Abstract
The Zaire Ebola virus (EBOV) protein VP35 is multifunctional; it inhibits IFN-α/β production and functions as a cofactor of the viral RNA polymerase. Mass spectrometry identified the double stranded RNA binding protein 76 (DRBP76/NFAR-1/NF90) as a cellular factor that associates with the VP35 C-terminal interferon inhibitory domain (IID). DRBP76 is described to regulate host cell protein synthesis and play an important role in host defense. The VP35-IID-DRBP76 interaction required the addition of exogenous dsRNA, but full-length VP35 associated with DRBP76 in the absence of exogenous dsRNA. Cells infected with a Newcastle disease virus (NDV)–expressing VP35 redistributed DRBP76 from the nucleus to the cytoplasm, the compartment in which EBOV replicates. Overexpression of DRBP76 did not alter the ability of VP35 to inhibit type I IFN production but did impair the function of the EBOV transcription/replication complex. These data suggest that DRBP76, via its association with VP35, exerts an anti-EBOV function.
The Ebola virus (EBOV) protein VP35 is of interest because it carries out multiple functions thought to be essential for virus replication and virulence. It functions as a viral polymerase cofactor, an inhibitor of IFN-α/β production, an inhibitor of protein kinase R (PKR) and an inhibitor of RNA interference [1–3]. VP35 inhibits IFNα/β production by impairing the RIG-I pathway [4–9], and studies on recombinant EBOVs encoding mutant VP35 proteins demonstrate that this IFN antagonist function is critical for efficient virus replication and virulence in vivo [9, 10]. Several mechanisms likely contribute to VP35 suppression of RIG-I signaling [5, 7, 8, 11]. Of note, VP35 binds dsRNA, and this activity correlates well with VP35 IFN antagonist function [5, 9, 11]. VP35 is also an essential component of the EBOV RNA polymerase complex [12–14]. The functional viral complex requires the EBOV nucleoprotein (NP), VP35, VP30 and the large protein (L), the catalytic subunit of the polymerase [12, 13]. In this complex, VP35 interacts with both L and NP, and these interactions are required for viral transcription and replication [15–17].
Recently, structural analysis identified multiple, functionally important regions within the VP35 carboxy-terminal domain, referred to as the interferon-inhibitory domain (IID). These include regions critical for VP35 interactions with dsRNA, inhibition of IFN-α/β production and for interaction with NP [11, 18, 19]. A “central basic patch” was found to make contacts with the phosphodiester backbone of dsRNA, and a hydrophobic pocket “end-caps” the blunt ends of dsRNA. Mutations within either the central basic patch or the end-cap abrogated VP35-dsRNA binding and severely attenuated VP35 inhibition of IFN-α/β production. These mutations did not, however, significantly alter VP35 polymerase co-factor function [9, 11]. In contrast, a separate basic patch, the “first basic patch,” was critical for VP35-NP interactions and for VP35 polymerase cofactor function but not IFN-antagonist function [11, 16, 18].
Several reports describe interactions between VP35 and host cell proteins. VP35 interacts with TBK-1 and IKKε, disrupting their interactions with IRF-3 and IRF-7 [8]. Separate studies found that VP35 interacts directly with IRF-7 and with PIAS-1 [7] and with the cytoplasmic dynein light chain 8 [20]. Because the VP35-IID carries out multiple critical functions, we sought to identify IID interacting cellular proteins. We demonstrate that VP35 associates, via its IID, with double stranded RNA binding protein 76 (DRBP76, also known as TCP80, MPP-4, NFAR-1 or NF90 [21]), one of several isoforms derived from the interleukin enhancer binding factor 3 (ILF3) gene [22, 23]. DRBP76 has been described to interact with viral proteins, with viral RNAs, with the interferon induced antiviral kinase PKR and to inhibit the replication of several viruses [21, 24–26]. Here DRBP76 is found capable of inhibiting EBOV polymerase function.
METHODS
Cells and Viruses
Both 293T and Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Sendai virus strain Cantell and recombinant Newcastle disease viruses expressing EBOV VP35 or firefly luciferase were propagated in 10-day old embyonating chicken eggs and have been previously described [27].
Maltose Binding Protein (MBP)–Fusion VP35 IID and VP35 IID Protein Expression and Purification
MBP-fusion VP35 IID proteins were expressed and purified as described previously [18].
Coprecipitations to Identify Protein Bands by Mass Spectrometry
MBP or MBP fused to IID were incubated with lysate prepared with 50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, and protease inhibitors [Complete; Roche]) from 1 × 107 293T cells or lysate mixed with polyI:C (pIC) overnight. MBP was then bound to amylose resin, washed three times, and eluted with 10mM maltose. A fraction was analyzed by protein gel, which was stained with Gel Code Blue (Pierce) and unique bands were excised and submitted for Mass Spectrometry.
