NF-κB transcription factor c-Rel. J Immunol. 2007;178:7097-109 [DOI] [PubMed] [Google Scholar]
- 23. Pham LV, Tamayo AT, Yoshimura L, Lin-Lee YC, Ford RJ. Constitutive NF-κB and NFAT activation in aggressive B-cell lymphomas synergistically activates the CD154 gene and maintains lymphoma cell survival. Blood. 2005;106:3940-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan Y, Rayet B, Gélinas C. Divergent C-terminal transactivation domains of Rel/NF-κB proteins are critical determinants of their oncogenic potential in lymphocytes. Oncogene. 2004;23:1030-42 [DOI] [PubMed] [Google Scholar]
- 25. Liou H-C, Hsia CY. Distinctions between c-Rel and other NF-κB proteins in immunity and disease. BioEssays. 2003;25:767-80 [DOI] [PubMed] [Google Scholar]
- 26. Liou H-C, Sha WC, Scott ML, Baltimore D. Sequential induction of NF-κB/Rel family proteins during B-cell terminal differentiation. Mol Cell Biol. 1994;14:5349-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carrasco D, Weih F, Bravo R. Developmental expression of the mouse c-rel proto-oncogene in hematopoietic organs. Development. 1994;120:2991-3004 [DOI] [PubMed] [Google Scholar]
- 28. Weih F, Carrasco D, Bravo R. Constitutive and inducible Rel/NF-κB activities in mouse thymus and spleen. Oncogene. 1994;9:3289-97 [PubMed] [Google Scholar]
- 29. Gerondakis S, Grumont R, Gugasyan R, et al. Unravelling the complexities of the NF-κB signaling pathway using mouse knockouts and transgenic models. Oncogene. 2006;25:6781-99 [DOI] [PubMed] [Google Scholar]
- 30. Hori S. c-Rel: a pioneer in directing regulatory T-cell lineage commitment? Eur J Immunol. 2010;40:664-7 [DOI] [PubMed] [Google Scholar]
- 31. Isomura I, Palmer S, Grumont RJ, et al. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med. 2009;206:3001-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor κB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921-31 [DOI] [PubMed] [Google Scholar]
- 33. Ruan Q, Kameswaran V, Tone Y, et al. Development of Foxp3(+) regulatory T cells is driven by the c-Rel enhancesome. Immunity. 2009;31:932-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vang KB, Yang J, Pagan AJ, et al. CD28 and c-Rel-dependent pathways initiate regulatory T cell development. J Immunol. 2010;184:4074-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Visekruna A, Huber M, Hellhund A, et al. c-Rel is crucial for the induction of Foxp3(+) regulatory CD4(+) T cells but not T(H)17 cells. Eur J Immunol. 2010;40:671-6 [DOI] [PubMed] [Google Scholar]
- 36. Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deenick EK, Elford AR, Pellegrini M, et al. c-Rel but not NF-κB1 is important for T regulatory cell development. Eur J Immunol. 2010;40:677-81 [DOI] [PubMed] [Google Scholar]
- 38. Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lio CW, Hsieh CS. A two step process for thymic regulatory T cell development. Immunity. 2008;28:100-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gerondakis S, Seibenlist U. Roles of the NF-κB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol. 2010;2:a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rao S, Gerondakis S, Woltring D, Shannon MF. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J Immunol. 2003;170:3724-31 [DOI] [PubMed] [Google Scholar]
- 42. Jones RG, Saibil SD, Pun JM, et al. NF-κB couples protein kinase B/Akt signaling to distinct survival pathways and the regulation of lymphocyte homeostasis in vivo. J Immunol. 2005;175:3790-9 [DOI] [PubMed] [Google Scholar]
- 43. Banerjee D, Liou H-C, Sen R. c-Rel-dependent priming of naïve T cells by inflammatory cytokines. Immunity. 2005;23:445-58 [DOI] [PubMed] [Google Scholar]
- 44. Grumont R, Lock P, Mollinari M, Shannon FM, Moore A, Gerondakis S. The mitogen-induced increase in T cell size involves PKC and NFAT activation of Rel/NF-κB-dependent c-myc expression. Immunity. 2004;21:19-30 [DOI] [PubMed] [Google Scholar]
- 45. Venkataraman L, Burakoff SJ, Sen R. FK506 inhibits antigen receptor-mediated induction of c-rel in B and T lymphoid cells. J Exp Med. 1995;181:1091-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Köntgen F, Grumont RJ, Strasser A, et al. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity and interleukin-2 expression. Genes Dev. 1995;9:1965-77 [DOI] [PubMed] [Google Scholar]
- 47. Mason NJ, Liou H-C, Hunter CA. T-cell intrinsic expression of c-Rel regulates Th1 responses essential for resistance to Toxoplasma gondii . J Immunol. 2004;172:3704-11 [DOI] [PubMed] [Google Scholar]
- 48. Harling-McNabb L, Deliyannis G, Jackson DC, Gerondakis S, Grigoriadis G, Brown LE. Mice lacking the transcription factor subunit Rel can clear an influenza infection and have functional anti-viral cytotoxic T cells but do not develop an optimal antibody response. Int Immunol. 1999;11:1431-9 [DOI] [PubMed] [Google Scholar]
- 49. Saibil SD, Jones RG, Deenick EK, et al. CD4+ and CD8+ T cell survival is regulated differentially by protein kinase Cθ, c-Rel and protein kinase B. J Immunol. 2007;178:2932-9 [DOI] [PubMed] [Google Scholar]
- 50. Zheng Y, Vig M, Lyons J, Van Parijs L, Beg AA. Combined deficiency of p50 and cRel in CD4+ T cells reveals an essential requirement for nuclear factor κB in regulating mature T cell survival and in vivo function. J Exp Med. 2003. 197:861-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Crotty S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol. 2011;29:621-63 [DOI] [PubMed] [Google Scholar]
