Abstract
In most mammalian species, a critical step of placenta development is the fusion of trophoblast cells into a multinucleated syncytiotrophoblast layer fulfilling essential fetomaternal exchange functions. Key insights into this process came from the discovery of envelope genes of retroviral origin, the syncytins, independently acquired by the human (syncytin-1 and -2), mouse (syncytin-A and -B), and rabbit (syncytin-Ory1) genomes, with fusogenic properties and placenta-specific expression. We previously showed that mouse syncytin-A is essential for the formation of one of the two syncytiotrophoblast layers and for embryo survival. Here, we have generated syncytin-B KO mice and demonstrate that syncytin-B null placenta displays impaired formation of syncytiotrophoblast layer II (ST-II), with evidence of unfused apposed cells, and enlargement of maternal lacunae disrupting the placenta architecture. Unexpectedly, syncytin-B null embryos are viable, with only limited late-onset growth retardation and reduced neonate number. Microarray analyses identified up-regulation of the connexin 30 gene in mutant placentae, with the protein localized at the fetomaternal interface, suggesting gap junction-mediated compensatory mechanisms. Finally, double-KO mice demonstrate premature death of syncytin-A null embryos if syncytin-B is deleted, indicating cooperation between ST-I and ST-II. These findings establish that both endogenous retrovirus-derived syncytin genes contribute independently to the formation of the two syncytiotrophoblast layers during placenta formation, demonstrating a major role of retroviral gene capture, through convergent evolution, to generate multiple placental structures. Although some are absolutely required for completion of pregnancy, others are still amenable to “epigenetic” compensations, thus illustrating the complexity of the molecular machinery that developed during placental evolution.
Keywords: cell–cell fusion, knockout mice, labyrinth
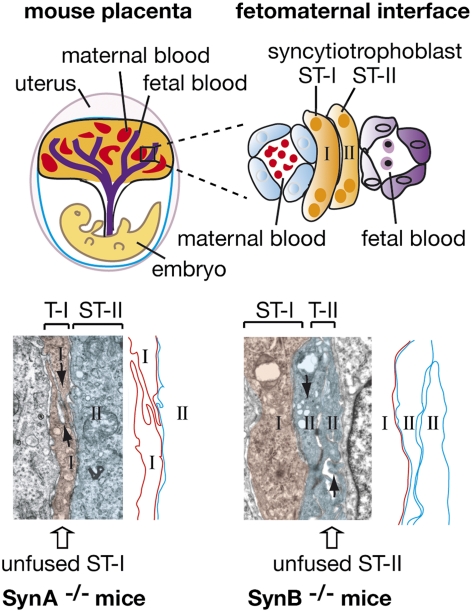
The placenta is an autonomous and transient organ of embryonic origin (same genotype as the embryo) essentially intended for feeding and oxygenating the fetus during intrauterine life. It originates from the outer layer of the implanting blastocyst, the trophoblast. In mammalian species with invasive placentation, including humans and mice, fusion of trophoblast cells into a multinucleated cell layer, called the syncytiotrophoblast, is a key process of placenta morphogenesis (1, 2). The syncytiotrophoblast is first formed during implantation as an invasive structure penetrating the uterine epithelium. It is then maintained throughout gestation as the main fetomaternal interface, mediating nutrient, gas, and waste exchanges between the maternal and fetal blood, along with hormone production and protection of the fetus against the maternal immune system (3–5). The understanding of the molecular mechanisms underlying the process of trophoblast cell–cell fusion came from the identification of envelope proteins encoded by endogenous retroviruses (ERVs), called syncytins. ERVs are the remnants of past infections of the germ line by ancestral retroviruses (6–8), and although the majority of these elements are defective, a few of them still contain intact ORFs, notably in envelope (env) genes. A systematic search through the murine genome has identified that the syncytin-A and -B env genes, which entered the rodent lineage 20 million years ago, are specifically expressed in the placenta and show in vitro cell–cell fusogenic activity involving a mechanism related to env-mediated viral cell entry (9). These genes share closely related functional properties, although they have a distinct origin, with two env genes previously identified in the human genome, syncytin-1 and -2 (10–12), and an env gene recently identified in the rabbit, syncytin-Ory1 (13). By generating KO mice for the syncytin-A gene, we previously showed that this gene is essential for placentation, with syncytin-A–deficient placentae disclosing defects in trophoblast fusion of syncytiotrophoblast layer I (ST-I) and overexpansion of trophoblast cells, ultimately leading to fetal growth retardation and death of the null embryos between 11.5 and 13.5 d of gestation (14). This demonstrated that env genes captured from ERVs have been “co-opted” by their host for a role in syncytiotrophoblast formation in placenta. Furthermore, in the sheep, ERV-derived env genes, clearly distinct from the syncytins, were demonstrated to be involved in periimplantation placental morphogenesis via loss-of-function experiments (15). Interestingly, this stochastic acquisition of genes of “exogenous” origin might be related to the unexpectedly large diversity observed in the structure and physiology of placentation in mammals (5, 16, 17). Whereas an invading syncytial structure is found in all species with interstitial implantation (invasive placentation), comparative studies revealed that the morphology of the definitive fetomaternal interface varies significantly from one type of placentation to another. The syncytiotrophoblast is found apposed to maternal blood vessels in endotheliochorial placenta (carnivores) or comes into direct contact with maternal blood in hemochorial placenta (mice and humans), whereas a hybrid fetomaternal syncytium is formed in synepitheliochorial placenta (ruminants). Even among species with hemochorial placentation, differences can be observed, with muroids having two layers of syncytiotrophoblast, whereas other species (primates and lagomorphs) only have a single layer (18). It is tempting to speculate that such differences could be attributable to differences in the intrinsic properties of the syncytin genes that have been captured in the different mammalian lineages, but experimental evidence was lacking. The mouse placenta provides a highly relevant model to address this hypothesis, given the unique process of trophoblast differentiation into two distinct and highly specialized syncytiotrophoblast layers (ST-I and ST-II) that takes place at the fetomaternal interface (18, 19) and the proviral capture of the syncytin-A and -B genes, which display related but clearly distinct properties (9, 20). Therefore, after demonstrating that syncytin-A is essential for ST-I formation and for embryo survival, we describe here the consequences of the deletion of the second murine syncytin-B gene to determine the respective roles of these two retroviral env genes in murine placentation and further assess the importance of syncytin gene capture in placenta diversity. Syncytin-B–deficient placenta shows defects in formation of ST-II (with evidence of unfused apposed cells), along with enlarged maternal lacunae disrupting the placenta architecture, demonstrating that syncytin-B is essential for syncytial fusion and placenta integrity. In contrast to the lethal effects of syncytin-A deletion, syncytin-B mutant (SynB−/−) neonates are viable, although they still display growth retardation and a reduced number at birth. The induction of connexin 30 expression in mutant placenta and the establishment of junctional complexes between unfused cells suggest compensatory mechanisms to the syncytial defects, probably rescuing SynB−/− embryos. Finally, double syncytin-A/syncytin-B KO mice demonstrate cooperative involvement of both ST-I and ST-II for the structural and functional integrity of the maternofetal interface. These findings provide evidence that genes captured from ancestral retroviruses have been pivotal in establishing diversified placental structures, in a process of convergent evolution and suggest that these structures have developed under different selective constraints during placenta evolution.