Protein Identification
The excised gel bands were washed with 250μl 1:1 acetonitrile:water for 5 minutes, 250μl of 50mM NH4HCO3 in 1:1 acetonitrile:water for 30 minutes, and a final 10mM NH4HCO3 in 1:1 acetonitrile:water wash for 30 minutes. The gels were then dried and rehydrated with 0.2μg of modified porcine trypsin (Promega) in 10mM NH4HCO3 solution and digested at 37°C for 18 hours. The LC-MS/MS analysis was performed on a Waters Q-TOF Ultima with a nanoACQUITY UPLC system. A Waters Symmetry® C18 180μm x 20mm trap column and a 1.7 μm, 75μm x 250mm UPLC™ column at 35°C was used for peptide separation. Trapping was done at 15μl/min, 99% Buffer A (0.1% formic acid) for 1 minute. Peptide separation was performed at 300 nl/min with Buffer A and Buffer B (0.075% formic acid in CH3CN). A linear gradient was run with 5% buffer B at initial conditions, 50% B at 50 minutes, and 85% B at 51 minutes. Data dependent acquisition was performed so that the mass spectrometer switched automatically from MS to 4 consecutive MS/MS modes. The LC-MS/MS data was processed using the Mascot Distiller peak picking and the Mascot protein database search algorithms (Matrix Science). Proteins were searched with a significance threshold of p < 0.5, a peptide tolerance of ±0.6 Da, MS/MS fragment tolerance of ±0.4 Da allowing 1 missed cleavage.
Coimmunoprecipitations and Western Blotting
Proteins expressed in 293T cells were immunoprecipitated (IP) with anti-Flag M2 monoclonal antibody (Sigma) overnight at 4°C, washed three times with lysis buffer and eluted with Flag peptide (Sigma). A fraction of the material was analyzed by western blot, probed with primary anti-Flag (Sigma) or anti-DRBP76 (BD Transduction labs) antibodies and a HRP-secondary antibody. Membranes were developed using a Western Lightning ECL kit (Perkin-Elmer) and Kodak BioMax film (Kodak).
Immunofluorescence
Vero cells were plated onto 12-mm-diameter glass coverslips. The next day, the cells were infected with NDV-VP35 or NDV-Luc. Twenty four hours postinfection, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton-X100 detergent for 10 minutes, blocked in PBS containing 4% normal goat serum and 2% BSA and then incubated with mouse anti-DRBP76 and a NDV polyclonal rabbit anti-NP for 1 hr. Next, the coverslips were incubated with a secondary rhodamine red-x-affinipure goat α-mouse IgG (Jackson), and an alexa 488 goat α-rabbit secondary (Invitrogen) for 30 minutes. To visualize nuclei, Hoechst 33342 (Molecular Probes) was added to one of the final washes. The coverslips were mounted with prolong gold antifade (Invitrogen) and imaged on a DM6000 microscope at the MSSM microscope facility.
IFN-β-luciferase Reporter Assay
Reporter gene assays to measure activation of the IFN-β promoter by SeV have been described [5, 11, 16].
EBOV Transcription/Replication Assay
The EBOV transcription/replication assay used has been previously described [9, 11, 16].
Stable Cell Lines and EBOV Infection
293T cells were transfected with a pKLO.1 lentivirus vector encoding either a scrambled shRNA (a gift from Tia Rai [28]) or a shRNA targeting the 3’ UTR of DRBP76 (Open biosystems, cat# RHS3979-9582267). Transfected cells were selected with and maintained in puromycin. Each cell line was infected with Zaire ebolavirus expressing GFP as indicated and GFP expression was quantified by flow cytometry.
RESULTS
The Interferon Inhibitory Domain (IID) of EBOV VP35 Associates With DRBP76 in the Presence of polyI:C (pIC)
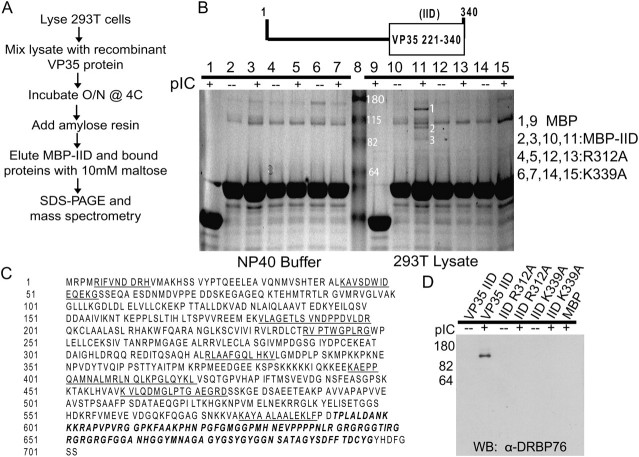
To identify proteins that associate with wild-type or mutant forms of EBOV VP35-IID, MBP alone or MBP-IIDs, purified as described [18], were incubated with lysates from 293T cells or with lysis buffer (Figure 1A). After washing, material was eluted and analyzed by SDS-PAGE and coomassie blue staining (Figure 1B). MBP-VP35-IID did not visibly associate with any cellular protein when compared to control lanes (Figure 1B compare lane 10 to lane 1, 2, and 9). Because VP35 binds dsRNA, MBP-VP35-IID’s were also incubated with cell lysates in the presence of pIC. This resulted in the coprecipitation of unique bands in the MBP-IID+pIC samples. These bands were not present in the MBP (+) pIC or MBP-IID (- -) pIC samples (compare lane 11 to lanes 9 and 10). Mass spectrometry identified the band labeled “3” as double stranded RNA binding protein 76 (DRBP76, also referred to as NF90 or NFAR-1, Figure 1B). Additionally, we identified RNA Helicase A, and nucleolin in this lane (Figure 1B, labeled as 1 and 2 respectively). The DRBP76 interaction was examined further because of its role in host defense against multiple viruses [21]. Figure 1C depicts the individual peptides identified from the mass spectrometry analysis from Figure 1B. The MBP-VP35-IID association with DRBP76 was also confirmed by western blot (Figure 1D).
