Abstract
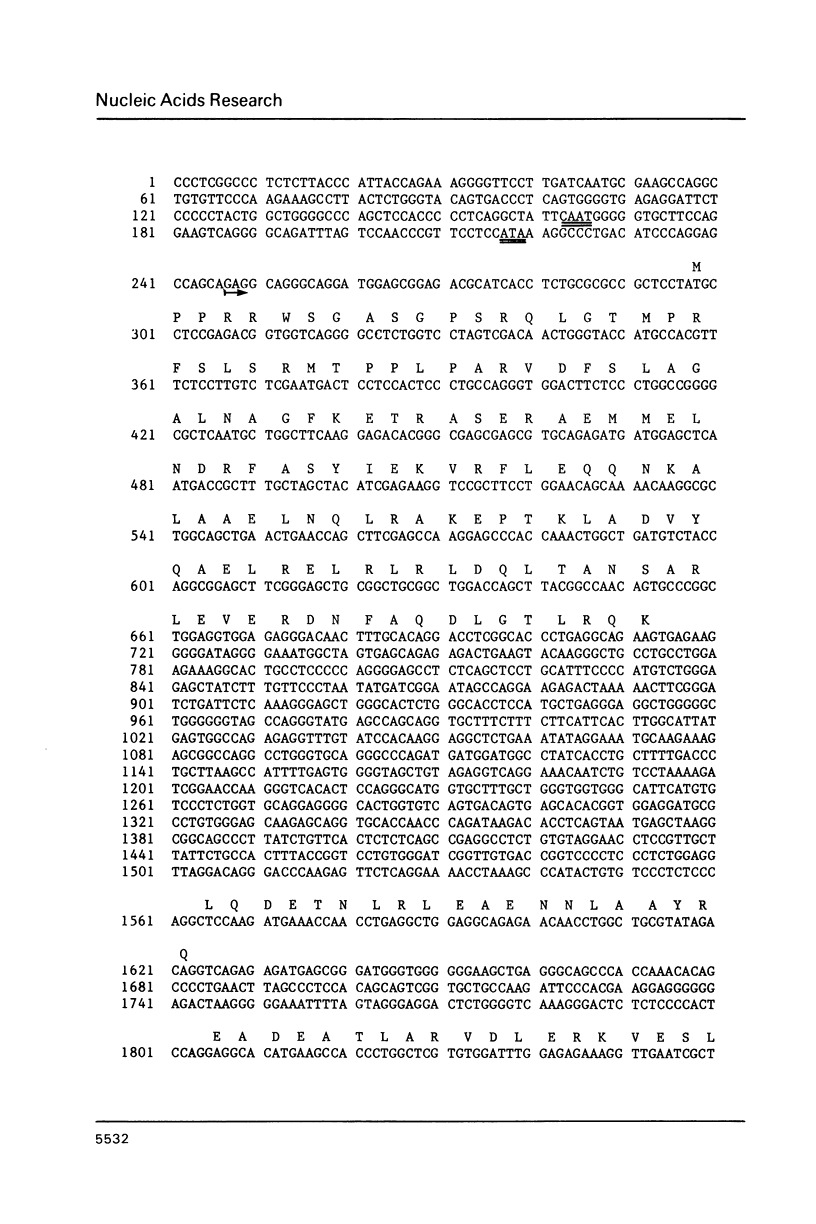
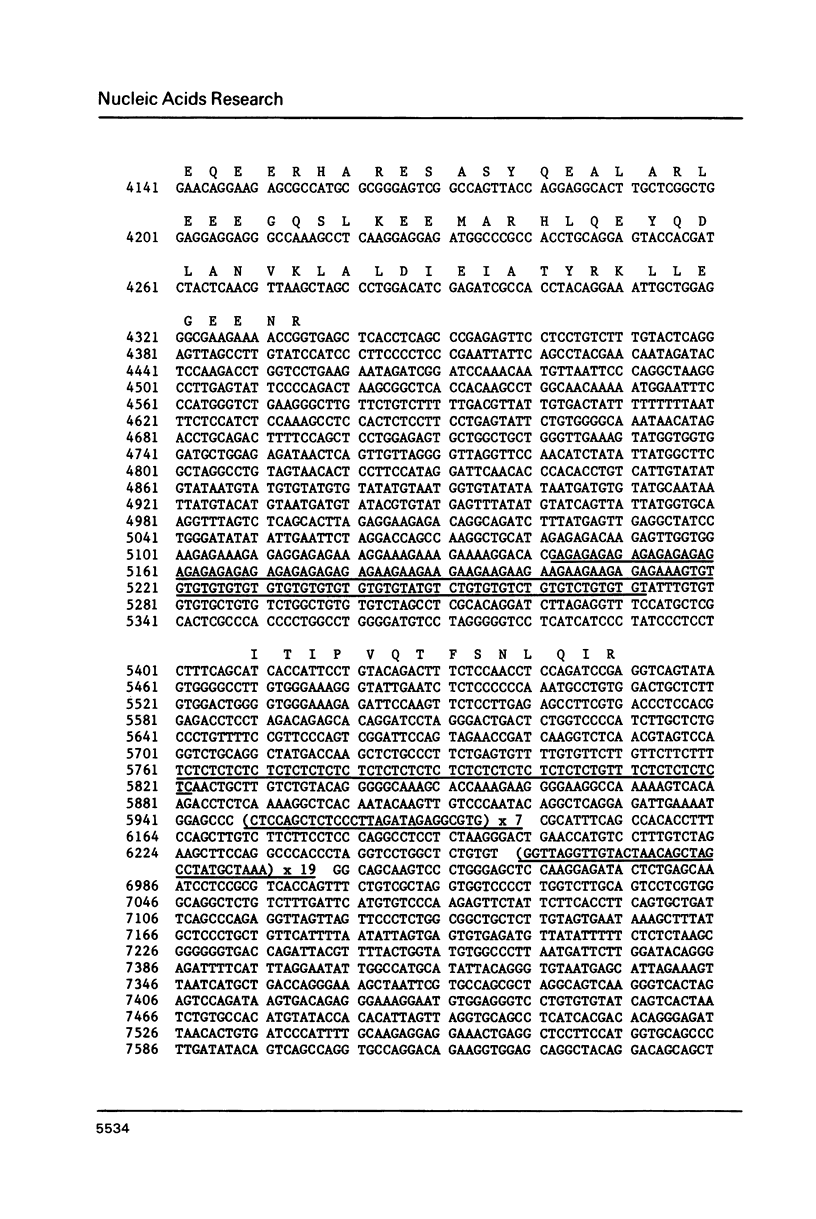
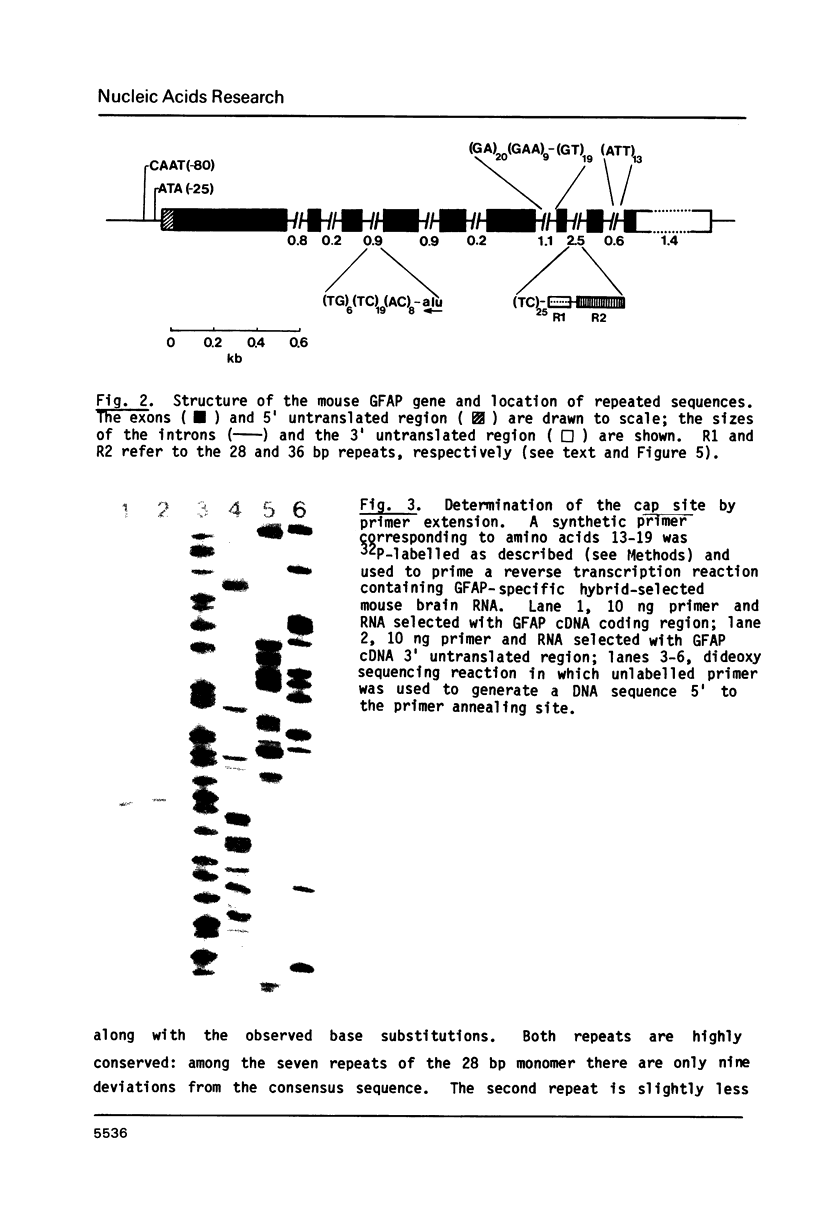
We report the complete sequence of the gene encoding mouse glial fibrillary acidic protein (GFAP), the intermediate filament (IF) protein specific to astrocytes. The 9.8 kb gene includes nine exons separated by introns ranging in size from 0.2 to 2.5 kb. A comparison of the organization of the GFAP gene with that of genes encoding other IF proteins reveals that the structure of IF genes is highly conserved in spite of considerable divergence at the amino acid level. Thus, most of the evolutionary events leading to the placement of introns in IF genes must have occurred prior to the duplication and subsequent divergence of IF genes from a presumptive common ancestral sequence. The conserved gene organization is unrelated to structural features of IF proteins. A curious feature of the GFAP gene is the large number of repeated sequences found in the introns. Six tracts of reiterated di- or trinucleotides are present, plus tandem repeats of two different novel sequences. One repeat is unique to the GFAP gene; the other occurs elsewhere in the mouse genome, although at relatively low frequency.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S. Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids Res. 1981 Jul 10;9(13):3015–3027. doi: 10.1093/nar/9.13.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artymiuk P. J., Blake C. C., Sippel A. E. Genes pieced together--exons delineate homologous structures of diverged lysozymes. Nature. 1981 Mar 26;290(5804):287–288. doi: 10.1038/290287a0. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignami A., Eng L. F., Dahl D., Uyeda C. T. Localization of the glial fibrillary acidic protein in astrocytes by immunofluorescence. Brain Res. 1972 Aug 25;43(2):429–435. doi: 10.1016/0006-8993(72)90398-8. [DOI] [PubMed] [Google Scholar]
- Craik C. S., Buchman S. R., Beychok S. Characterization of globin domains: heme binding to the central exon product. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1384–1388. doi: 10.1073/pnas.77.3.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Hanukoglu I. Unraveling the structure of the intermediate filaments. Cell. 1983 Sep;34(2):332–334. doi: 10.1016/0092-8674(83)90367-7. [DOI] [PubMed] [Google Scholar]
- Fuchs E., Marchuk D. Type I and type II keratins have evolved from lower eukaryotes to form the epidermal intermediate filaments in mammalian skin. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5857–5861. doi: 10.1073/pnas.80.19.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler N., Plessmann U., Weber K. Related amino acid sequences in neurofilaments and non-neural intermediate filaments. Nature. 1982 Apr 1;296(5856):448–450. doi: 10.1038/296448a0. [DOI] [PubMed] [Google Scholar]
- Geisler N., Weber K. The amino acid sequence of chicken muscle desmin provides a common structural model for intermediate filament proteins. EMBO J. 1982;1(12):1649–1656. doi: 10.1002/j.1460-2075.1982.tb01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Hall J. L., Dudley L., Dobner P. R., Lewis S. A., Cowan N. J. Identification of two human beta-tubulin isotypes. Mol Cell Biol. 1983 May;3(5):854–862. doi: 10.1128/mcb.3.5.