Results
Late-Onset Embryonic Growth Retardation and Altered Viability of SynB−/− Embryos.
The mouse syncytin-B ORF (carried by a single 1.8-kb exon) was deleted by homologous recombination using a strategy based on the Cre/LoxP recombination system for generating KO mice (Fig. 1). Mice bearing the conditional alleles, as well as heterozygous SynB+/− animals, obtained after crossing with mice expressing the Cre recombinase under control of the early-acting PGK-1 promoter (Fig. 1), were viable and fertile and exhibited no obvious phenotypic defects. Therefore, heterozygosity of syncytin-B is phenotypically unapparent. SynB+/− mice were then intercrossed, and the neonate progeny (1–3 d after birth) were analyzed. At variance with syncytin-A mutant mice, viable syncytin-B homozygous mutants were obtained, although they displayed growth retardation, showing, on average, 82% of the weight of wild-type (WT) neonates (i.e., 1,790 ± 68 mg, n = 47 for WT vs. 1,469 ± 68 mg, n = 37 for mutants; P < 0.0001) (Fig. 2). This body weight reduction is independent of the sex of the neonates, whereas in adults (6–8 wk old), a significant body weight reduction could still be observed but only in male individuals, which showed 89% of the weight of their WT male littermates (P < 0.02). Correlatively, viable SynB−/− neonates were obtained in a slightly reduced proportion with regard to the expected Mendelian ratio (approximately 20% deficit; 116 null neonates were obtained instead of the 144 expected) (Fig. 2). We then performed timed matings from embryonic day (E) 10.5 to E18.5 to determine the onset of embryonic growth retardation and showed that it became evident at E18.5, with SynB−/− embryos displaying 90% of the weight of their WT littermates (P < 0.03) (Fig. 2). Syncytin-B deletion therefore results in altered viability and late-onset growth retardation of embryos, demonstrating that the gene is required for normal prenatal development.
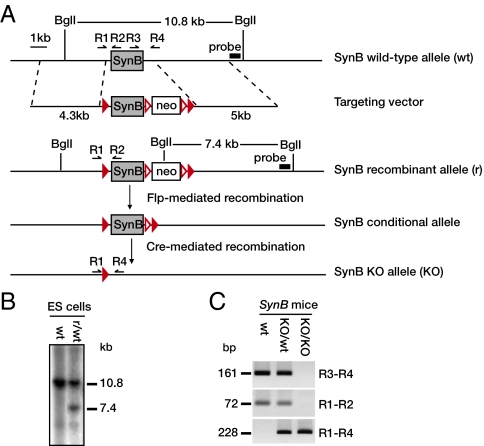
Fig. 1.
Targeted disruption of the single exon syncytin-B gene. (A) Structure of the WT (wt) locus, the targeting vector, the targeted recombinant allele (r), the conditional allele, and the deleted KO syncytin-B allele. The loxP and FRT recombination sites (filled and empty red triangles, respectively), the BglI restriction sites, the probe for Southern blot analysis, and the R1–R4 primers for PCR genotyping are indicated. (B) Southern blot analysis of the DNA from wt ES cells and from the recombinant (r/wt) clone used to establish the recombinant mouse line. BglI-restricted DNA yielded 10.8-kb and 7.4-kb bands for the WT and recombinant alleles, respectively, with the probe in A. (C) PCR-based genotyping of mice, with the R3–R4, R1–R2, and R1–R4 primer pairs in A (fragments <500 bp shown).
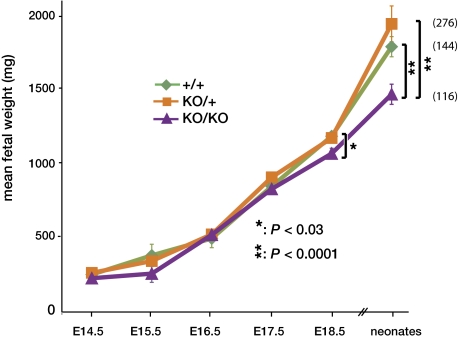
Fig. 2.
Late-onset growth retardation of SynB−/− fetuses. Mean fetal weight of embryos at different embryonic days (E), or of neonates 1–3 d after birth, depending on their genotypes [WT (+/+), heterozygous (KO/+), or syncytin-B null (KO/KO)], is shown. Values (mg) are means ± SEM of a minimum of four living embryos (heart beating): Significant differences between WT and syncytin-B null were observed for E18.5 embryos and neonates only. The number of neonates from each genotype is indicated to the right.
SynA−/−/SynB−/− Double Mutants Die Between E9.5 and E10.5.
Unlike syncytin-A, whose deletion leads to 100% midgestation embryonic death (14), inactivation of both syncytin-B alleles results in low-penetrance embryonic lethality occurring perinatally. Given the fact that syncytin-B is expressed at early stages in the placenta, with the same kinetics as syncytin-A (9, 20), we hypothesized that its deletion may nevertheless provoke subtle changes in early embryonic development, which could only be revealed in a context in which embryonic and placenta development is altered. We therefore generated double-mutant embryos lacking both syncytin-A and syncytin-B genes and tested whether the severity of the syncytin-A phenotype [i.e., embryonic lethality between 11.5 and 13.5 d of gestation (14)] is increased in double mutants. To do so, we generated mice homozygous for syncytin-B deletion and heterozygous for syncytin-A deletion (SynB−/−/SynA+/−) by appropriate crosses between SynA+/− and SynB−/− mice. SynB−/−/SynA+/− mice were then intercrossed, and the viability of the embryos (as judged from the absence of heart beats) was evaluated from E9.5 to E13.5 (Table 1). At E9.5, viable fetuses were recovered at the expected Mendelian ratio, whereas at E10.5, the proportion of double-homozygous null fetuses was about one-third of that expected from a Mendelian distribution. Finally, no viable double-homozygote mutants were found among fetuses at E11.5, E12.5, and E13.5 (Table 1). Thus, SynA−/−/SynB−/− mutants die between E9.5 and E10.5. To check for the onset of lethality in control SynA−/− embryos in this experiment, intercrosses between mice heterozygous for syncytin-A alone (SynA+/−) were carried out in parallel. As previously observed, viable homozygous SynA−/− mutant embryos were still detected at E11.5, E12.5, and E13.5, although with a progressive decrease in their frequency (Table 1) consistent with 100% of syncytin-A null fetuses being dead at E14.5 (14). Thus, the combined loss of syncytin-B and syncytin-A results in embryonic death occurring earlier than that caused by the syncytin-A homozygous mutation alone (i.e., E9.5 vs. E11.5). This indicates that an effect of syncytin-B deletion can be detected as early as E9.5 under conditions in which syncytin-A is knocked out, thus suggesting that syncytin-B plays a role in early embryonic development.
Table 1.