Figure 1.
The interferon inhibitory domain (IID) of EBOV VP35 binds to the endogenous double stranded RNA binding protein 76 (DRBP76) in the presence of dsRNA. A, Experimental outline of MBP-VP35-IID pull down. B, Recombinant MBP or MBP fused to wild-type or mutant VP35 IID was incubated overnight with NP40 buffer or 293T cell lysate in the presence (+) or absence (−) of pIC. Unique bands (labeled 1, 2, and 3) were excised and analyzed by mass spectrometry. Results from the protein database search indicated that protein bands 1, 2, and 3 corresponded to RNA Helicase A, nucleolin, and DRBP76 (also known at NFAR-1 and NF90), respectively. C, Primary sequence of the DRBP76 isoform described previously [26]. The underlined regions indicate the enzymatically digested peptides identified by tandem mass spectrometry. The bold-italic region represents the peptide sequence used to generate a DRBP76 specific antibody used in (D) that is common to 2 of the 6 isoforms. D, Western blot analysis from the same material used in (B) probed with an anti-DRBP76 antibody.
To further address the role of dsRNA in the IID-DRBP76 interaction, pull-downs were also performed with two IID central basic patch mutants, R312A and K339A, which lack dsRNA binding activity. These mutations abolished the association between the MBP-IID and DRBP76 in the presence of pIC (Figure 1B, lanes 12–15) further supporting a role for dsRNA binding in the VP35-IID -DRBP76 association.
Longer VP35 Constructs Bind DRBP76 in the Absence of dsRNA
We next tested this interaction with a MBP fused to VP35 residues 56–340, which includes both the IID and the coiled-coil domain of VP35 (Figure 2A). DRBP76 associated with this larger MBP-VP35 fusion in the absence of pIC (Figure 2B). Additional pull downs were performed with MBP-fusions to either full-length VP35 constructs or to constructs encoding amino acids 58–340 (Figure 2A) that had mutations in either the central basic patch or first basic patch (Figure 2C). The first basic patch mutants (R225A, K248A, K251A), which retain the ability to bind dsRNA, retained interaction with DRBP76. Interestingly, a full length construct with a mutation in the central basic patch that abrogates dsRNA binding (VP35 R312A) was still able to associate with DRBP76. However, two other central basic patch mutants (R322A and K339A) either had reduced or no association with DRBP76 (Figure 2C). We also transfected a full length plasmid expressing Flag tagged VP35 and could detect interactions with full length DRBP76, but not with a Flag-GFP (Figure 2D). This suggests that DRBP76 may not require dsRNA binding to interact with VP35, but that dsRNA binding likely contributes to the interaction.
Figure 2.