- 52. Chen G, Hardy K, Bunting K, Daley S, Ma L, Shannon MF. Regulation of the IL-21 gene by the NF-κB transcription factor c-Rel. J Immunol. 2010;185:2350-9 [DOI] [PubMed] [Google Scholar]
- 53. Banerjee A, Grumont R, Gugasyan R, White C, Strasser A, Gerondakis S. NF-κB1 and c-Rel cooperate to promote the survival of TLR4-activated B cells by neutralizing Bim via distinct mechanisms. Blood. 2008;112:5063-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grossmann M, O’Reilly LA, Gugasyan R, Strasser A, Adams JM, Gerondakis S. The anti-apoptotic activities of Rel and RelA required during B-cell differentiation involve the regulation of Bcl-2 expression. EMBO J. 2000;19:6351-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grumont RJ, Gerondakis S. The subunit composition of NF-κB complexes changes during B-cell development. Cell Growth Differ. 1994;5:1321-31 [PubMed] [Google Scholar]
- 56. Grumont RJ, Rourke IJ, O’Reilly LA, et al. B lymphocytes differentially use the Rel and NF-κB1 transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J Exp Med. 1998;187:663-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cheng S, Hsia CY, Leone G, Liou H-C. Cyclin E and Bcl-XL cooperatively induce cell cycle progression in c-Rel-/- B cells. Oncogene. 2003;171:4886-92 [DOI] [PubMed] [Google Scholar]
- 58. Feng B, Cheng S, Hsia CY, King LB, Monroe JG, Liou H-C. NF-κB inducible genes BCL-X and cyclin E promote immature B-cell proliferation and survival. Cell Immunol. 2004;232:9-20 [DOI] [PubMed] [Google Scholar]
- 59. Grumont RJ, Strasser A, Gerondakis S. B cell growth is controlled by phosphatidylinositoal 3-kinase-dependent induction of Rel/NF-κB regulated c-myc transcription. Mol Cell. 2002;10:1283-94 [DOI] [PubMed] [Google Scholar]
- 60. Grumont RJ, Rourke IJ, Gerondakis S. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 1999;13:400-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zong WX, Edelstein LC, Chen C, Bash J, Gélinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNFα-induced apoptosis. Genes Dev. 1999;13:382-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen C, Edelstein LC, Gélinas C. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-xL . Mol Cell Biol. 2000;20:2687-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaku H, Horikawa K, Obata Y, et al. NF-κB is required for CD38-mediated induction of C1 germline transcripts in murine B lymphocytes. Int Immunol. 2002;14:1055-64 [DOI] [PubMed] [Google Scholar]
- 64. Agresti A, Vercelli D. c-Rel is a selective activator of a novel IL-4/CD40 responsive element in the Ig γ4 germline promoter. Mol Immunol. 2002;38:849-59 [DOI] [PubMed] [Google Scholar]
- 65. Zelazowski P, Shen Y, Snapper CM. NF-κB/p50 and NF-κB/c-Rel differentially regulate the activity of the 3′αE-hsl,2 enhancer in normal murine B cells in an activation-dependent manner. Int Immunol. 2000;12:1167-72 [DOI] [PubMed] [Google Scholar]
- 66. Mani P, Yadav VK, Das SK, Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS ONE. 2009;4:e4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gregersen PK, Amos CI, Lee AT, et al. REL, encoding a member of the NF-κB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41:820-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Eyre S, Hinks A, Flynn E, et al. Confirmation of association of the REL locus with rheumatoid arthritis susceptibility in the UK population. Ann Rheum Dis. 2010;69:1572-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Varadé J, Palomino-Morales R, Ortego-Centeno N, et al. Analysis of the REL polymorphism rs13031237 in autoimmune diseases. Ann Rheum Dis. 2011;70:711-2 [DOI] [PubMed] [Google Scholar]
- 70. Trynka G, Zhernakova A, Romanos J, et al. Coeliac disease– associated risk variants in TNFAIP3 and REL implicate altered NF-κB signaling. Gut. 2009;1078-83 [DOI] [PubMed] [Google Scholar]
- 71. Strange A, Capon F, Spencer CC, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-c and ERAP1. Nat Genet. 2010;42:985-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140:1704-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Janse M, Lamberts LE, Franke L, et al. Three ulcerative colitis susceptibility loci are associated with primary sclerlosing cholangitis and indicate a role for IL2, REL, and CARD9. Hepatology. 2011;53:1977-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Enciso-Mora V, Broderick P, Ma Y, et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat Genet. 2010;42:1126-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Campbell IK, Gerondakis S, O’Donnell K, Wicks IP. Distinct roles for the NF-κB1 (p50) and c-Rel transcription factors in inflammatory arthritis. J Clin Invest. 2000;105:1799-806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gugasyan R, Voss A, Varigos G, et al. The transcription factors c-rel and RelA control epidermal development and homeostasis in embryonic and adult skin via distinct mechanisms. Mol Cell Biol. 2004;24:5733-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ellinghaus E, Ellinghaus D, Stuart PE, et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat Genet. 2010. 42:991-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gilmore TD. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene. 1999;18:6925-37 [DOI] [PubMed] [Google Scholar]