Genotypes of offspring from intercrosses between mice deleted for syncytin-A and/or syncytin-B allele
| Living embryos with indicated genotype |
||||||
| SynB−/− |
SynB+/+ |
|||||
| Day of gestation | SynA+/+ | SynA+/− | SynA−/− | SynA+/+ | SynA+/− | SynA−/− |
| E9.5 | 10 | 15 | 7 | nd | nd | nd |
| E10.5 | 11 | 21 | 3 | nd | nd | nd |
| E11.5 | 7 | 20 | 0 | 6 | 5 | 5 |
| E12.5 | 10 | 18 | 0 | 4 | 10 | 3 |
| E13.5 | 7 | 10 | 0 | 6 | 10 | 1 |
nd, not determined.
Maternal Vascular Abnormalities in SynB−/− Placental Labyrinth.
Syncytin-B expression takes place in the placenta and not in the developing embryo (9), thus suggesting that alteration of syncytin-B null embryo development is caused by defects in placentation. To analyze placental defects, we performed histological analyses of WT and SynB−/− placentae between E11.5 and E18.5. Hemalun-eosin-saffron (HES) staining of paraffin sections from the placentae of SynB−/− embryos exhibited normal overall histological organization, with the maternal decidua, spongiotrophoblast, giant cell, and labyrinth layers correctly positioned (Fig. 3 A and B), as also observed for syncytin-A–deficient placentae. Quantitative RT-PCR further showed unmodified expression of markers of the spongiotrophoblast and the giant cells, the tpbpa/4311 and prl3d1/mPL-I genes, respectively, indicating that these tissues are not affected in mutant placentae (Fig. S1). To examine embryonic vascularization in the labyrinth area, endothelial cells lining the lumen of the fetal blood vessels were stained for CD34 (a cell surface marker strongly expressed in vascular endothelial cells and their progenitors) by immunochemistry (Fig. 3 C and D). CD34-positive cells can be found all over the labyrinth of WT and mutant placentae, indicating that invasion and branching of the fetal vessels occurred normally during the course of labyrinth formation in SynB−/− placentae, as also observed for SynA−/− placentae. However, whereas the fetal vessel space was significantly reduced in SynA−/− placenta because of accumulating labyrinthine cells, the fetal blood vessels in SynB−/− placenta are regularly distributed and not compressed (Fig. 3D), suggesting that overexpansion of trophoblast cells does not occur. The most striking feature of SynB−/− placenta was found to concern the maternal vascular network. As clearly shown in semithin placental sections, the labyrinth region of normal placenta contains narrow maternal lacunae filled with small anucleated red blood cells (Fig. 3E, arrows). In contrast, these lacunae appear dilated and/or more frequent in mutant placenta, with evidence of abnormal blood stasis (Fig. 3E). This was quantified by analyses of E11.5 and E13.5 semithin placental sections (Materials and Methods). Although individual variations in the severity of the phenotype have been observed, an average fivefold increase in the area of maternal lacunae was observed in mutant vs. WT placental labyrinth (2,016 ± 349 μm2 vs. 378 ± 33 μm2; P < 0.0001). In contrast, the area of the fetal vessels was not systematically modified (mutant vs. WT: 479 ± 41 μm2 vs. 584 ± 70 μm2) (Fig. 3E), although enlargement could be observed in some mutant placentae. These vascular abnormalities are moderate in E11.5 to E15.5 placentae and become clearly more important at E16.5 and E18.5. As shown in Fig. 3 F and G, blood sinuses are considerably enlarged and extend from the spongiotrophoblast throughout the entire labyrinthine zona as a continuous blood pool, thus strongly disrupting the architecture of the mutant labyrinth layer (Fig. 3F). Closer examination of E17.5 semithin placental sections (Fig. 3G) revealed dilated maternal blood lacunae whose walls are extremely thin and about to rupture. Anastomosis of these lacunae under high blood pressure may account for the formation of the dramatically enlarged maternal lacunae (that we named giant maternal lacunae) observed at late gestational stages. Moreover, signs of degradation, including breaks and vacuolization, are frequently observed in the mutant placenta labyrinth at late gestational stages (see below). TUNEL assays performed on the placentae of WT and living mutant E18.5 embryos did not show any significant increase in apoptotic cells, at variance with SynA−/− placentae, suggesting that apoptosis was not involved in these defects. These abnormalities of placental vascularization may cause placental dysfunction, such as incomplete material transport between the maternal and fetal blood, presumably leading to the late fetal growth retardation in the SynB−/− embryos.
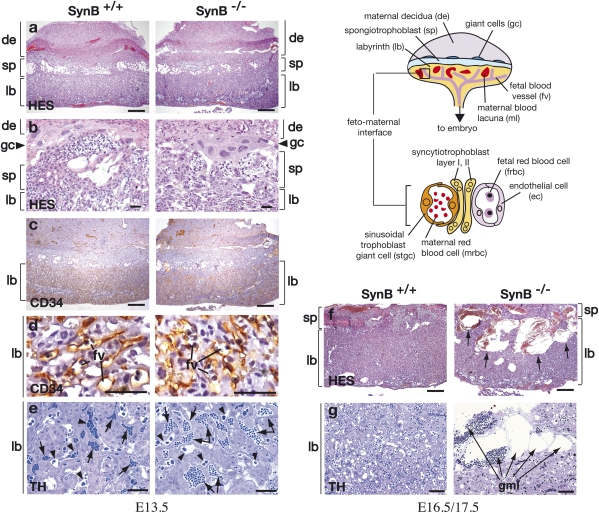
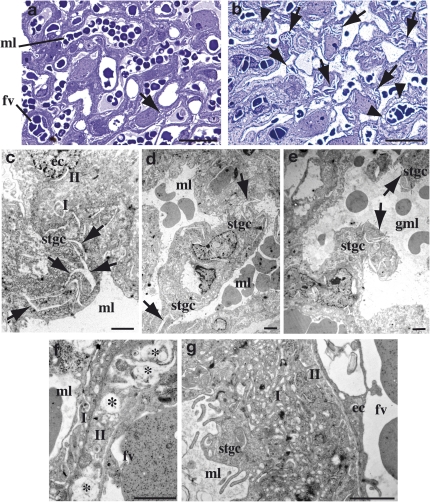
Fig. 3.
Placental anomalies in the labyrinth of syncytin-B null embryos. (Scheme) Schematic representation of the mouse placenta showing the labyrinth (lb), spongiotrophoblast (sp), and giant cells (gc) adjacent to the maternal decidua (de). In the lb (enlarged lower), maternal blood lacunae containing maternal red blood cells (mrbc) are separated from fetal blood vessels (fv) containing nucleated fetal red blood cells (frbc) by a barrier composed of mononuclear STGCs (stgc) and two syncytiotrophoblast layers (ST-I and ST-II), with the latter apposed to the fetal endothelial cells (ec). Light microscopy analysis of WT and mutant placental tissues from E13.5 (A–E), E16.5 (F), and E17.5 (G) embryos. (A and B) HES staining of 7-μm placental sections with the lb, sp, and de delineated, and with the gc shown in the enlarged view in B. The four indicated layers are organized normally in the placenta of SynB−/− mice. (C and D) CD34 histochemical analysis of placental sections showing normal invasion of CD34-positive fbv through the lb; the enlarged view in D shows that the fv are regularly distributed and not squeezed. (E) Thionin (TH) staining of semithin 1-μm placenta Epon sections; in mutant placenta, the maternal blood lacunae (arrows) containing anucleated red blood cells are numerous and significantly enlarged, with evidence of blood stasis. The area of fv (arrowheads) is not significantly modified. (F) HES staining of 7-μm placental sections; the lb of SynB−/− placenta contains dramatically enlarged maternal blood lacunae (arrows) extending from the sp throughout the entire labyrinthine zona. (G) TH staining of semithin 1-μm placenta Epon sections showing, in mutant placenta, several juxtaposed dilated maternal lacunae (arrows) separated by thin walls about to rupture. (Scale bars: A, C, and F, 400 μm; B, D, E, and G, 50 μm.)