EBOV VP35 amino acids 56–340 bind endogenous DRBP76 in the absence of dsRNA. A, Top panel. Diagram depicting different regions of VP35 IID fused to MBP. B, The VP35-IID constructs were incubated with cell lysate in the presence (+) or absence (−) of pIC. The larger VP35 constructs were only incubated with lysate in the absence of pIC. A fraction of the eluate was analyzed by either coomassie blue stain (bottom) or probed with an anti-DRBP76 antibody (top). C, Characterizing the MBP-VP35 first and central basic patch mutants interaction with DRBP76 in the absence of pIC. The first basic patch mutants used were MBP-VP35 constructs with mutations at amino acids 225, 248, 251. The central basic patch mutants were MBP-VP35 constructs with mutations at amino acids 312, 322, 339. D, A plasmid expressing Flag-VP35 interacts with DRBP76 in 293T cells in the absence of pIC. Western blots against Flag and DRBP76 were perfomed on IP’d material and on whole cell extracts (WCE)
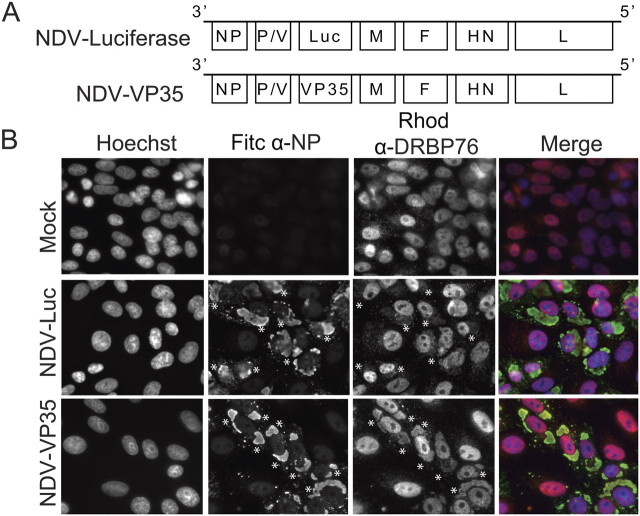
Newcastle Disease Virus–Expressing VP35 Relocates Endogenous DRPB76 from the Nucleus to the Cytoplasm
While VP35 and the remainder of the EBOV replication machinery localize to the cytoplasm, DRBP76 is predominately nuclear [26]. To address the possible impact of VP35 on DRBP76 subcellular localization, Vero cells were infected with recombinant Newcastle disease viruses that express either luciferase (NDV-Luc) or VP35 (NDV-VP35) [27] (Figure 3A) at a multiplicity of infection of 0.5 and then stained for the NDV nucleoprotein, DRBP76 and nuclei at 24 hours post-infection (Figure 3). In mock infected cells DRPB76 was predominately nuclear with punctuate cytoplasmic staining, consistent with previous reports [26]. Cells infected with NDV-Luc showed modest relocalization of DRBP76 from the nucleus to the cytoplasm compared to mock infected cells (Figure 3B). In contrast, NDV-VP35 infected cells triggered an additional increase in the amount of cytoplasmic DRBP76 (Figure 3B). This redistribution appears to require infection as well as VP35 expression since transfection of a VP35 expression plasmid did not result in reproducible relocalization of the endogenous protein (data not shown).
Figure 3.
Infection with NDV-VP35 relocates endogenous DRBP76 from the nucleus to the cytoplasm in Vero cells. A, Schematic of the NDV-VP35 and NDV-Luc viruses used in this experiment. B, Vero cells were infected with NDV-Luc or NDV-VP35 at an MOI of 0.5. Twenty-five hours postinfection, cells were fixed and stained for the NP protein (FITC), DRBP76 (rhodamine) and nuclei (Hoechst). DRPB76 relocalized to the cytoplasm more dramatically in cells infected with NDV-VP35 compared to NDV-Luc. Stars indicate infected cells/DRBP76 relocalization.
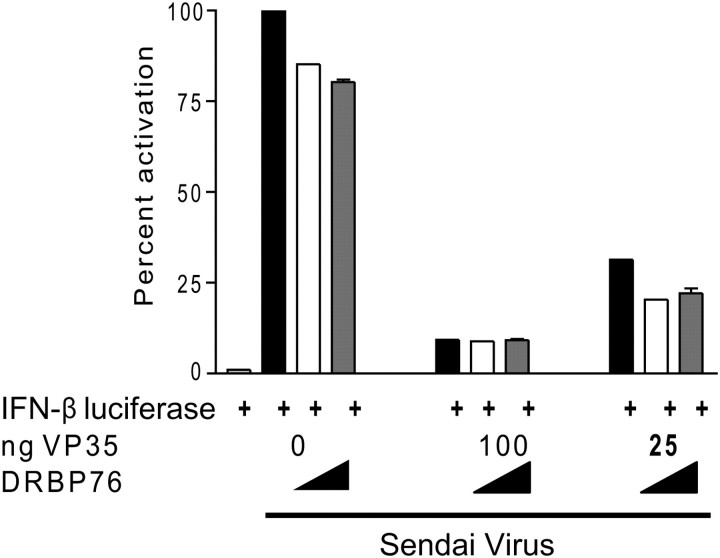
Overexpressed DRBP76 Does Not Alter the Ability of VP35 to Inhibit IFN-β Promoter Activity
VP35 inhibits RIG-I dependent signaling via its IID [5, 7, 11]. Because DRBP76 associates with the IID domain of VP35 (Figures 1 and 2), we tested if DRBP76 perturbs the ability of VP35 to antagonize IFN-β promoter activation. SeV-induced IFN-β promoter driven reporter gene expression is potently block in the presence of VP35 (Figure 4). When a Flag-tagged full length DRBP76 was overexpressed, VP35 inhibition of IFN-β promoter activation was unaffected, even at low concentrations of a VP35 plasmid (25ng) relative to higher concentrations of DRBP76 (750ng) (Figure 4).
Figure 4.