- 79. Gilmore TD, Cormier D, Jean-Jacques J, Gapuzan M-E. Malignant transformation of primary chicken spleen cells by human transcription factor c-Rel. Oncogene. 2001;20:7098-103 [DOI] [PubMed] [Google Scholar]
- 80. Gilmore TD, Kalaitzidis D, Liang M-C, Starczynowski DT. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene. 2004;23:2275-86 [DOI] [PubMed] [Google Scholar]
- 81. Kamens J, Richardson P, Mosialos G, Brent R, Gilmore T. Oncogenic transformation by vrel requires an amino-terminal activation domain. Mol Cell Biol. 1991;10:2840-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fan Y, Gélinas C. An optimal range of transcription potency is necessary for efficient cell transformation by c-Rel to ensure optimal nuclear localization and gene-specific activation. Oncogene. 2007;26:4033-8 [DOI] [PubMed] [Google Scholar]
- 83. Chin M, Herscovitch M, Zhang N, Waxman DJ, Gilmore TD. Overexpression of an activated REL mutant enhances the transformed state of the human B-lymphoma BJAB cell line and alters its gene expression profile. Oncogene. 2009;28:2100-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Thompson RC, Herscovitch I, Zhao I, Ford TJ, Gilmore TD. NF-κB down-regulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway. J Biol Chem. 2011;286:1675-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fukuhara N, Tagawa H, Kameoka Y, et al. Characterization of target genes at the 2p15-16 amplicon in diffuse large B-cell lymphoma. Cancer Sci. 2006;97:499-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Reader JC, Zhao XF, Butler MS, Rapoport AP, Ning Y. REL-positive double minute chromosomes in follicular lymphoma. Leukemia. 2006;20:1624-6 [DOI] [PubMed] [Google Scholar]
- 87. Mader A, Bruderlein S, Wegener S, et al. U-HO1, a new cell line derived from a primary refractory classical Hodgkin lymphoma. Cytogenet Genome Res. 2007;119:204-10 [DOI] [PubMed] [Google Scholar]
- 88. Staudt LM. Oncogenic activation of NF-κB. Cold Spring Harb Perspect Biol. 2010;2:a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Courtois G, Gilmore TD. Mutations in the NF-κB signaling pathway: implications for human disease. Oncogene. 2006;25:6831-43 [DOI] [PubMed] [Google Scholar]
- 90. Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large B-cell lymphoma. N Engl J Med. 2002;346:1937-47 [DOI] [PubMed] [Google Scholar]
- 91. Bea S, Zettl A, Wright G, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B-cell differentiation. Nat Rev Immunol. 2002;2:920-32 [DOI] [PubMed] [Google Scholar]
- 93. Houldsworth J, Olshen AB, Cattoretti G, et al. Relationship between REL amplification, REL function, and clinical and biologic features in diffuse large B-cell lymphoma. Blood. 2004;103:1862-8 [DOI] [PubMed] [Google Scholar]
- 94. Jardin F, Jais JP, Molina TJ, et al. Diffuse large B-cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R-CHOP treatment: a GELA study. Blood. 2010;116:1092-104 [DOI] [PubMed] [Google Scholar]
- 95. Bash J, Zong W-X, Gélinas C. c-Rel arrests the proliferation of HeLa cells and affects critical regulators of the G1/S-phase transition. Mol Cell Biol. 1997;17:6526-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kabrun N, Bumstead N, Hayman MJ, Enrietto PJ. Characterization of a novel promoter insertion in the c-rel locus. Mol Cell Biol. 1990;10:4788-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hansen GM, Skapura D, Justice MJ. Genetic profile of insertion mutations in mouse leukemias and lymphoma. Genome Res. 2000;10:237-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166-74 [DOI] [PubMed] [Google Scholar]
- 99. Lu D, Thompson JD, Gorski GK, Rice NR, Mayer MG, Yunis JJ. Alterations at the rel locus in human lymphoma. Oncogene. 1991;6:1235-41 [PubMed] [Google Scholar]
- 100. Kalaitzidis D, Gilmore TD. Genomic organization and expression of the REL proto-oncogene in the human B-cell lymphoma cell line RC-K8. Genes Chromosomes Cancer. 2002;34:129-35 [DOI] [PubMed] [Google Scholar]
- 101. Barth TF, Martin-Subero JI, Joos S, et al. Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin’s lymphoma. Blood. 2003;101:3681-86 [DOI] [PubMed] [Google Scholar]
- 102. Martin-Subero JI, Klapper W, Sotnikova A, et al. Chromosomal breakpoints affecting immunoglobulin loci are recurrent in Hodgkin and Reed-Sternberg cells of classical Hodgkin lymphoma. Cancer Res. 2006;66:10332-8 [DOI] [PubMed] [Google Scholar]
- 103. Luo WJ, Takakuwa T, Ham MF, et al. Epstein-Barr virus is integrated between REL and BCL-11A in American Burkitt lymphoma cell line (NAB-2). Lab Invest. 2004;84:1193-9 [DOI] [PubMed] [Google Scholar]
- 104. Starczynowski DT, Reynolds JG, Gilmore TD. Mutations of tumor necrosis factor α-responsive serine residues within the C-terminal transactivation domain of human transcription factor REL enhance its in vitro transforming ability. Oncogene. 2005;24:7355-68 [DOI] [PubMed] [Google Scholar]
- 105. Kalaitzidis D, Davis RE, Rosenwald A, Staudt LM, Gilmore TD. The human B-cell lymphoma cell line RC-K8 has multiple genetic alterations that dysregulate the Rel/NF-κB signal transduction pathway. Oncogene. 2002;21:8759-68 [DOI] [PubMed] [Google Scholar]