Impaired Syncytiotrophoblast Formation in SynB−/− Placenta.
To characterize the labyrinth defects in syncytin-B null placenta further, we then examined the fine structure of the interface between maternal and fetal blood compartments (Fig. 3, Scheme) of E11.5, E12.5, and E13.5 placentae by electron microscopy (EM). As illustrated in Fig. 3 (Scheme) and shown in Fig. 4 A and B, the labyrinth from E12.5 WT placenta showed the typical interhemal barrier separating fetal blood vessels and maternal lacunae, which consists of a layer of mononuclear sinusoidal trophoblast giant cells (STGCs) and two layers of syncytiotrophoblast, ST-I and ST-II, with the latter apposed to the fetal vessel endothelial cells. The two syncytiotrophoblast layers arise from the fusion of trophoblast cells during labyrinth formation. They differ by their cellular composition, with ST-II containing abundant lipid inclusions (Fig. 4B, asterisk), and tightly adhere to each other through frequent desmosomes and gap junctions for intercellular transport (Fig. 4B, arrow and arrowhead). We previously demonstrated that ST-I (but not ST-II) is specifically altered and not syncytial in the placenta of syncytin-A KO mice (14), indicating that syncytin-A is a direct effector in the ST-I fusion process. In syncytin-B null placenta, the interhemal barrier shows normal STGCs and fetal endothelial cells, whereas ST-I appears normal, with a continuous regular membrane structure consistent with a syncytial nature (Fig. 4 C and D). The second layer displays ST-II features (i.e., elongated shape, interaction with ST-I and with the fetal endothelium, presence of lipid droplets) (Fig. 4D). However, on close examination along its entire length, cell membranes extending within the layer itself can be found [Fig. 4 E (Scheme), G, and H]. They are cytoplasmic membranes of unfused apposed cells, indicating impaired syncytialization of ST-II (henceforth designated T-II for trophoblast layer II). In agreement with impaired formation of ST-II but preservation of ST-I in syncytin-B null mice, syncytin-B transcripts were previously detected, by in situ hybridization on normal placenta sections, within the labyrinth close to the fetal vessels, consistent with ST-II, whereas syncytin-A transcripts were detected close to the maternal lacunae, consistent with ST-I (20). Interestingly, in mutant placenta, electron-opaque “dark” cellular junctions are frequently observed joining the lateral edges of two adjacent unfused T-II cells (Fig. 4 E and F). They are concentrated near the maternal blood side or ST-I side (Fig. 4F). Repeatedly, in favorable planes of thin sections, an ordered series of three different types of cellular junctions could be distinguished (Fig. 4E, enlarged in Fig. 4F). Just adjacent to ST-I is a junction reminiscent of a tight junction, with very close apposition of the cell membranes (Fig. 4F, arrow 1). It is followed at a short distance by an intercellular junction with separated cell membranes and filaments inserted, suggesting an adherent junction (Fig. 4F, arrow 2). Finally, more basally, a desmosome-like structure with a wider intercellular space and typical plaque-forming dense structures is observed (Fig. 4F, arrow 3). Association of these three intercellular junctions is highly reminiscent of the junctional complexes that connect the apical side of specialized epithelial cells (21). These structures, which are still present at E17.5, were not seen in WT placenta. Finally, some section planes revealed the apposition of more than two T-II cells, typically three or four, with junctional complexes also connecting them (Fig. 4 G and H). Altogether, these data indicate impaired syncytialization of ST-II, demonstrating that syncytin-B is a direct effector of ST-II fusion, and suggest a compensatory mechanism for the syncytial defects via junctional complex formation (see below).
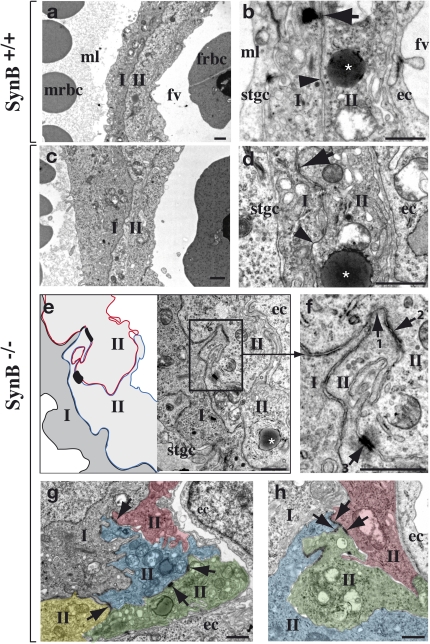
Fig. 4.
Electron micrographs of the labyrinth from E12.5 placentae. (A) Interhemal barrier of WT labyrinth at the maternofetal interface [maternal blood lacuna (ml) with maternal red blood cells (mrbc) and fetal blood vessel (fv) with a nucleated fetal red blood cell (frbc)] with ST-I and ST-II delineated. (B) Enlarged view of the interhemal barrier, with a gap junction (arrowhead) and a desmosome (arrow) between ST-I and ST-II, an endothelial cell (ec), a mononuclear STGC (stgc), and an ST-II–specific lipid inclusion (asterisk). (C and D) Same as A and B for a nonperturbed interhemal zona of SynB−/− placenta, with a gap junction (arrowhead) and a desmosome (arrow) between the two layers and a lipid inclusion (asterisk) shown. (E) Abnormal interhemal interface in SynB−/− placenta, showing two unfused T-II layer cells. (Left) Apposed membranes from T-II cells are schematized. (F) Enlarged view of the area boxed in E, showing the three ordered intercellular junctions connecting two unfused T-II layer cells: tight junction (1), adherent junction (2), and desmosome (3). Section planes showing three (G) and four (H) artificially colored and apposed T-II cells connected by intercellular junctions (arrows). (Scale bars: 1 μm.)
Ultrastructural Defects in the Labyrinth of Late-Gestation SynB−/− Placenta.
