Cellular senescence has been thought to inhibit formation of iPSCs from somatic cells. Lemaitre et al. demonstrate that senescent cells from very old patients do not prohibit reprogramming of somatic cell into iPSCs. In addition, the authors show that many of the old-age-related changes in cells, such as telomere length and gene expression profiles, are reversed in reprogrammed pluripotent cells, which has important implications for regenerative medicine.
Keywords: senescence, aging, reprogramming, iPSC, rejuvenation
Abstract
Direct reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) provides a unique opportunity to derive patient-specific stem cells with potential applications in tissue replacement therapies and without the ethical concerns of human embryonic stem cells (hESCs). However, cellular senescence, which contributes to aging and restricted longevity, has been described as a barrier to the derivation of iPSCs. Here we demonstrate, using an optimized protocol, that cellular senescence is not a limit to reprogramming and that age-related cellular physiology is reversible. Thus, we show that our iPSCs generated from senescent and centenarian cells have reset telomere size, gene expression profiles, oxidative stress, and mitochondrial metabolism, and are indistinguishable from hESCs. Finally, we show that senescent and centenarian-derived pluripotent stem cells are able to redifferentiate into fully rejuvenated cells. These results provide new insights into iPSC technology and pave the way for regenerative medicine for aged patients.
The recent discovery of induced pluripotent stem cells (iPSCs) by Yamanaka's group (Takahashi et al. 2007) and rapid progress in this technology have opened up a new avenue in autologous regenerative medicine whereby patient-specific pluripotent cells could potentially be derived from adult cells. iPSCs have been reproducibly obtained in different cell types by forced expression of the OCT4, SOX2, KLF4, and c-MYC (Takahashi et al. 2007) transcription factor cocktail (OSKM) or by an alternative combination of four factors (OSNL), substituting KLF4 and c-MYC with NANOG and LIN28 (Yu et al. 2007). However, the reprogramming into iPSCs still suffers from several hurdles that have to be overcome for their practical application (Hanna et al. 2010). Among them, cellular senescence increasing with the age was thought to be a critical issue. Senescence is linked to physiological aging, and is characterized by an irreversible cell cycle arrest in response to various forms of stress, including activation of oncogenes, shortened telomeres (replicative senescence), DNA damage, oxidative stress, and mitochondrial dysfunction (Campisi and d'Adda di Fagagna 2007). A common feature is the activation of the p53/p21CIP1 and p16INK4A/pRb pathways, alteration of cell morphology and metabolism, increase in senescence-associated β-galactosidase (SA-β-Gal) activity, and formation of senescence-associated heterochromatic foci (SAHF) (Narita et al. 2003).
It has been reported by several groups that cellular senescence is a barrier to reprogramming due to up-regulation of p53, p16INK4A, and p21CIP1, which are also induced by some of the iPSC reprogramming factors (Banito et al. 2009; Kawamura et al. 2009; Li et al. 2009; Marion et al. 2009a; Utikal et al. 2009). This suggested that cellular aging might be an important limitation for the derivation of iPSCs for therapeutic purposes from elderly individuals. Here we demonstrate that a specific six-factor gene cocktail causes efficient reversing of cellular senescence and reprogramming into iPSCs and that reprogrammed iPSCs generated from senescent and centenarian fibroblasts are indistinguishable from human embryonic stem cells (hESCs). Finally, we demonstrate that redifferentiation led to rejuvenated cells with a reset cellular physiology, defining a new paradigm for cell rejuvenation.