- 106. Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. 2009;459:717-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kato M, Sanada M, Kato I, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712-6 [DOI] [PubMed] [Google Scholar]
- 108. Garbati MR, Alço G, Gilmore TD. Histone acetyltransferase p300 is a coactivator for transcription factor REL and is C-terminally truncated in the human diffuse large B-cell lymphoma cell line RC-K8. Cancer Lett. 2010;291:237-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Garbati MR, Thompson RC, Haery L, Gilmore TD. A rearranged EP300 gene in the human B-cell lymphoma cell line RC-K8 encodes a disabled transcriptional co-activator that contributes to cell growth and oncogenicity. Cancer Lett. 2010;302:76-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Mining for regulatory programs in the cancer transcriptome. Nat Genet. 2005;37:579-83 [DOI] [PubMed] [Google Scholar]
- 111. Ruiz-Ballesteros E, Mollejo M, Rodriguez A, et al. Splenic marginal zone lymphoma: proposal of new diagnostic and prognostic markers identified after tissue and cDNA microarray analysis. Blood. 2005;106:1831-38 [DOI] [PubMed] [Google Scholar]
- 112. Tian W, Liou H-C. RNAi-mediated c-Rel silencing leads to apoptosis of B cell tumor cells and suppresses antigenic immune response in vivo. PLoS ONE. 2009;4:e5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kluiver J, Kok K, Pfeil I, et al. Global correlation of genome and transcriptome changes in classical Hodgkin lymphoma. Hematol Oncol. 2007;25:21-9 [DOI] [PubMed] [Google Scholar]
- 114. Curry CV, Ewton AA, Olsen RJ, et al. Prognostic impact of C-REL expression in diffuse large B-cell lymphoma. J Hematop. 2009;2:20-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Xiao Q, Shen N, Hedvat CV, et al. Differential expression patterns of c-REL protein in classic and nodular lymphocyte predominant Hodgkin lymphoma. Appl Immunohistochem Mol Morphol. 2004;12:211-5 [DOI] [PubMed] [Google Scholar]
- 116. Garcia JF, Mollejo M, Fraga M, et al. Large B-cell lymphoma with Hodgkin’s features. Histopathology. 2005;47:101-10 [DOI] [PubMed] [Google Scholar]
- 117. Savage KJ, Monti S, Kutok JL, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871-9 [DOI] [PubMed] [Google Scholar]
- 118. Feuerhake F, Kutok JL, Monti S, et al. NFκB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes. Blood. 2005;106:1392-99 [DOI] [PubMed] [Google Scholar]
- 119. Rodig SJ, Savage KJ, Nguyen V, et al. TRAF1 expression and c-Rel activation are useful adjuncts in distinguishing classical Hodgkin lymphoma from a subset of morphologically or immunophenotypically similar lymphomas. Am J Surg Pathol. 2005;29:196-203 [DOI] [PubMed] [Google Scholar]
- 120. Rodig SJ, Savage KJ, Lacasce AS, et al. Expression of TRAF1 and nuclear c-Rel distinguishes primary mediastinal large cell lymphoma from other types of diffuse large B-cell lymphoma. Am J Surg Pathol. 2007;31:106-12 [DOI] [PubMed] [Google Scholar]
- 121. Boehm JS, Zhao JJ, Yao J, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065-79 [DOI] [PubMed] [Google Scholar]
- 122. Romieu-Mourez R, Kim DW, Shin SM, et al. Mouse mammary tumor virus c-rel transgenic mice develop mammary tumors. Mol Cell Biol. 2003;23:5738-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cha JD, Kim HJ, Cha IH. Genetic alterations in oral squamous cell carcinoma progression detected by combining array-based comparatic genomic hybridization and multiplex ligation-dependent probe amplification. Oral Surg Oral Med Pathol Oral Radiol Endod. 2011;111:594-607 [DOI] [PubMed] [Google Scholar]
- 124. Liu CJ, Lin SC, Chen YJ, Chang K, Chang KW. Array-comparative genomic hybridization to detect genomewide changes in microdissected primary and metastatic oral squamous cell carcinoma. Mol Carcinog. 2006;45:721-31 [DOI] [PubMed] [Google Scholar]
- 125. Boelens MC, Kok K, van der Viles P, et al. Genomic alterations in squamous cell lung carcinoma related to lymph node or distant metastasis. Lung Cancer. 2009;66:372-8 [DOI] [PubMed] [Google Scholar]
- 126. Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kallifatidis G, Labsch S, Rausch V, et al. Sulforaphane increases drug-mediated cytotoxicity toward cancer stem-like cells of the pancreas and prostate. Mol Ther. 2011;19:188-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Pallares J, Martinez-Guitarte JL, Dolcet X, et al. Abnormalities in the NF-κB family and related proteins in endometrial carcinoma. J Pathol. 2004;204:569-77 [DOI] [PubMed] [Google Scholar]
- 129. Qu Y, Zhou F, Dai X, et al. Clinicopathologic significances of nuclear expression of nuclear factor–κB transcription factors in retinoblastoma. J Clin Pathol. 2011;64:695-700 [DOI] [PubMed] [Google Scholar]
- 130. Yang X, Lu H, Yan B, Romano RA, et al. ΔNp63 versatility regulates a broad NF-κB gene program and promotes squamous epithelia proliferation, migration, and inflammation. Cancer Res. 2011;71:3688-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Keino H, Watanabe T, Sato Y, Nakura M, Wada Y, Okada A. Therapeutic effect of the potent IL-12/IL-23 inhibitor STA-5326 on experimental autoimmune uveoretinitis. Arthritis Res Ther. 2008;10:R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sen J, Venkataraman L, Shinkai Y, et al. Expression and induction of nuclear factor-κB-related proteins in thymocytes. J Immunol. 1995;154:3213-21 [PubMed] [Google Scholar]