To characterize the placental defects observed in SynB−/− placentae at late-gestation stages (i.e., formation of giant maternal lacunae), we examined the fine structure of the E17.5 labyrinth on Epon-embedded placental sections (Fig. 5). As revealed by semithin (1 μm) sections (Fig. 5 A and B), the labyrinth of mutant placentae has a much more porous aspect than that of the WT labyrinth, with the presence of numerous small “comma-like” cytoplasmic breaks (Fig. 5 A and B, arrows) as well as clusters of small vacuoles (Fig. 5 A and B, arrowheads). EM analysis of ultrafine sections (80 nm) further localized the breaks essentially to the cytoplasm of the STGCs (Fig. 5C) that directly line the maternal lacunae (Fig. 3, Scheme, and Fig. 4 B–E). Disruption of the cytoplasm of STGCs at several places (Fig. 5 D and E, arrows) probably leads to STGC rupture and, finally, to maternal blood lacuna anastomosis, as observed in Fig. 5E. In addition to STGC alterations, other signs of degeneration of the interhemal layer were observed in mutant placenta. The lipid droplets present in the T-II layer (Fig. 4 B and D) are no longer filled but translucent and empty, suggesting lipid resorption (Fig. 5F, asterisk), and ST-I has become highly vacuolated (Fig. 5G), with both phenomena contributing to the observed increased porosity of the mutant labyrinth. It should be noted that the integrity of the interhemal zona is preserved in some large areas, such that fetomaternal exchanges probably still occur in some places. All these phenomena may contribute to the disruption of the maternal vascular network observed in late gestation and most likely alter maternal-fetal exchanges, resulting in reduced viability of SynB−/− embryos in late gestation. Finally, quantitative RT-PCR analyses of mutant vs. WT whole-placenta tissues at different gestational stages show no quantitative changes in the expression of a series of trophoblast markers (Fig. S1). Notably, normal expression of markers of the STGC layer and ST-I, the ctsq (22) and syncytin-A genes (20), respectively, confirms normal trophoblast differentiation of both layers in SynB−/− placenta. Expression of the gcm1 gene, a marker of ST-II (20), is not modified either, indicating that its expression is independent of the syncytial status of ST-II, consistent with gcm1 being an upstream regulator of syncytin-B (Discussion).
Fig. 5.
Structure of the labyrinth from E17.5 placentae. Thionin staining of semithin 1-μm section showing numerous intracellular “comma-like” breaks (arrows) and vacuoles (arrowheads) in the mutant placenta (B) but scarce in WT placenta (A). (C–G) Electron microscopic observations of ultrathin (80 nm) sections of mutant placentae. (C) Enlarged view of an STGC (stgc) showing numerous breaks inside the cytoplasm (arrows). Overview of areas with maternal lacunae (ml) separated from each other by stgc, with an arrow indicating breaks that occur within their cytoplasm (D), which will lead to stgc disruption and formation of giant maternal lacunae (gml) as observed (E). Degenerate interhemal barrier of SynB−/− placenta showing lipid resorption in T-II (F, asterisk) and abundant vacuoles in ST-I (G). (Scale bars: A and B, 50 μm; C–G, 2 μm.)
Microarrays Revealed Induction of connexin 30 in the Labyrinth of Mutant Placenta.
To investigate the alterations in the placenta of SynB−/− mice further, we searched for genes whose expression is modified in syncytin-B null placenta compared with WT by performing microarray analyses. RNAs were isolated from WT and mutant placentae of embryos from the same litters and were processed, either pooled or individually, for labeling and hybridization on cDNA microarrays. This was done for three litters at E12.5 and for one litter at E14.5, thus providing four WT/KO hybridization pairs (array data have been deposited in the European Bioinformatics Institute (EBI) database under accession no. E-TABM-1188). We then listed the genes that were found to have statistically significant changes in expression (2-fold or greater, increase or decrease) in at least two WT/KO pairs (Table 2). Only 11 genes were found to have their expression modified. Two striking features of the microarray results were noted. First, the majority of the listed genes (9 of 11) are down-regulated in the mutant placentae, with a 2.5- to 4.4-fold decrease at E12.5. Interestingly, 4 of these genes (apolipoprotein F, apolipoprotein A-II, albumin, and arylacetamide deacetylase) encode proteins involved in lipid metabolism, a fact that would be consistent with the observed growth retardation and late gestational loss of SynB−/− embryos. Second, the most highly regulated gene is the connexin 30/gjb6 gene, coding for an intercellular gap junction protein (23), which displays a 7.7-fold increase in mutant placenta. Among the deregulated genes, connexin 30 is the only one whose alteration is reproducibly observed for the four litters analyzed, at both E12.5 and E14.5 (Table 2). In fact, expression of the 9 down-regulated genes is statistically altered at E12.5 but not at E14.5 (Table 2), indicating that these modifications are only transient and probably compensated for, without excluding their possible involvement in SynB−/− embryo growth retardation, whereas the up-regulated 0610042G0Rik gene is not reproducibly overexpressed in the placenta of every mutant embryo when assayed by quantitative RT-PCR analysis as described below.
Table 2.
List of genes with significant expression changes in syncytin-B KO placenta as determined by microarrays
| Gene name | ID | Gene locus | Fold change (mean) | Function* | Positive WT/KO pair† E12.5 | Positive WT/KO pair† E14.5 |
| Connexin 30/gjb6 (Gap junction protein, beta6) | AK134617 | 14-C3 | +7.7 | Cell communication | 3/3 | 1/1 |
| 0610042G04Rik | AK002909 | 9 | +2.7 | Unknown | 2/3 | 1/1 |
| Apof (Apolipoprotein F) | BC022795 | 10-D3 | −4.4 | Lipid transport | 2/3 | 0/1 |
| Cps1 (Carbamoyl-phosphate synthetase 1) | AK028683 | 1-C3 | −3.2 | Urea cycle | 2/3 | 0/1 |
| ApoH (Apolipoprotein H) | S70439 | 11D | −3 | Secreted plasma glycoprotein | 2/3 | 0/1 |
| Aldob (Aldolase B, fructose-bisphosphate) | AK149598 | 4-B1 | −2.95 | Gluconeogenesis | 2/3 | 0/1 |
| Apoa2 (Apolipoprotein A-II) | CA478728 | 1-H3 | −2.8 | Lipid transport | 2/3 | 0/1 |
| Alb (Albumin) | AJ011413 | 5-E1 | −2.7 | Lipid binding | 2/3 | 0/1 |
| Aadac (Arylacetamide deacetylase) | AK008244 | 3-D3 | −2.68 | Lipase activity | 2/3 | 0/1 |
| Suv39h2 (Suppressor of variegation 3–9 homolog 2) | BC032960 | 2A | −2.6 | Histone methyl transferase | 2/3 | 0/1 |
| Serpina1b (Serine/cysteine peptidase inhibitor, clade A, member 1B | BC037008 | 12E | −2.5 | Serine-type endopeptidase activity | 2/3 | 0/1 |
*From Gene Ontology (www.geneontology.org).
†Proportion of hybridization pairs displaying significant changes in gene expression.
