Abstract
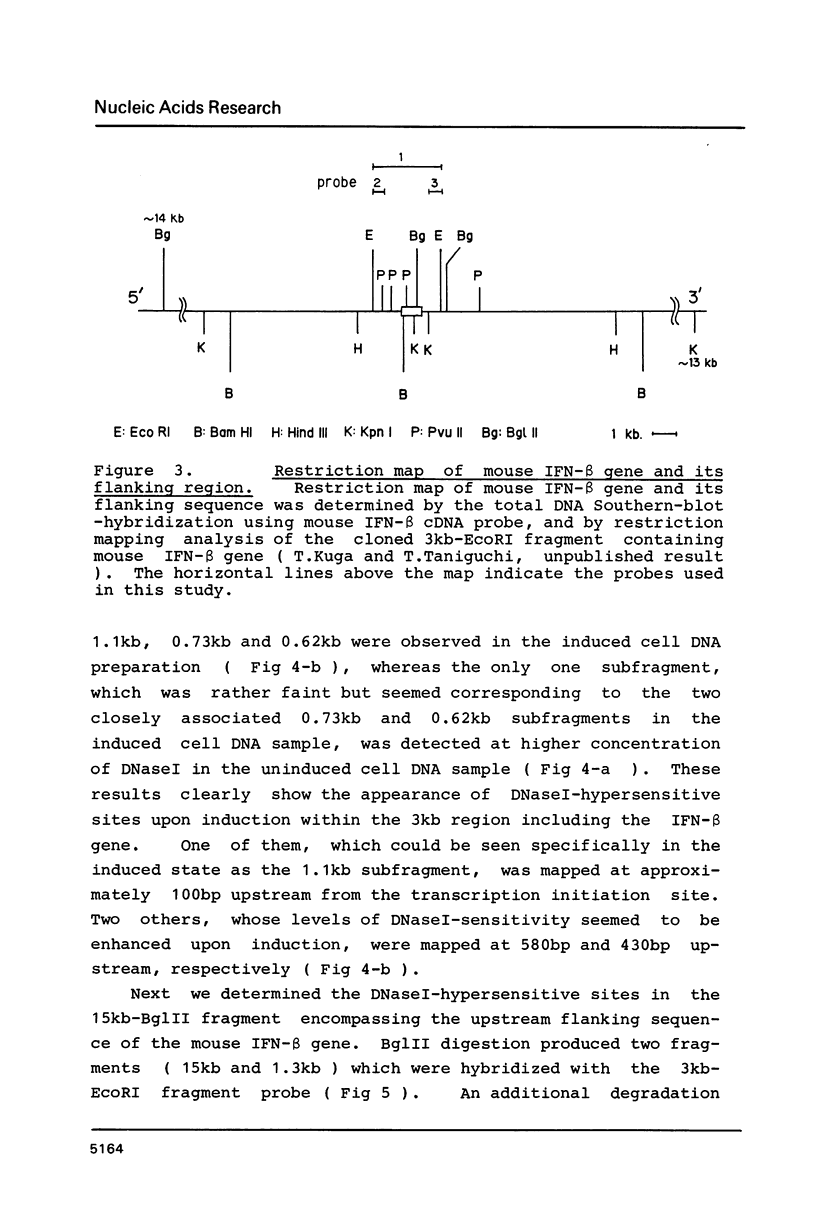
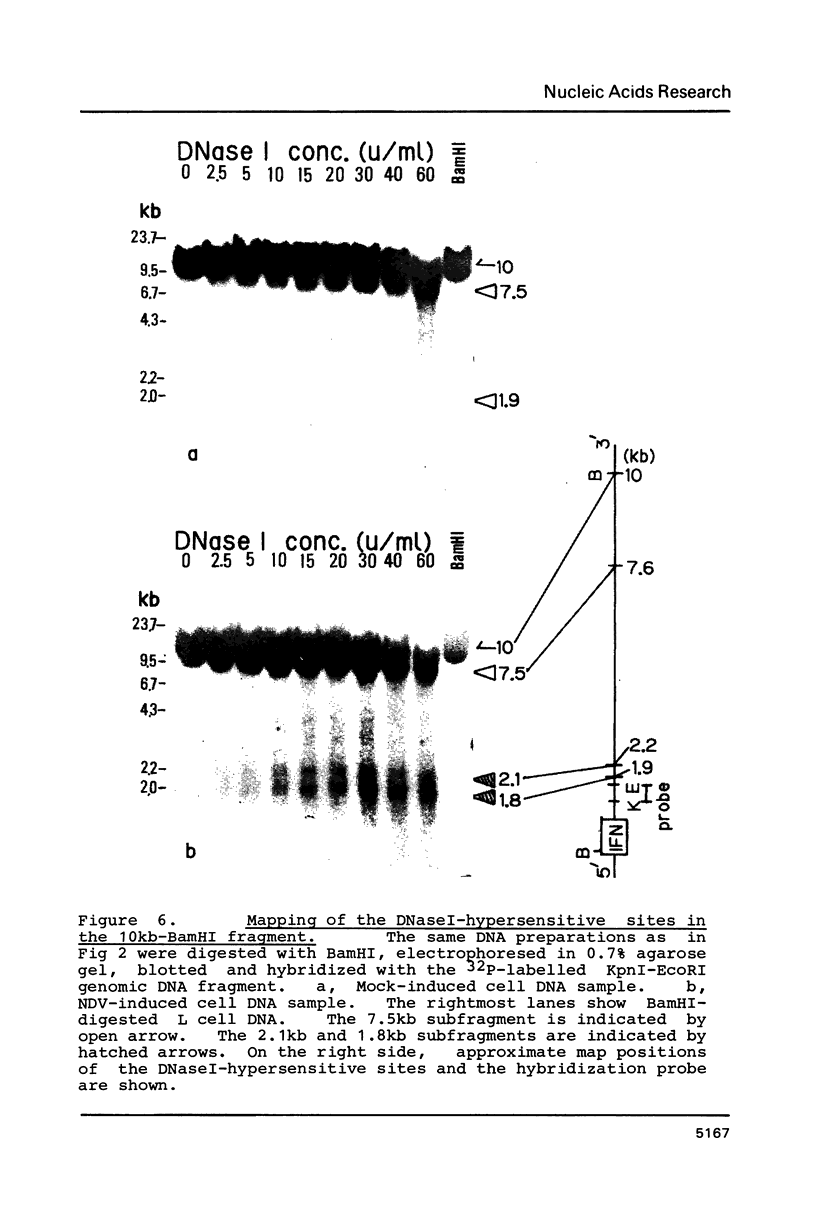
Changes of chromatin structure around the mouse interferon-beta gene associated with induction were examined by the use of sensitivity to digestion with DNaseI. Upon induction, the DNaseI-sensitivity of mouse interferon-beta gene was increased and seven DNaseI-hypersensitive sites were mapped. One of them, which was located at approximately 100bp upstream and was insensitive in the uninduced state, showed a marked sensitivity upon induction. Four others, located at 580bp and 430bp upstream, and 1.9kb and 2.2kb downstream, whose levels of DNaseI-sensitivity were very low in the uninduced state, showed significant sensitivity in the induced state. Two remaining sites, located at 7.5kb upstream and 7.6kb downstream, had relatively high DNaseI-sensitivities even in the uninduced state and slightly increased their sensitivities upon induction.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chung S. Y., Folsom V., Wooley J. DNase I-hypersensitive sites in the chromatin of immunoglobulin kappa light chain genes. Proc Natl Acad Sci U S A. 1983 May;80(9):2427–2431. doi: 10.1073/pnas.80.9.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coveney J., Scott G., King R., Burke D. C., Skup D. Changes in the conformation of the interferon beta gene during differentiation and induction. Biochem Biophys Res Commun. 1984 May 31;121(1):290–296. doi: 10.1016/0006-291x(84)90721-6. [DOI] [PubMed] [Google Scholar]
- Cremisi C. The appearance of DNase I hypersensitive sites at the 5' end of the late SV40 genes is correlated with the transcriptional switch. Nucleic Acids Res. 1981 Nov 25;9(22):5949–5964. doi: 10.1093/nar/9.22.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Gray P. W., Goeddel D. V. Cloning and expression of murine immune interferon cDNA. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5842–5846. doi: 10.1073/pnas.80.19.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y., Sokawa Y., Watanabe Y., Kawade Y., Ohno S., Takaoka C., Taniguchi T. Structure and expression of a cloned cDNA for mouse interferon-beta. J Biol Chem. 1983 Aug 10;258(15):9522–9529. [PubMed] [Google Scholar]