- 133. Cha-Molstad H, Young DP, Kushner I, Samols D. The interaction of c-Rel with C/EBPβ enhances C/EBPβ binding to the C-reactive protein gene promoter. Mol Immunol. 2007;44:2933-42 [DOI] [PubMed] [Google Scholar]
- 134. Liu J, Ma X. Interferon regulatory factor 8 regulates RANTES gene transcription in cooperation with interferon regulatory factor-1, NF-κB, and PU.1. J Biol Chem. 2006;281:19188-95 [DOI] [PubMed] [Google Scholar]
- 135. Sif S, Gilmore TD. Interaction of the v-Rel oncoprotein with cellular transcription factor Sp1. J Virol. 1994;68:7131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Loizou L, Andersen KG, Betz AG. Foxp3 interacts with c-Rel to mediate NF-κB repression. PLoS ONE. 2011;6:e18670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Mukhopadhyay NK, Ferdinand AS, Mukhopadhyay L, et al. Unraveling androgen receptor interactomes by an array-based method: discovery of proto-oncoprotein c-Rel as a negative regulator of androgen receptor. Exp Cell Res. 2006;312:3782-95 [DOI] [PubMed] [Google Scholar]
- 138. Gailien R, Garcia T. Estrogen receptor impairs interleukin-6 expression by preventing protein binding on the NF-κB site. Nucleic Acids Res. 1997;25:2424-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kalaitzidis D, Gilmore TD. Transcription factor cross-talk: the estrogen receptor and NF-κB. Trends Endocrinol Metab. 2005;16:46-52 [DOI] [PubMed] [Google Scholar]
- 140. Xu X, Prorock C, Ishikawa H, Maldonado E, Ito Y, Gélinas C. Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol Cell Biol. 1993;13:6733-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Dutta J, Fan G, Gélinas C. CAPERα is a novel REL-TAD-interacting factor that inhibits lymphocyte transformation by the potent Rel/NF-κB oncoprotein Rel. J Virol. 2008;82:10792-802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Chen E, Li CC. Association of Cdk2/cyclin E and NF-κB complexes at G1/S phase. Biochem Biophys Res Commun. 1998;249:728-34 [DOI] [PubMed] [Google Scholar]
- 143. van Essen D, Zhu Y, Saccani S. A feed-forward circuit controlling inducible NF-κB target gene activation by promoter histone demethylation. Mol Cell. 2010;39:750-60 [DOI] [PubMed] [Google Scholar]
- 144. Fagerlund R, Melén K, Cao X, Julkumen I. NF-κB p52, RelB and c-Rel are transported into the nucleus via a subset of importin alpha molecules. Cell Signal. 2008;20:1442-51 [DOI] [PubMed] [Google Scholar]
- 145. Fu L, Lin-Lee Y-C, Pham LV, Tamayo AT, Yoshimura LC, Ford RJ. BAFF-R promotes cell proliferation and survival through interaction with IKKβ and NF-κB/c-Rel in the nucleus of normal and neoplastic B-lymphoid cells. Blood. 2009;113:4627-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Antonsson A, Hughes K, Edin S, Grundström T. Regulation of c-Rel nuclear localization by binding of Ca2+/calmodulin. Mol Cell Biol. 2003;23:1418-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Khan N, Rahim SS, Goddupalli CS, et al. Hydrogen peroxide inhibits IL-12/p40 induction in macrophages by inhibiting c-rel translocation to the nucleus through activation of calmodulin protein. Blood. 2006;107:1513-20 [DOI] [PubMed] [Google Scholar]
- 148. Knuefermann P, Chen P, Misra A, Shi SP, Abdellatif M, Sivasubramanian N. Myotrophin/V-1, a protein up-regulated in the failing human heart and in postnatal cerebellum, converts NF-κB p50-p65 heterodimers to p50-p50 and p65-p65 homodimers. J Biol Chem. 2002;277:23888-97 [DOI] [PubMed] [Google Scholar]
- 149. Bouwmeester T, Bauch A, Ruffner H, et al. A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat Cell Biol. 2004;6:97-105 [DOI] [PubMed] [Google Scholar]
- 150. Ress A, Moelling K. Interaction partners of the PDZ domain of erbin. Protein Pept Lett. 2006;13:877-81 [DOI] [PubMed] [Google Scholar]
- 151. Meyer CF, Wang X, Chang C, Templeton D, Tan TH. Interaction between c-Rel and the mitogen-activated kinase kinase kinase 1 signaling cascade in mediating κB enhancer activation. J Biol Chem. 1996;271:8971-6 [DOI] [PubMed] [Google Scholar]
- 152. Sánchez-Valdepeñas C, Punzón C, San-Antonio B, Martin AG, Fresno M. Differential regulation of p65 and c-Rel NF-κB transactivating activity by Cot, protein kinase Cζ and NIK protein kinases in CD3/CD28 activated T cells. Cell Signal. 2007;19:528-37 [DOI] [PubMed] [Google Scholar]
- 153. Yu SH, Chiang WC, Shih HM, Wu KJ. Stimulation of c-Rel transcriptional activity by PKA catalytic subunit beta. J Mol Med. 2004;82:621-8 [DOI] [PubMed] [Google Scholar]
- 154. Hu MC, Tang-Oxley Q, Qiu WR, et al. Protein phosphatase X interacts with c-Rel and stimulates c-Rel/nuclear factor κB activity. J Biol Chem. 1998;273:33561-5 [DOI] [PubMed] [Google Scholar]
- 155. Fan G, Fan Y, Gupta N, et al. Peptidyl-prolyl isomerase Pin1 markedly enhances the oncogenic activity of the rel proteins in the nuclear factor–κB family. Cancer Res. 2009;69:4589-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Suzuki T, Hirai H, Yoshida M. Tax protein of HTLV-1 interacts with the Rel homology domain of NF-κB p65 and c-Rel proteins bound to the NF-κB binding site and activates transcription. Oncogene. 1994;9:3099-105 [PubMed] [Google Scholar]
- 157. Grumont RJ, Richardson IB, Gaff C, Gerondakis S. rel/NF-κB nuclear complexes that bind κB sites in the murine c-rel promoter are required for constitutive c-rel transcription in B-cells. Cell Growth Differ. 1993;4:731-43 [PubMed] [Google Scholar]