Results and Discussion
Efficient iPSC generation from senescent fibroblasts using a six-factor gene cocktail
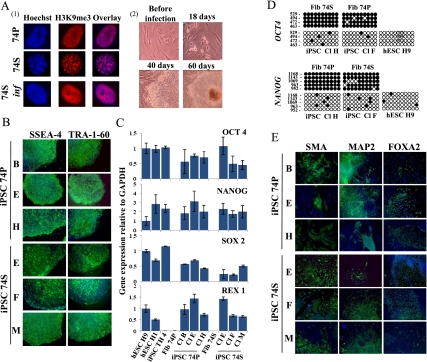
Because iPSC generation from senescent cells was only assayed using the initial gene set OSKM, we decided to investigate the effect of a six-factor combination containing NANOG and LIN28 in addition. Thus, proliferative human diploid fibroblasts from a 74-yr-old donor, which had undergone 12 population doublings (PDs) in culture after the biopsy (hereafter 74P), were induced into replicative senescence by serial passaging (hereafter 74S). Senescence was reached after an additional 39 PDs (51 PDs total) and assessed by FACS analysis, showing a permanent cell cycle arrest (Supplemental Fig. 1A,B), increase in SA-β-Gal activity, up-regulation of p16INK4A and p21CIP1, and formation of SAHF (Fig. 1A, panel 1; Supplemental Fig. 1C,D). These senescent cells were also maintained for >2 mo in culture without any detectable increase in the cell number. Next, 74P and 74S cells were infected with a mix of the individual lentiviruses carrying each of the six genes to further enhance the efficiency of iPSC generation (Liao et al. 2008). One week after infection, we observed first the disappearance of SAHFs in 74S senescent-infected cells (Fig. 1A, panel 1), and then a restored proliferation after 18–20 d. Colonies resembling hESCs appeared at 35–40 d post-infection (Fig. 1A, panel 2), with a mean reprogramming efficiency of 0.06%, similar to 74P infected under the same conditions (Supplemental Fig. 2). We randomly selected six iPSC colonies from proliferative (iPSC 74P) and senescent (iPSC 74S) fibroblasts and further characterized three clones, which were successfully maintained in culture during >35 passages on either hESC feeders or in feeder-free culture conditions. The continued presence of stem cell surface markers SSEA-4 and TRA-1-60 (Fig. 1B), as well as reexpression of endogenous OCT4, SOX2, NANOG, and REX1 pluripotent marker genes, compared with the parental fibroblasts and with H1 and H9 hESCs and IMR90 TH 4 iPSCs (Yu et al. 2007) used as pluripotent control cell lines (Fig. 1C), assessed the successful reprogramming. Reactivation of endogenous pluripotency genes in either iPSCs from senescent (iPSC 74S Cl F) or proliferative (iPSC 74P Cl H) cells was also confirmed by DNA demethylation in previously described CpG-rich regions of the OCT4 and NANOG promoters highly methylated in fibroblasts (Fig. 1D). To exclude any cell type-specific effects, we repeated the same protocol using the human embryonic fibroblast IMR90 induced into replicative senescence, and we also achieved efficient reprogramming from senescent (IMR90S) or proliferative (IMR90P) fibroblasts with the six-factor gene cocktail (Supplemental Fig. 3).
Figure 1.
Induction of pluripotency in proliferative and senescent 74-yr-old-derived cells. (A, panel 1) Detection of SAHF by indirect immunofluorescence of H3K9me3 (red) and Hoechst (blue) in proliferative (74P), senescent (74S), and transduced senescent 74-yr-old cells (74S inf) by the six factors. Seven days after transduction, no SAHF were detected. (Panel 2) Eighteen days after transduction, proliferation of infected 74S cells was observed (74S inf). Around day 40, distinct colonies were observed. Representative phase-contrast images are shown. (B) Immunodectection of surface markers TRA-1-60 and SSEA-4 on iPSCs colonies derived from 74S- and 74P-year-old cells (three independent clones). (C) Quantitative RT–PCR of expression levels for endogenous pluripotency factors in the iPSCs from 74P and 74S and their parental fibroblasts. H1 and H9 hESCs and iPSC TH 4 were used as controls. Transcript levels were normalized to GAPDH expression. Error bars indicate standard deviations from duplicate experiments. (D) Bisulfite sequencing analysis of OCT4 and NANOG promoter regions showing demethylation in iPSCs from 74P and 74S, as in H9 hESCs, compared with parental fibroblasts. Each column of circles for a given amplicon represents the methylation status of CpG dinucleotides in one clone for that region. Open circles are unmethylated CpGs and closed circles methylated ones. The left numbers of each column indicate CpG localization relative to the transcriptional start site. (E) In vitro differentiation experiments of iPSCs reveal their potential to generate cell derivatives of all three primary germ cell layers. Immunodetection of SMA, MAP2, and FOXA2 markers specific for endoderm, ectoderm, and mesoderm, respectively. Nuclei are stained with Hoechst (blue). Three independent clones are shown.