- Kawade Y., Yamamoto Y. Induction and production of L cell interferon. Methods Enzymol. 1981;78(Pt A):139–143. doi: 10.1016/0076-6879(81)78107-2. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Nagata S., Mantei N., Weissmann C. The structure of one of the eight or more distinct chromosomal genes for human interferon-alpha. Nature. 1980 Oct 2;287(5781):401–408. doi: 10.1038/287401a0. [DOI] [PubMed] [Google Scholar]
- Nagata S., Taira H., Hall A., Johnsrud L., Streuli M., Ecsödi J., Boll W., Cantell K., Weissmann C. Synthesis in E. coli of a polypeptide with human leukocyte interferon activity. Nature. 1980 Mar 27;284(5754):316–320. doi: 10.1038/284316a0. [DOI] [PubMed] [Google Scholar]
- Ohno S., Taniguchi T. Structure of a chromosomal gene for human interferon beta. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5305–5309. doi: 10.1073/pnas.78.9.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Chromatin changes accompany immunoglobulin kappa gene activation: a potential control region within the gene. Nature. 1982 Sep 30;299(5882):449–451. doi: 10.1038/299449a0. [DOI] [PubMed] [Google Scholar]
- Ragg H., Weissmann C. Not more than 117 base pairs of 5'-flanking sequence are required for inducible expression of a human IFN-alpha gene. Nature. 1983 Jun 2;303(5916):439–442. doi: 10.1038/303439a0. [DOI] [PubMed] [Google Scholar]
- Shaw G. D., Boll W., Taira H., Mantei N., Lengyel P., Weissmann C. Structure and expression of cloned murine IFN-alpha genes. Nucleic Acids Res. 1983 Feb 11;11(3):555–573. doi: 10.1093/nar/11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., Ohno S., Fujii-Kuriyama Y., Muramatsu M. The nucleotide sequence of human fibroblast interferon cDNA. Gene. 1980 Jun;10(1):11–15. doi: 10.1016/0378-1119(80)90138-9. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell. 1984 Aug;38(1):17–27. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- Wu C., Wong Y. C., Elgin S. C. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979 Apr;16(4):807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]
- van den Berg J., van Ooyen A., Mantei N., Schamböck A., Grosveld G., Flavell R. A., Weissmann C. Comparison of cloned rabbit and mouse beta-globin genes showing strong evolutionary divergence of two homologous pairs of introns. Nature. 1978 Nov 2;276(5683):37–44. doi: 10.1038/276037a0. [DOI] [PubMed] [Google Scholar]