- 158. Lee H, Arsura M, Wu M, Duyao M, Buckler AJ, Sonenshein GE. Role of Rel-related factors in control of c-myc gene transcription in receptor-mediated apoptosis of the murine B cell WEHI 231 line. J Exp Med. 1995;181:1169-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Grumont RJ, Gerondakis S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor κB. J Exp Med. 2000;191:1281-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. De Seirvi A, De Luca P, Moila C, et al. Identification of new Rel/NF-κB regulatory networks by focused genome location analysis. Cell Cycle. 2009:8:2093-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Tolnay M, Vereshchagina LA, Tsokos GC. NF-κB regulates the expression of the human complement receptor 2 gene. J Immunol. 2002;169:6236-43 [DOI] [PubMed] [Google Scholar]
- 162. Brettingham-Moore KH, Rao S, Juelich R, Shannon MF, Holloway AF. GM-CSF promoter chromatin remodeling and gene transcription display distinct signal and transcription factor requirements. Nucleic Acids Res. 2005;33:225-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Himes SR, Coles LS, Reeves R, Shannon MF. High mobility group protein I(Y) is required for function and for c-Rel binding to the CD28 response elements within the GM-CSF and IL-2 promoters. Immunity. 1996;5:479-89 [DOI] [PubMed] [Google Scholar]
- 164. Li-Weber M, Giasi M, Krammer PH. Involvement of Jun and Rel proteins in up-regulation of interleukin-4 gene activity by the T cell accessory molecule CD28. J Biol Chem. 1998;273:32460-6 [DOI] [PubMed] [Google Scholar]
- 165. Heckman CA, Mehew JW, Boxer LM. NF-κB activates Bcl-2 expression in t(14;18) lymphoma cells. Oncogene. 2002;21:3898-908 [DOI] [PubMed] [Google Scholar]
- 166. Wissink S, van de Stolpe A, Caldenhoven E, Koenderman L, van der Saag PT. NF-κB/Rel family members regulating the ICAM-1 promoter in moncytic THP-1 cells. Immunobiology. 1997;198:50-64 [DOI] [PubMed] [Google Scholar]
- 167. Schreiber J, Jenner RG, Murray HL, Gerber GK, Gifford DK, Young RA. Coordinated binding of NF-κB family members in the response of cells to lipopolysaccharide. Proc Natl Acad Sci USA. 2006;103:5899-904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Fu T, Li P, Wang H, et al. c-Rel is a transcriptional repressor of EPHB2 in colorectal cancer. J Pathol. 2009;219:103-13 [DOI] [PubMed] [Google Scholar]
- 169. Grumont R, Hochrein H, O’Keeffe M, et al. c-Rel regulates interleukin 12 p70 expression in CD8(+) dendritic cells by specifically inducing p35 gene transcription. J Exp Med. 2001;194:1021-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Carmody RJ, Ruan Q, Liou H-C, Chen YH. Essential roles of c-Rel in TLR-induced IL-23 p19 gene expression in dendritic cells. J Immunol. 2007;178:186-91 [DOI] [PubMed] [Google Scholar]
- 171. Fu L, Lin-Lee YC, Pham LV, Tamayo A, Yoshimura L, Ford RJ. Constitutive NF-κB and NFAT activation leads to stimulation of the BlyS survival pathway in aggressive B-cell lymphomas. Blood. 2006;107:4540-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Son JS, Sahoo A, Chae CS, Hwang JS, Park ZY, Im SH. JunB and c-Rel cooperatively enhance Foxp3 expression during induced regulatory T cell differentiation. Biochem Biophys Res Commun. 2011;407:141-7 [DOI] [PubMed] [Google Scholar]
- 173. Kenneth NS, Mudie S, Rocha S. IKK and NF-κB-mediated regulation of Claspin impacts ATR checkpoint function. EMBO J. 2010;29:2966-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Barré B, Perkins ND. The Skp2 promoter integrates signaling through the NF-κB, p53, and Akt/GSK3β pathways to regulate autophagy and apoptosis. Mol Cell. 2010;38:524-38 [DOI] [PubMed] [Google Scholar]
- 175. Jordan KA, Dupont CD, Tait ED, Liou H-C, Hunter CA. Role of the NF-κB transcription factor c-Rel in the generation of CD28+ T-cell responses to Toxoplasma gondii . Int Immunol. 2010;22:851-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Courtine E, Pène F, Cagnard N, et al. Critical role of cRel subunit of NF-κB in sepsis survival. Infect Immun. 2011;79:1848-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Liou H-C, Smith KA. The roles of c-Rel and interleukin-2 in tolerance: a molecular explanation of self-nonself discrimination. Immunol Cell Biol. 2011;89:27-32 [DOI] [PubMed] [Google Scholar]
- 178. Boffa DJ, Feng B, Sharma V, et al. Selective loss of c-Rel compromises dendritic cell activation of T lymphocytes. Cell Immunol. 2003;222:105-15 [DOI] [PubMed] [Google Scholar]
- 179. O’Keeffe M, Grumont RJ, Hochrein H, et al. Distinct roles for the NF-κB1 and c-Rel transcription factors in the differentiation and survival of plasmacytoid and conventional dendritic cells activated by TLR-9 signals. Blood. 2005;106:3457-64 [DOI] [PubMed] [Google Scholar]
- 180. Lamhamedi-Cherradi SE, Zheng S, Hilliard BA, et al. Transcriptional regulation of type I diabetes by NF-κB. J Immunol. 2003;171:4886-92 [DOI] [PubMed] [Google Scholar]
- 181. Wang Y, Rickman BH, Poutahidis T, et al. c-Rel is essential for the development of innate and T cell-induced colitis. J Immunol. 2008;180:8118-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Gieling RG, Eisharkawy AM, Caamaño JH, et al. The c-Rel subunit of nuclear factor κB regulates murine liver inflammation, wound-healing, and hepatocyte proliferation. Hepatology. 2010;51:922-31 [DOI] [PubMed] [Google Scholar]