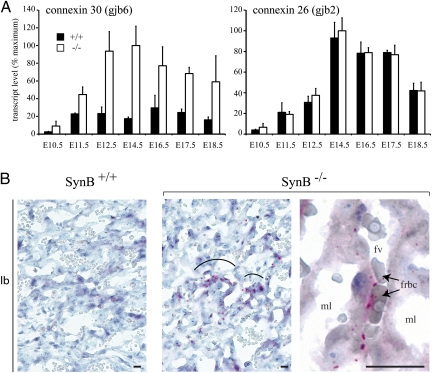
Change in connexin 30 expression was further confirmed by quantitative RT-PCR analysis of RNA extracted form WT and mutant placentae at different embryonic stages from E10.5 to E18.5. The connexin 26/gjb2 gene, which has been shown to be expressed in murine placenta labyrinth (24), was tested as a control (Fig. 6A). A four- to fivefold increase in the expression of connexin 30 was confirmed for embryonic stages E11.5 to E18.5, whereas the connexin 26 transcript levels remained unchanged (Fig. 6A). Localization of connexin 30 was carried out by immunohistochemical staining of placenta cryostat sections. It showed a signal strictly restricted to the labyrinth in mutant placenta (Fig. 6B), whereas no immunoreactivity was observed in WT placenta (Fig. 6B). At higher magnification, positive labeling for connexin 30 was seen as distinctive dot-shaped structures located at the interhemal membrane between the maternal lacunae and fetal capillaries (Fig. 6B), and apparently closer to fetal blood spaces than to maternal blood spaces. These data are consistent with connexin 30 being localized between ST-I and unfused T-II cells, or at the junction between two unfused T-II cells. This could contribute to increase the extension of gap junction areas and the exchange of molecules in mutant placenta through a compensatory mechanism.
Fig. 6.
Induced expression of connexin 30 in the labyrinth of SynB−/− mice. (A) Real-time quantitative RT-PCR analysis of connexin 30 and connexin 26 gene expression in the placenta from WT (+/+) and mutant (−/−) embryos at different embryonic days (E). Values (transcript levels expressed as percent of maximum, after normalization with 18S RNA) are means ± SEM of three to nine living embryos. A significant induction of connexin 30 expression is observed in SynB−/− placenta, whereas the expression of connexin 26 is not modified. (B) Immunohistochemical staining for Connexin 30 on cryostat sections of WT (SynB+/+) or SynB−/− placenta. The labyrinth (lb) region shows positive cells in SynB−/− placenta (Center) but not in WT placenta (Left). (Right) Enlarged view showing Connexin 30-positive signals at the interhemal layer between maternal blood lacunae (ml) and a fetal blood vessel (fv), with the labeling being closer to fetal blood spaces than to maternal blood spaces. frbc, fetal red blood cells. (Scale bar: 25 μm.)
Discussion
During labyrinth formation in the mouse, trophoblast cells fuse to form two syncytiotrophoblast layers, ST-I and ST-II, that separate fetal blood vessels from maternal blood lacunae and mediate fetomaternal exchanges. We previously demonstrated that one of the two murine syncytin genes, syncytin-A, is a direct effector of the formation of ST-I, facing the maternal lacunae (14). Here, to elucidate the role of the second murine syncytin-B gene, we generated mice deficient for syncytin-B by gene targeting. SynB−/− embryos were found at birth, although in reduced proportion (20% neonate lethality), whereas the survivors displayed a significant 10–18% pre- and postnatal growth retardation. The cause of embryonic growth retardation and lethality was found to be defective development of the placenta, consistent with our previous observations that syncytin-B is expressed exclusively in this tissue, being silent in both the developing embryo and adult nonplacental tissues (9). In the labyrinth of syncytin-B null embryos, we showed (i) defects in syncytial formation of the interhemal ST-II, facing the embryonic vessels, with evidence for unfused apposed cells; (ii) signs of degeneration of the interhemal trophoblastic cells; and (iii) progressive anastomosis of maternal blood lacunae, leading to late disruption of the architecture of the labyrinth layer. Although not formally demonstrated, it could be speculated that shutoff of ST-II syncytialization perturbs the interactions and functions of neighboring trophoblast cells (e.g., by disturbing nutrient or waste transfer), thus potentially contributing to the observed degeneration of the interhemal layer and to disorganization of the maternal blood network architecture. This would finally lead to late fetal growth retardation and decreased neonate viability. Although the physiological consequences of syncytin-B deletion became evident in late gestation only, the defects they arise from most probably take place very early in placenta formation, as shown by experiments with double-KO mice, demonstrating premature death of SynA−/−/SynB−/− embryos as soon as E9.5.
Here, we have therefore demonstrated that syncytin-B is essential for the formation of ST-II and for the integrity of the labyrinth architecture, further providing evidence that syncytins have been captured on several independent occasions for a convergent physiological role involved in placenta formation. However, at variance with what is observed in the case of the SynA−/− embryos, deletion of syncytin-B does not result in complete abortion but in a reduced proportion of SynB−/− individuals at birth (summary of the syncytin KO phenotypes is presented in Table 3). Whereas differences in the intrinsic properties of ST-I and ST-II progenitor cells could account for this phenotypic difference (19), compensatory mechanisms are likely involved. Interestingly, microarray analysis of genes differentially expressed in WT vs. mutant placenta revealed a strong induction of the connexin 30 gene in SynB−/− placenta. Expression of this gene is almost undetectable in normal placenta, consistent with connexin 30-deficent mice having no placental defect and being viable (25). In SynB−/− placentae, immunohistochemical experiments further localized the connexin 30 protein at the level of the syncytial interhemal barrier, close to the fetal vessels. Connexin proteins are known to form channels between adjacent cells for the exchange of small molecules and ions (24); thus, connexin 30 molecules may form channels between two unfused T-II cells or between unfused T-II cells and ST-I, with the latter expressing connexin-26, which has been shown to form functional heteromeric gap junctions with connexin 30 (23). This may compensate for altered fetomaternal exchanges or make the T-II unfused mononuclear cells functionally equivalent to a syncytial layer by allowing molecules to pass between cells. The importance of connexin 30 in compensating for the loss of syncytin-B could be definitively assessed by the analysis of mice that are deficient for both syncytin-B and connexin 30 (25). In addition to a compensatory process related to cell–cell communication, refined electron microscopy revealed the occurrence of intercellular junctions tightly associating the plasma membranes of adjacent unfused T-II cells and displaying all the hallmarks of junctional complexes. By analogy with the depicted functions of junctional complexes in specialized epithelial cells (21), the connection of T-II interhemal cells by junctional complexes would form a sealed barrier limiting the passage of cells from mother to fetus and maintain polarized maternofetal transport of molecules by preventing the diffusion of membrane proteins between apical and basal surfaces, finally restoring a syncytium-like function. We therefore propose that in SynB−/− mice, defects in syncytium formation are counteracted by at least two processes, one mediated by gap junctions and the other by junctional complexes, rescuing, at least in part, syncytin-B null embryos.
Table 3.