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982 Jul 22;298(5872):396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Hamada H., Petrino M. G., Kakunaga T. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6465–6469. doi: 10.1073/pnas.79.21.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W. R., Schmid C. W. Repetitive sequences in eukaryotic DNA and their expression. Annu Rev Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- Kenney S., Natarajan V., Strike D., Khoury G., Salzman N. P. JC virus enhancer-promoter active in human brain cells. Science. 1984 Dec 14;226(4680):1337–1339. doi: 10.1126/science.6095453. [DOI] [PubMed] [Google Scholar]
- Lazarides E. Intermediate filaments: a chemically heterogeneous, developmentally regulated class of proteins. Annu Rev Biochem. 1982;51:219–250. doi: 10.1146/annurev.bi.51.070182.001251. [DOI] [PubMed] [Google Scholar]
- Lewis S. A., Balcarek J. M., Krek V., Shelanski M., Cowan N. J. Sequence of a cDNA clone encoding mouse glial fibrillary acidic protein: structural conservation of intermediate filaments. Proc Natl Acad Sci U S A. 1984 May;81(9):2743–2746. doi: 10.1073/pnas.81.9.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S. A., Cowan N. J. Genetics, evolution, and expression of the 68,000-mol-wt neurofilament protein: isolation of a cloned cDNA probe. J Cell Biol. 1985 Mar;100(3):843–850. doi: 10.1083/jcb.100.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchuk D., McCrohon S., Fuchs E. Complete sequence of a gene encoding a human type I keratin: sequences homologous to enhancer elements in the regulatory region of the gene. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1609–1613. doi: 10.1073/pnas.82.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchuk D., McCrohon S., Fuchs E. Remarkable conservation of structure among intermediate filament genes. Cell. 1984 Dec;39(3 Pt 2):491–498. doi: 10.1016/0092-8674(84)90456-2. [DOI] [PubMed] [Google Scholar]