Overexpressed DRBP76 does not alter the ability of VP35 to inhibit IFN-beta reporter activity. An IFN-β promoter-firefly luciferase, and a CMV promoter-renilla luciferase were cotransfected with empty plasmid, a VP35 expression plasmid, or a DRBP76 expression plasmid. The next day, cells were either mock infected or infected with Sendai Virus (SeV) for 18 hours. Cells were lysed and the ratio of firefly luciferase to Renilla luciferase was determined. The positive control (SeV+empty plasmid) was set to 100%.
Overexpressed DRBP76 Inhibits the EBOV Polymerase
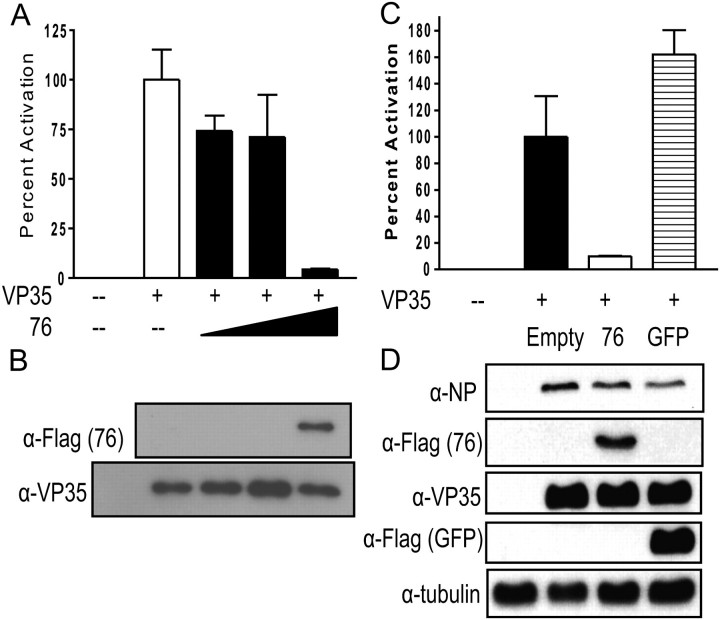
VP35 also functions as a viral polymerase cofactor. To determine if DRBP76 affects EBOV transcription/replication, DRBP76 was overexpressed in the context of a well characterized EBOV polymerase (minigenome) assay [12]. Figure 5A illustrates the effect of DRBP76 on EBOV minigenome activity. At very low doses of DRBP76 (10 and 50 ng) there is a minor inhibition of the EBOV minigenome polymerase activity, but at 250ng of DRBP76 there is potent inhibition of activity as indicated by the dramatic and specific decrease in expression of viral polymerase-dependent reporter gene expression (Figure 5A). Representative western blots indicate that VP35 expression levels are similar for each group (Figure 5B). Overexpressed GFP had no effect on activity, demonstrating the specificity of the DRBP76 effect (Figure 5C), and both VP35 and NP levels remain constant in the presence of overexpressed DRBP76 (Figure 5D).
Figure 5.
DRBP76 overexpression suppresses EBOV minigenome activity. A, Plasmids encoding T7 polymerase, T7 promoter-expressed EBOV proteins VP30, VP35, NP, L and a model genome with the 3’ and 5’ noncoding region of EBOV flanking a GFP/CAT reporter were transfected into 293T cells. A firefly luciferase with a CMV promoter was cotransfected as a transfection efficiency control. These plasmids alone, or 10, 50, or 250 ng of CMV driven Flag-DRBP76 were cotransfected with the minigenome constructs. At 44 hours post transfection, cells were lysed and CAT activity was measured and normalized to firefly luciferase activity. Error bars represent the mean and standard error of the mean of triplicate samples. B, Western blot analysis from representative samples from (A) to test for expression levels of T7 driven VP35 and CMV driven Flag-DRPB76. C, Similar experiment as in (A), but an irrelevant Flag-GFP plasmid at the same concentration as Flag-DRBP76 was transfected (250ng of each). D, Western blot analysis from representative samples from (C) to measure protein expression levels of a subset of the transfected plasmids.
Overexpressed DRBP76 Disrupts VP35-NP Interaction
In the viral replication complex, VP35 must interact with NP and L. To test if DRBP76 disrupts VP35-NP interaction, the viral polymerase complex was reconstituted by transfection in the presence of overexpressed DRBP76, and the status of VP35-NP interaction was assessed. Overexpression of DRBP76 once again inhibited EBOV minigenome activity, but the GFP control did not (Figure 6A). Immunoprecipitation of VP35 with a VP35 monoclonal antibody pulled down roughly equivalent amounts of VP35 in each group (Figure 6B). However, the amount of NP that co-immunoprecipitated with VP35 was drastically reduced in the presence of overexpressed levels of DRBP76 (Figure 6B). This is in contrast to the VP35/NP ratio in the mingenome sample transfected with GFP alone. Figure 6C shows that the total levels of both VP35 and NP are similar in all groups. While there may be a slight reduction in VP35 and NP protein levels in the sample transfected with DRBP76, the VP35/NP ratio in the whole cell extract is similar between all the groups tested (Figure 6C). These data indicate that overexpression of DRBP76 disrupts VP35-NP interactions which correlates with impaired EBOV minigenome function.