Summary of fetoplacental phenotypes from syncytin KO mice
| syncytin-A KO | syncytin-B KO |
| 100% embryonic lethality (midgestation) | 20% reduced proportion of neonates (late gestation) |
| Growth retardation (early gestation) | Growth retardation (late gestation) |
| Trophoblast overexpansion | No trophoblast accumulation |
| Squeezed fetal vessels | Enlarged maternal lacunae |
| Unfused ST-I | Unfused ST-II |
The establishment of compensation processes to restore ST-II structure/function in syncytin-B mutant mice, as well as the embryonic lethal consequence of ST-I disruption in syncytin-A mutant mice, does argue for a fundamental selective advantage of the syncytiotrophoblast structures, which may be better suited for carrying out the multifaceted requirements of a placenta than would be a mononucleated cell layer system. Furthermore, crosses resulting in double-KO genotypes, demonstrating an enhanced and more premature death of syncytin-A null embryos if syncytin-B is deleted, strongly suggest that both layers are not redundant but cooperate for the structural and functional integrity of the maternofetal interface.
However, the importance of the syncytiotrophoblast formed at the early implantation stages, when it invades the uterine epithelium, has long been an open question. In fact, comparative studies in humans, as well as in other species with invasive placentation, showing that invading cells are invariably multinucleated syncytiotrophoblasts (16, 17) have suggested that syncytial fusion is required for successful trophoblast invasion. However, experimental support for this hypothesis was lacking because of the unavailability of suitable animal models in which the two processes can be dissociated. In mice, invasive branching morphogenesis is initiated by ST-II that has formed at the tip of the villi from chorionic trophoblast cells (19, 20, 26). Unexpectedly, we showed, using CD34 labeling and ultrastructural analyses, that the process of interstitial invasion, i.e. development of chorionic villi and of the fetal vasculature, occurs normally in syncytin-B mutant placentae, although the syncytial differentiation of ST-II was blocked. Thus, syncytin-B–deficient mice demonstrate that syncytial fusion of trophoblast cells is not a prerequisite for the invasive process and that both differentiation pathways, although taking place simultaneously, are independent. This could be explained by the presence in syncytin genes of regulatory responsive elements for transcription factors involved in the trophoblast invasion pathways. In this respect, the Gcm1 transcription factor, which has been shown to regulate both human syncytin-1 (27) and murine syncytin-B (20) expression and to be required for both trophoblast invasion and syncytial differentiation in mutant mice (28, 29), may have been independently “recruited” by syncytin genes in different species via convergent evolution.
In summary, syncytin-B KO mice are invaluable tools to study placental development and provide important insights into the syncytial differentiation processes. Altogether, the present results now establish that the two syncytin-A and -B genes contribute independently to the formation of the two syncytiotrophoblast layers in the mouse. Considering the high variability of the placental structures in mammals, with, among those with hemochorial placentation, some having one syncytial interhemal layer (humans and rabbits) and others having two (mice) (16–18), our results strongly support the view that the large structural diversity observed among the placentae of different orders of mammals results from the intrinsic properties of the syncytin genes that have been stochastically captured in the course of evolution and that parameters such as the intrinsic level of fusogenicity of the envelope proteins and the cellular expression pattern of env genes, and of their receptor, control and finely tune the syncytial placentation process. Interestingly, our findings show that among the different syncytial structures generated by independent acquisition of retroviral sequences, some of them have become absolutely, and irreversibly, required for completion of pregnancy, whereas others still seem to be amenable to “epigenetic” compensations, thus probably illustrating the complexity and versatility of the molecular machinery that developed in the course of placental evolution.
Materials and Methods
Gene Targeting.
A 129/SvJ ES cell mouse BAC genomic library (Mouse BAC DNA pools; Invitrogen) was screened by PCR assay using primers specific for syncytin-B to obtain the BAC clone C9-1 containing the complete genomic sequence of this gene. The BAC DNA was used to generate by PCR the 4.3-kb 5′-arm and the 5-kb 3′-arm of the targeting vector, corresponding to sequences bracketing the syncytin-B ORF and a 2.6-kb syncytin-B ORF-containing fragment preceded by an upstream LoxP site, using primers listed in Table S1. The targeting vector (Fig. 1) contains the syncytin-B and neomycin resistance (neo) genes flanked by FRT and LoxP recombination sites, allowing their conditional excision. Syncytin-B recombinant mouse lines were established at the Mouse Clinical Institute (Institut Clinique de la Souris, Illkirch, France) using 129S2/SvPas mouse ES cells. After G418 selection, targeted ES clones were identified by PCR assay and further confirmed by Southern blot analysis with a 3′ external probe (Fig. 1). Positive ES cell clones were injected into C57BL/6J blastocysts. Male chimeras positive for germline transmission were used to establish independent recombinant mouse lines. The syncytin-B unique ORF was deleted through breeding with 129/Sv mice expressing the Cre recombinase under control of the early-acting PGK-1 promoter (30) (a gift from M. Cohen-Tannoudji, Institut Pasteur, Paris, France). Syncytin-B conditional and mutant mice were maintained by crossing with 129/Sv mice.
Histological Analyses.
Freshly collected placentae were fixed in 4% (wt/vol) paraformaldehyde at 4 °C and embedded in paraffin, and serial sections (7 μm) were stained with HES. For CD34 immunohistochemistry, paraffin sections were processed for heat-induced antigen retrieval and incubated first with a rat anti-mouse CD34 antibody diluted 1:20 (Hycult Biotechnology) and then with a rabbit anti-rat antibody diluted 1:400 (Southern Biotech). Staining was visualized using the peroxidase/DAB Rabbit PowerVision kit (ImmunoVision Technologies). Slides were counterstained with Mayer's hematoxylin and mounted (Pertex). For connexin 30 immunochemistry, placenta cryostat sections (4 μm) were fixed in acetone and incubated overnight with rabbit anti-mouse connexin 30 antibody (Invitrogen) diluted 1:100 in blocking reagent (ImmunoVision Technologies). Slides were then incubated with the alkaline phosphatase Rabbit PowerVision kit for 30 min and then with Permanent Red Substrate (Dako) with levamisole diluted 1:200 for 25 min. Slides were counterstained with Mayer's hematoxylin and mounted (Aqueous Mount).
EM.
Placentae were fixed with 4% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer for 36 h at 4 °C. Specimens were postfixed with 2% (wt/vol) osmium tetroxide in 0.1 M cacodylate buffer for 2 h and stained with 2% (wt/vol) uranyl acetate in 0.1 M cacodylate buffer-30% (vol/vol) methanol for 1 h. Samples were then dehydrated through a graded series of 30–100% (vol/vol) ethanol and 100% (vol/vol) propylene oxide and embedded in Epon 812 (Electron Microscopy Sciences, Hatfield, PA). Ultrathin sections (80 nm) were contrasted with 4% (wt/vol) uranyl acetate and 0.25% (wt/vol) lead citrate and then examined with an FEI Technai 12 microscope operated at 80 kV. For optical observation, semithin sections (1 μm) were incubated in sodium hydroxide-saturated ethanol for 20 min, stained in 1% thionin for 30 min at 56 °C, and mounted.
RNA Isolation and Gene Expression Profiling.