- Milner R. J., Bloom F. E., Lai C., Lerner R. A., Sutcliffe J. G. Brain-specific genes have identifier sequences in their introns. Proc Natl Acad Sci U S A. 1984 Feb;81(3):713–717. doi: 10.1073/pnas.81.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncz M., Solowiejczyk D., Ballantine M., Schwartz E., Surrey S. "Nonrandom" DNA sequence analysis in bacteriophage M13 by the dideoxy chain-termination method. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4298–4302. doi: 10.1073/pnas.79.14.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax W., Egberts W. V., Hendriks W., Quax-Jeuken Y., Bloemendal H. The structure of the vimentin gene. Cell. 1983 Nov;35(1):215–223. doi: 10.1016/0092-8674(83)90224-6. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rogers J. Molecular biology. CACA sequences - the ends and the means? Nature. 1983 Sep 8;305(5930):101–102. doi: 10.1038/305101a0. [DOI] [PubMed] [Google Scholar]
- Saffer J. D., Lerman M. I. Unusual class of Alu sequences containing a potential Z-DNA segment. Mol Cell Biol. 1983 May;3(5):960–964. doi: 10.1128/mcb.3.5.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Singer M. F., Thayer R. E., Grimaldi G., Lerman M. I., Fanning T. G. Homology between the KpnI primate and BamH1 (M1F-1) rodent families of long interspersed repeated sequences. Nucleic Acids Res. 1983 Aug 25;11(16):5739–5745. doi: 10.1093/nar/11.16.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]