Figure 6.
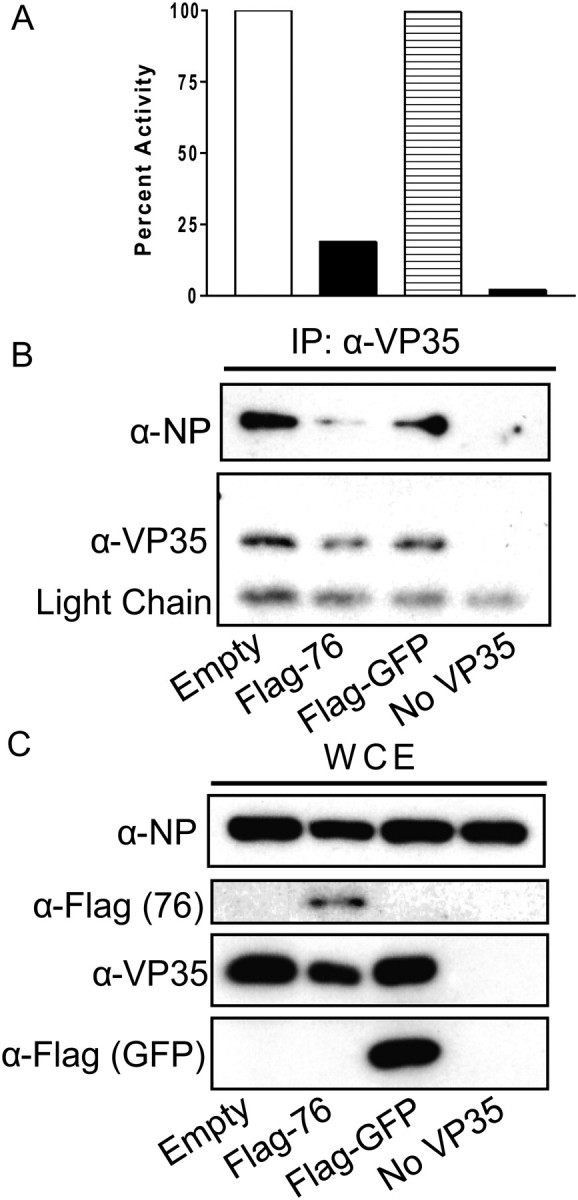
The presence of overexpressed DRBP76 disrupts VP35-NP interaction. A, Graph depicting polymerase activity from the EBOV minigenome co-transfected with empty plasmid, DRBP76-Flag, GFP-Flag or no VP35 (negative control). Each bar represents duplicate samples. B, Fifty percent of the total material from each well in (A) was immunoprecipitated with an anti-VP35 antibody and measured by western blot with antibodies to VP35 or NP. C, Western blot of the total cell extract from the same samples as (B).
Impact of DRBP76 Knockdown on EBOV Replication
To determine if endogenous DRBP76 modulates EBOV replication, we generated a 293T cell-based cell line stably incorporating a lentiviral vector expressing shRNA to DRBP76 (Figure 7A). Cells transfected with the shRNA to DRBP76 had reduced DRBP76 expression levels compared to scrambled shRNA control cells (Figure 7B). Both cell lines were infected with an EBOV expressing GFP at a multiplicity of infection (MOI) of 0.04 or 1.00, and virus growth was monitored by GFP expression. DRBP76 knockdown slightly increased virus replication at a MOI of 0.04, although this increase was small and could be overcome by increasing the MOI to 1.00. Even at a low MOI, the GFP expression in the DRBP76 knockdown cells eventually reached the same level as the control cell line by day 4 postinfection (Figure 7C).
Figure 7.
Impact of DRBP76 knockdown in 293T cells on EBOV replication. A, Schematic of DRBP76/NF90 and the related isoform NF110. The location of the sequence targeted by the shRNA is indicated. B, Western blot depicting levels of DRBP76 in both the DRBP76 knockdown and scrambled control cell line. C, Growth of EBOV in the scrambled and DRBP76 knockdown cell lines, measured by the percentage of GFP positive cells.
DISCUSSION
The VP35 IID is critical for the EBOV VP35 interaction with dsRNA and with NP, interactions important for its IFN antagonist and polymerase cofactor functions, respectively [9, 11, 16]. Because host cell factors that modulate EBOV replication are not well characterized, this study sought to identify host cell proteins that associate with the VP35-IID. DRBP76/NF90/NFAR-1 was thereby identified as a VP35-IID interactor.