- 183. Levenson JM, Choi S, Lee SY, et al. A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel. J Neurosci. 2004;24:3933-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Ahn HJ, Hernandez CM, Levenson JM, Lubin FD, Liou H-C, Sweatt JD. c-Rel, an NF-κB family transcription factor, is required for hippocampal long-term synaptic plasticity and memory formation. Learn Mem. 2008;15:539-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Carrasco D, Cheng J, Lewin A, et al. Multiple hemopoietic defects and lymphoid hyperplasia in mice lacking the transcriptional activation domain of the c-Rel protein. J Exp Med. 1998;187:973-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Pohl T, Gugasyan R, Grumont TJ, et al. The combined absence of NF-κB1 and c-Rel reveals that overlapping roles for these transcription factors in the B cell lineage are restricted to the activation and function of mature cells. Proc Natl Acad Sci USA. 2002;99:4514-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Grossman M, Metcalf D, Merryfull J, Beg A, Baltimore D, Gerondakis S. The combined absence of the transcription factors Rel and RelA leads to multiple hemopoietic cell defects. Proc Natl Acad Sci USA. 1999;96:11848-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Joos S, Granzow M, Holtgreve-Grez H, et al. Hodgkin’s lymphoma cell lines are characterized by frequent aberrations on chromosomes 2p and 9p including REL and JAK2. Int J Cancer. 2003;103:489-95 [DOI] [PubMed] [Google Scholar]
- 189. Joos S, Menz CK, Wrobel G, et al. Classical Hodgkin lymphoma is characterized by recurrent copy number gaoins of the short arm of chromosome 2. Blood. 2002;99:1381-7 [DOI] [PubMed] [Google Scholar]
- 190. Martin-Subero JI, Gesk S, Harder L, et al. Recurrent involvement of REL and BCL11A loci in classical Hodgkin lymphoma. Blood. 2002;99:1474-7 [DOI] [PubMed] [Google Scholar]
- 191. Martin-Subero JI, Knippschild U, Harder L, et al. Segmental chromosomal aberrations and centrosome amplifications: pathogenetic mechanisms in Hodgkin and Reed-Sternberg cells of classical Hodgkin’s lymphoma? Leukemia. 2003;17:2214-9 [DOI] [PubMed] [Google Scholar]
- 192. Steidl C, Telenius A, Shah SP, et al. Genome-wide copy number analysis of Hodgkin Reed-Sternberg cells identifies recurrent imbalances with correlations to treatment outcome. Blood. 2010;116:418-27 [DOI] [PubMed] [Google Scholar]
- 193. Werner CA, Dohner H, Joos S, et al. High-level DNA amplifications are common genetic aberrations in B-cell neoplasms. Am J Pathol. 1997;151:335-42 [PMC free article] [PubMed] [Google Scholar]
- 194. Barth TFE, Döhner H, Werner CA, et al. Characteristic pattern of chromosomal gains and losses in primary large B-cell lymphomas of the gastrointestinal tract. Blood. 1998;91:4321-30 [PubMed] [Google Scholar]
- 195. Barth TF, Bentz M, Leithauser F, et al. Molecular-cytogenetic comparison of mucose-associated marginal zone B-cell lymphoma and large B-cell lymphoma arising in the gastrointestinal tract. Genes Chromosomes Cancer. 2001;31:316-25 [DOI] [PubMed] [Google Scholar]
- 196. Rao PH, Houldsworth J, Dyomina K, et al. Chromosomal and gene amplification in diffuse large B-cell lymphoma. Blood. 1998;92:234-40 [PubMed] [Google Scholar]
- 197. Goff LK, Neat MJ, Crawley CR, et al. The use of real-time quantitative polymerase chain reaction and comparative genomic hybridization to identify amplification of the REL gene in follicular lymphoma. Br J Haematol. 2000;111:618-25 [DOI] [PubMed] [Google Scholar]
- 198. Yamato H, Ohshima K, Suzumiya J, Kikuchi M. Evidence for local immunosuppression and demonstration of c-myc amplification in pyothorax-associated lymphoma. Histopathology. 2001;39:163-71 [DOI] [PubMed] [Google Scholar]
- 199. Palanisamy N, Abou-Elella AA, Chaganti SR, et al. Similar patterns of genomic alterations characterize primary mediastinal large-B-cell lymphoma and diffuse large-B-cell lymphoma. Genes Chromosomes Cancer. 2002;33:114-22 [DOI] [PubMed] [Google Scholar]
- 200. Houldsworth J, Mathew S, Rao PH, et al. REL proto-oncogene is frequently amplified in extranodal diffuse large cell lymphoma. Blood. 1996;87:25-9 [PubMed] [Google Scholar]
- 201. Martinez-Climent JA, Alizadeh AA, Segraves R, et al. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with a heterogeneous set of DNA copy number and gene expression alterations. Blood. 2003;101:3109-17 [DOI] [PubMed] [Google Scholar]
- 202. Wessendorf S, Schwaenen C, Kohlhammer H, et al. Hidden gene amplifications in aggressive B-cell non-Hodgkin lymphomas detected by microarray-based comparative genomic hybridization. Oncogene. 2003;22:1425-9 [DOI] [PubMed] [Google Scholar]
- 203. Bea S, Colomo L, López-Guillermo A, et al. Clinicopathologic significance and prognostic value of chromosomal imbalances in diffuse large B-cell lymphomas. J Clin Oncol. 2004;22:3498-506 [DOI] [PubMed] [Google Scholar]
- 204. Tagawa H, Suguro M, Tsuzuki S, et al. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood. 2005;106:1770-7 [DOI] [PubMed] [Google Scholar]