Total RNAs were extracted from whole placenta using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The quantity and purity of the extracted RNAs were evaluated using a NanoDrop spectrophotometer (Thermo Scientific, Gometz le Chatel, France), and their integrity was measured using an Agilent Bioanalyzer (Agilent Technologies). For microarray experiments, we compared RNAs from WT and mutant placenta from embryos of the same litter; in each litter, RNAs from individuals of the same genotype were either pooled or analyzed individually (array data have been deposited in the EBI database under accession number E-TABM-1188). For microarray hybridizations, 500 ng of total RNA from each RNA sample was amplified and labeled with two fluorescent dyes (Cy5 and Cy3) using the Quick Amp Labeling two-color kit (Agilent Technologies) following the manufacturer's protocol. Cy3-labeled and Cy5-labeled cRNAs were hybridized to the Agilent Mouse Whole Genome Oligo Microarray format 4 × 44K (Agilent Technologies) before washing and scanning. Data were then extracted from scanned images using Feature Extraction software (v10.5.1.1; Agilent Technologies) with default settings. Data from all hybridizations were analyzed with Rosetta Resolver software (Rosetta Biosoftware, Ceiba Solutions, Inc). Dye-swapped arrays were combined using an error-weighted average. Differentially expressed transcripts were then extracted for each hybridization using the following criteria: an absolute fold change >2, a P value calculated by the Rosetta Resolver software <1.10−5, and a log10 (mean of red and green intensities) >1.69. For real-time quantitative RT-PCR, RT was performed with 1 μg of DNase-treated RNA as reported by de Parseval et al. (31). The PCR assay was with 5 μL of cDNA diluted 1:10 in a final volume of 25 μL using SYBR Green PCR Master Mix (or Taqman Universal PCR Master Mix for 18S rRNA detection) (Applied Biosystems) and the primers 5′-TTTTTAATGACTGGCCTTCC and 5′-TTGTCCAATGAACACATCCC for connexin 30 and 5′-GCTTCAGACCTGCTCCTTAC and 5′-ATCTCCCCACACCTCCTTT for connexin 26. The transcript levels were normalized relative to the amount of 18S rRNA (as determined with the primers and Taqman probe from Applied Biosystems).
Statistical Tests.
A two-tailed unpaired t test was used for comparison between mutant and WT embryos or neonate weights. For maternal and fetal vascular area measurement, one WT and two null mutants at E11.5 and E13.5 were examined at a magnification of 400×. At least five fields were scanned at random across the labyrinth, and a two-tailed unpaired t test was used for comparison between WT and mutant. Differences were considered statistically significant when P < 0.05.
Supplementary Material
Acknowledgments
We acknowledge M. Cohen-Tannoudji (Institut Pasteur) for providing the mouse BAC genomic library and help in its screening, for the gift of plasmids and Cre recombinase transgenic mice, and for helpful discussions; O. Bawa and S. Souquere-Besse for technical support; and the Institut Gustave Roussy Service Commun d'Expérimentation Animale for animal care. We are grateful to D. Hernandez-Verdun for valuable discussions. We thank C. Lavialle for comments and critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique and the Ligue Nationale Contre le Cancer (Equipe “labellisée”) as well as by “taxe d'apprentissage” funding from the Institut Gustave Roussy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray expression data have been deposited in the European Bioinformatics Institute (EBI) database (accession no. E-TABM-1188).
See Author Summary on page 18591.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112304108/-/DCSupplemental.
References
- 1.Huppertz B, Tews DS, Kaufmann P. Apoptosis and syncytial fusion in human placental trophoblast and skeletal muscle. Int Rev Cytol. 2001;205:215–253. doi: 10.1016/s0074-7696(01)05005-7. [DOI] [PubMed] [Google Scholar]
- 2.Bischof P, Irminger-Finger I. The human cytotrophoblastic cell, a mononuclear chameleon. Int J Biochem Cell Biol. 2005;37:1–16. doi: 10.1016/j.biocel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Watson ED, Cross JC. Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 2005;20:180–193. doi: 10.1152/physiol.00001.2005. [DOI] [PubMed] [Google Scholar]
- 4.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 5.Wooding P, Burton GJ. Comparative Placentation: Structures, Functions and Evolution. Berlin, Heidelberg: Springer; 2008. [Google Scholar]
- 6.Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- 7.de Parseval N, Heidmann T. Human endogenous retroviruses: From infectious elements to human genes. Cytogenet Genome Res. 2005;110:318–332. doi: 10.1159/000084964. [DOI] [PubMed] [Google Scholar]
- 8.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 9.Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blond JL, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 12.Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new “syncytin” in a third order of mammals. Retrovirology. 2009;6:107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunlap KA, et al. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA. 2006;103:14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leiser R, Kaufmann P. Placental structure: In a comparative aspect. Exp Clin Endocrinol. 1994;102:122–134. doi: 10.1055/s-0029-1211275. [DOI] [PubMed] [Google Scholar]
- 17.Carter AM, Enders AC. Comparative aspects of trophoblast development and placentation. Reprod Biol Endocrinol. 2004;2:46. doi: 10.1186/1477-7827-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez-Verdun D. Morphogenesis of the syncytium in the mouse placenta. Ultrastructural study. Cell Tissue Res. 1974;148:381–396. doi: 10.1007/BF00224265. [DOI] [PubMed] [Google Scholar]
- 20.Simmons DG, et al. Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development. 2008;135:2083–2091. doi: 10.1242/dev.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch S, Nusrat A. Dynamic regulation of epithelial cell fate and barrier function by intercellular junctions. Ann N Y Acad Sci. 2009;1165:220–227. doi: 10.1111/j.1749-6632.2009.04025.x. [DOI] [PubMed] [Google Scholar]
- 22.Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev Biol. 2007;304:567–578. doi: 10.1016/j.ydbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Dahl E, et al. Molecular cloning and functional expression of mouse connexin-30, a gap junction gene highly expressed in adult brain and skin. J Biol Chem. 1996;271:17903–17910. doi: 10.1074/jbc.271.30.17903. [DOI] [PubMed] [Google Scholar]
- 24.Kibschull M, Gellhaus A, Winterhager E. Analogous and unique functions of connexins in mouse and human placental development. Placenta. 2008;29:848–854. doi: 10.1016/j.placenta.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Teubner B, et al. Connexin30 (Gjb6)-deficiency causes severe hearing impairment and lack of endocochlear potential. Hum Mol Genet. 2003;12:13–21. doi: 10.1093/hmg/ddg001. [DOI] [PubMed] [Google Scholar]
- 26.Cross JC, Nakano H, Natale DR, Simmons DG, Watson ED. Branching morphogenesis during development of placental villi. Differentiation. 2006;74:393–401. doi: 10.1111/j.1432-0436.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- 27.Yu CY, et al. GCMa regulates the syncytin-mediated trophoblastic fusion. J Biol Chem. 2002;277:50062–50068. doi: 10.1074/jbc.M209316200. [DOI] [PubMed] [Google Scholar]
- 28.Anson-Cartwright L, et al. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet. 2000;25:311–314. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber J, et al. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol Cell Biol. 2000;20:2466–2474. doi: 10.1128/mcb.20.7.2466-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lallemand Y, Luria V, Haffner-Krausz R, Lonai P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 1998;7:105–112. doi: 10.1023/a:1008868325009. [DOI] [PubMed] [Google Scholar]
- 31.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]