DRBP76 is a member of the dsRNA binding protein (DRBP) family. Additional family members include NF110/NFAR-2, PKR, PACT, ADAR, and DICER. DRBP76 regulates translation of host cell mRNAs, and has important functions in host defense for multiple viruses [21, 24, 29–36]. DRBP76 binds both dsRNA as well as ssRNA with secondary structure and preferentially binds RNA encoding AU-rich elements (ARE’s) [37]. While dsRNA enhanced DRBP association with MBP-VP35-IID, MBP-VP3556–340 or a full-length VP35 pulled down DRBP76 in the absence of dsRNA. The role of dsRNA in this interaction requires further study. VP35 self associates to form oligomers through a coiled-coil domain located near its N-terminus (a.a. 82–118) [38]. It is possible that the truncated VP35-IID construct is unable to multimerize as the purified IID was monomeric in solution [18]. However, in vitro studies demonstrate that the IID forms tetramers on dsRNA [11, 19]. Therefore, it is possible that dsRNA promotes VP35-DRBP76 interaction via multimerization.
Given that EBOV replicates entirely in the cytoplasm, it was interesting to consider how VP35 would interact with a protein that is predominantly (but not exclusively) nuclear. We observed a VP35 dependent relocalization from the nucleus to the cytoplasm. The mechanism by which VP35 modulates DRBP76 cellular distribution remains to be defined. Based on a model by Harashima et al, activated PKR phosphorylates DRBP76 (and NFAR-2), resulting in its dislocation from NF45 and the sequestration of DRBP76 in the cytoplasm [29]. Therefore, NDV-induced activation of PKR could explain the modest redistribution of DRBP76 seen in the NDV-Luc–infected cells. However, given that VP35 is reported to inhibit PKR, it seems unlikely that PKR mediates the enhanced redistribution seen with the NDV-VP35 virus. Alternative hypotheses include a model where VP35 associates with newly synthesized or shuttling DRBP76 and prevents its nuclear transport. It will be of interest to determine whether EBOV infection also affects DRBP76 localization and whether this has implications for virus replication or host response to infection.
Overexpression of DRBP76 inhibited EBOV polymerase activity. Furthermore, DRBP76 overexpression perturbed the VP35-NP interaction required for transcription/replication [16]. Thus, DRBP76 might exert an EBOV antiviral mechanism or modulate viral polymerase function. Interestingly, partial knockdown of DRBP76 in 293T cells very slightly enhanced EBOV growth at a low MOI of 0.04. This difference was overcome at a higher MOI of 1.00. It is possible that a more complete DRBP76 knockdown would result in a more dramatic effect. Furthermore, knockdown of the NF110 isoform may also be required for a more dramatic effect to be observed, since the antiviral activity of DRBP76/NF90 and NF110 occurs by a similar mechanism [25, 29]. Therefore, the role of endogenous DRBP76 on EBOV replication requires further study.
Funding
This work is supported by NIH grants (grant 1R56AI089547 to C. F. B. and G. K. A., grant 1F32AI084324 to D. W. L., grants R01AI059536 and AI057158 [Northeast Biodefense Center-Lipkin] to C. F. B.; Northeast Biodefense Center Proteomics Core-Lipkin to E. E. G.; 5F32AI084453 to R. S. S., and R01AI081914 to G. K. A.), an MRCE developmental grant (grant U54AI057160-Virgin to G. K. A.), and the Roy J. Carver Charitable Trust (grant 09-3271 to G. K. A.).
Acknowledgments
We would like to thank Dr Ganes Sen for the DRBP76 expression plasmid and Kathleen Prins for technical assistance.
References
- 1.Basler CF, Wang X, Mühlberger E, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci U S A. 2000;97:12289–94. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schumann M, Gantke T, Muhlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. 10.1128/JVI.00523-09. J Virol. 2009;83:8993–7. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler CF, Mikulasova A, Martinez-Sobrido L, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. 10.1128/JVI.77.14.7945-7956.2003. J Virol. 2003;77:7945–56. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardenas WB, Loo YM, Gale M, Jr., et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits Alpha/Beta interferon production induced by RIG-I signaling. 10.1128/JVI.02199-05. J Virol. 2006;80:5168–78. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartman AL, Towner JS, Nichol ST. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–84. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Chang TH, Kubota T, Matsuoka M, et al. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prins KC, Cardenas WB, Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKK{varepsilon} and TBK-1. 10.1128/JVI.01875-08. J Virol. 2009;83:3069–77. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prins KC, Delpeut S, Leung DW, et al. Mutations abrogating VP35 interaction with double-stranded RNA Render Ebola virus avirulent in Guinea Pigs. 10.1128/JVI.02459-09. J Virol. 2010;84:3004–15. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman AL, Bird BH, Towner JS, Antoniadou ZA, Zaki SR, Nichol ST. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of Ebola virus. 10.1128/JVI.02344-07. J Virol. 2008;82:2699–704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leung DW, Prins KC, Borek DM, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. 2010;17:165–72. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muhlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–42. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muhlberger E, Lotfering B, Klenk HD, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are Sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol. 1998;72:8756–64. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moller P, Pariente N, Klenk HD, Becker S. Homo-oligomerization of Marburg virus VP35 is essential for its function in replication and transcription. 10.1128/JVI.79.23.14876-14886.2005. J Virol. 2005;79:14876–86. doi: 10.1128/JVI.79.23.14876-14886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker S, Rinne C, Hofsäss U, Klenk HD, Mühlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology. 1998;249:406–17. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 16.Prins KC, Binning JM, Shabman RS, Leung DW, Amarasinghe GK, Basler CF. Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. 10.1128/JVI.00925-10. J Virol. 2010;84:10581–91. doi: 10.1128/JVI.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiCarlo A, Moller P, Lander A, Kolesnikova L, Becker S. Nucleocapsid formation and RNA synthesis of Marburg virus is dependent on two coiled coil motifs in the nucleoprotein. Virol J. 2007;4:105. doi: 10.1186/1743-422X-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung DW, Ginder ND, Fulton DB, et al. Structure of the Ebola VP35 interferon inhibitory domain. 10.1073/pnas.0807854106. Proc Natl Acad Sci U S A. 2009;106:411–6. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimberlin CR, Bornholdt ZA, Li S, Woods VL, MacRae IJ, Saphire EO. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. 10.1073/pnas.0910547107. Proc Natl Acad Sci U S A. 2010;107:314–9. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubota T, Matsuoka M, Chang TH, et al. Ebolavirus VP35 interacts with the cytoplasmic dynein light chain. 8 10.1128/JVI.00480-09. J Virol. 2009;83:6952–6. doi: 10.1128/JVI.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barber GN. The NFAR's (nuclear factors associated with dsRNA) RNA Biol. 2009;6:35–9. doi: 10.4161/rna.6.1.7565. [DOI] [PubMed] [Google Scholar]
- 22.Duchange N, Pidoux J, Camus E, Sauvaget D. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene. 2000;261:345–53. doi: 10.1016/s0378-1119(00)00495-9. [DOI] [PubMed] [Google Scholar]
- 23.Saunders LR, Jurecic V, Barber GN. The 90- and 110-kDa human NFAR proteins are translated from two differentially spliced mRNAs encoded on chromosome 19p13. Genomics. 2001;71:256–9. doi: 10.1006/geno.2000.6423. [DOI] [PubMed] [Google Scholar]
- 24.Wang P, Song W, Mok BW, et al. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. 10.1128/JVI.00735-09. J Virol. 2009;83:7850–61. doi: 10.1128/JVI.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeifer I, Elsby R, Fernandez M, et al. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc Natl Acad Sci U S A. 2008;105:4173–8. doi: 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel RC, Vestal DJ, Xu Z, et al. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. 10.1074/jbc.274.29.20432. J Biol Chem. 1999;274:20432–7. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- 27.Leung LW, Park MS, Martinez O, Valmas C, Lopez CB, Basler CF. Ebolavirus VP35 suppresses IFN production from conventional but not plasmacytoid dendritic cells. Immunol Cell Biol. 2011:1–11. doi: 10.1038/icb.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rai T, Mosoian A, Resh MD. Annexin 2 is not required for human immunodeficiency virus type 1 particle production but plays a cell type-dependent role in regulating infectivity. J Virol. 2010;84:9783–92. doi: 10.1128/JVI.01584-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harashima A, Guettouche T, Barber GN. Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. 10.1101/gad.1965010. Genes Dev. 2010;24:2640–53. doi: 10.1101/gad.1965010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the Rhinovirus type 2 internal ribosome entry site. 10.1128/JVI.00243-06. J Virol. 2006;80:6936–42. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrill MK, Dobrikova EY, Gromeier M. Cell-type-specific repression of internal ribosome entry site activity by double-stranded RNA-binding protein. 76 10.1128/JVI.80.7.3147-3156.2006. J Virol. 2006;80:3147–56. doi: 10.1128/JVI.80.7.3147-3156.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin HJ, Kim SS, Cho YH, Lee SG, Rho HM. Host cell proteins binding to the encapsidation signal epsilon in hepatitis B virus RNA. Arch Virol. 2002;147:471–91. doi: 10.1007/s007050200001. [DOI] [PubMed] [Google Scholar]
- 33.Isken O, Baroth M, Grassmann CW, et al. Nuclear factors are involved in hepatitis C virus RNA replication. 10.1261/rna.594207. RNA. 2007;13:1675–92. doi: 10.1261/rna.594207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isken O, Grassmann CW, Sarisky RT, et al. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. 2003;22:5655–65. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao HJ, Kobayashi R, Mathews MB. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc Natl Acad Sci U S A. 1998;95:8514–9. doi: 10.1073/pnas.95.15.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agbottah E, Traviss C, McArdle J, Karki S, St Laurent GC, Kumar A. Nuclear Factor 90(NF90) targeted to TAR RNA inhibits transcriptional activation of HIV-1. Retrovirology. 2007;4:41. doi: 10.1186/1742-4690-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuwano Y, Pullmann R, Marasa BS, et al. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. 10.1093/nar/gkp861. Nucleic Acids Res. 2010;38:225–38. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reid SP, Cárdenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–89. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]