- 205. Chen W, Houldsworth J, Olshen AB, et al. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood. 2006;107:2477-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Davies AJ, Rosenwald A, Wright G, et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma proceeds by distinct oncogenic mechanisms. Br J Haematol. 2007;136:286-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Jardin F, Ruminy P, Kerckaaert JP, et al. Detection of somatic quantitative genetic alterations by multiplex polymerase chain reaction for the prediction of outcome in diffuse large B-cell lymphoma. Haematologica. 2008;93:543-50 [DOI] [PubMed] [Google Scholar]
- 208. Bentz M, Werner CA, Döhner H, et al. High incidence of chromosomal imbalances and gene amplifications in the classical follicular variant of follicle center lymphoma: high incidence of chromosomal imbalances and gene amplifications in the classical follicular variant of follicle center lymphoma. Blood. 1996;88:1437-44 [PubMed] [Google Scholar]
- 209. Avet-Loiseau H, Vigier M, Moreau A, et al. Comparative genomic hybridization detects genomic abnormalities in 80% of follicular lymphomas. Br J Haematol. 1997;97:119-22 [DOI] [PubMed] [Google Scholar]
- 210. Neat MJ, Foot N, Jenner M, et al. Localisation of a novel region of recurrent amplification in follicular lymphoma to an approximately 6.8 Mb region of 13q32-33. Genes Chromosomes Cancer. 2001;32:236-43 [DOI] [PubMed] [Google Scholar]
- 211. Ferreira BI, Garcia JF, Suela J, et al. Comparative genome profiling across subtypes of low-grade B-cell lymphoma identifies type-specific and common aberrations that target genes with a role in B-cell neoplasia. Haematologica. 2008;93:670-8 [DOI] [PubMed] [Google Scholar]
- 212. Schwaenen C, Viardot A, Berger H, et al. Microarray-based genomic profiling reveals novel genomic aberrations in follicular lymphoma which associate with patient survival and gene expression status. Genes Chromosomes Cancer. 2009;48:39-54 [DOI] [PubMed] [Google Scholar]
- 213. Joos S, Otaño-Joos MI, Ziegler S, et al. Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood. 1996;87:1571-8 [PubMed] [Google Scholar]
- 214. Weniger MA, Gesk S, Ehrlich S, et al. Gains of REL in primary mediastinal B-cell lymphoma coincide with nuclear accumulation of REL protein. Genes Chromosomes Cancer. 2007;46:406-15 [DOI] [PubMed] [Google Scholar]
- 215. Dijkman R, Tensen CP, Jordanova ES, et al. Array-based comparative genomic hybridization analysis reveals recurrent chromosomal alterations and prognostic parameters in primary cutaneous large B-cell lymphoma. J Clin Oncol. 2006;24:296-305 [DOI] [PubMed] [Google Scholar]
- 216. Pfeifer D, Pantic M, Skatulla I, et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202-10 [DOI] [PubMed] [Google Scholar]
- 217. Deambrogi C, De Paoli L, Fangazio M, et al. Analysis of the REL, BCL11A and MYCN proto-oncogenes belonging to the 2 p amplicon in chronic lymphocytic leukemia. Am J Hematol. 2010;85:541-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Jarosova M, Urbankova H, Plachy R, et al. Gain of chromosome 2p in chronic lymphocytic leukemia: significant heterogeneity and a new recurrent dicentric rearrangement. Leuk Lymphoma. 2010;51:304-13 [DOI] [PubMed] [Google Scholar]
- 219. Shao L, Kang SH, Li J, et al. Array comparative genomic hybridization detects chromosomal abnormalities in hematological cancers that are not detected by conventional cytogenetics. J Mol Diagn. 2010;12:670-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Wessendorf S, Barth TF, Viardot A, et al. Further delineation of chromosomal consensus regions in primary mediastinal B-cell lymphomas: an analysis of 37 tumor samples using high-resolution genomic profiling (array-CGH). Leukemia. 2007;21:2463-9 [DOI] [PubMed] [Google Scholar]
- 221. Toujani S, Dessen P, Ithzar N, et al. High resolution genome-wide analysis of chromosomal alterations in Burkitt’s lymphoma. PLoS ONE. 2009;4:e7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Hartmann S, Gesk S, Scholtysik R, et al. High resolution SNP array genomic profiling of peripheral T cell lymphomas, not otherwise specified, identifies a subgroup with chromosomal aberrations affecting the REL locus. Br J Haematol. 2010;148:402-12 [DOI] [PubMed] [Google Scholar]
- 223. Mao X, Orchard G, Lillington DM, Russell-Jones R, Young BD, Whittaker S. Genetic alterations in primary cutaneous CD30+ anaplastic large cell lymphoma. Genes Chromosomes Cancer. 2003;37:176-85 [DOI] [PubMed] [Google Scholar]
- 224. Mao X, Onadim Z, Price EA, et al. Genomic alterations in blastic natural killer/extranodal natural killer-like T cell lymphoma with cutaneous involvement. J Invest Dermatol. 2003;121:618-27 [DOI] [PubMed] [Google Scholar]
- 225. Wang W, Tam HF, Hughes CC, Rath S, Sen R. c-Rel is a target of pentoxifylline-mediated inhibition of T lymphocyte activation. Immunity. 1997;6:165-74 [DOI] [PubMed] [Google Scholar]
- 226. Liang M-C, Bardhan S, Porco JA, Jr, Gilmore TD. The synthetic epoxyquinoids jesterone dimer and epoxyquinone A monomer induce apoptosis and inhibit REL (human c-Rel) DNA binding in an IκBα-deficient diffuse large B-cell lymphoma cell line. Cancer Lett. 2006;24:69-78 [DOI] [PubMed] [Google Scholar]
- 227. Sperandeo MP, Tosco A, Izzo V, et al. Potential celiac patients: a model of celiac disease pathogenesis. PLoS ONE. 2011;6:e21281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Ruan Q, Want T, Kameswaran V, et al. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic β cell death. Proc Natl Acad Sci USA. 2011;108:12030-5 [DOI] [PMC free article] [PubMed] [Google Scholar]