Next we assessed the pluripotent state of the derived iPSCs by evaluating their capacity to differentiate into the three embryonic lineages. All iPSCs clones generated from 74S or 74P were able to differentiate efficiently into endoderm, ectoderm, and mesoderm, as shown by immunodetection of SMA, MAP2, and FOXA2 proteins, respectively (Fig. 1E). We obtained similar results with proliferative and senescent IMR90 (data not shown).
Six-factor but not four-factor combination allows iPSC generation from senescent fibroblasts
NANOG overexpression has been described to facilitate reprogramming in a predominantly cell division rate-independent manner, and LIN28 overexpression, similar to inhibition of the p53/p21CIP1 pathway, increased the cell division rate, resulting in accelerated kinetics of iPSC production (Hanna et al. 2009; Yu et al. 2009). This suggested that the addition of the combination of NANOG and LIN28 might be essential for generating iPSCs from senescent cells. So, we decided to use first the other described combination, OSNL (Yu et al. 2007), on senescent cells, but after 40 d, we did not observe restarted proliferation or formation of hESCs resembling colonies when compared with proliferative cells. Additional treatments (VPA, 5-aza-dC, Wnt3A, or BIO) previously described to enhance the reprogramming efficiency (Feng et al. 2009) also failed to induce iPSCs, as confirmed by the absence of detectable expression of endogenous pluripotency genes in the infected cell population (Supplemental Fig. 4). These results demonstrate that the OSNL combination is not able to reverse the replicative senescence state to generate iPSCs, as also previously described for the OSKM combination.
Altogether, these results indicate that the combination of the six transcription factors (OSKMNL) is a key determinant for a successful and efficent reprogramming reversing cellular senescence to derive iPSCs, without any direct suppression of senescence effectors contributing as safeguards of the genome.
Generation of iPSCs from centenarian fibroblasts using the six-factor combination
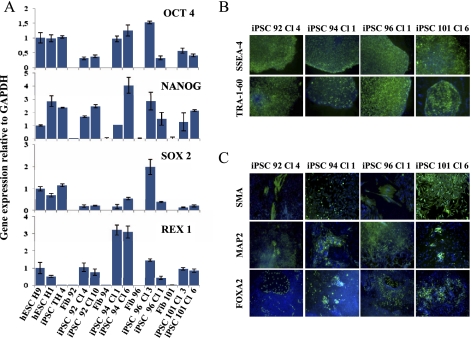
The number of senescent cells increases in the human body during aging, and this is thought to impair tissue homeostasis (Campisi 2011). Increased expression of p16INK4A and p21CIP1 also occurs in cells from aged donors; this correlates with decreased proliferation capacity and an increased ability to activate cell-intrinsic programs such as senescence and might impair their ability to be reprogrammed. To explore whether it might be a limitation, we investigated the possibility of deriving iPSCs for clinical purposes from very old humans on fibroblasts of extremely aged 92-, 94-, 96-, and 101-yr-old donors using our six-factor cocktail. We successfully derived iPSCs from all of the aged donor fibroblasts with efficiency similar to those obtained with senescent fibroblasts. All iPSC clones generated re-expressed endogenous pluripotency genes OCT4, SOX2, NANOG, and REX1 (Fig. 2A); underwent demethylation of CpG in the OCT4 and NANOG promoter regions (Supplemental Fig. 5); and re-expressed the pluripotency cell surface markers SSEA-4 and TRA-1-60 (Fig. 2B; Supplemental Fig. 6A). Finally, we demonstrated the capacity of iPSCs from very old donors to differentiate into the three embryonic lineages as shown previously (Fig. 2C; Supplemental Fig. 6B). These results demonstrate that our procedure efficiently reinstates self-renewal capacity and pluripotency from centenarian fibroblasts, and thus that cellular aging is definitely not a barrier to reprogramming.
Figure 2.
Induction of pluripotency in centenarian-derived cells. (A) Quantitative RT–PCR of expression levels of endogenous pluripotency factors in the indicated donor fibroblasts and iPSC lines. H1 and H9 hESCs as well as iPSC IMR90 TH Cl 4 cells were used as control. Transcript levels were normalized to GAPDH expression. Error bars indicate standard deviations from duplicate experiments. (B) Immunodetection of pluripotent cell surface markers TRA-1-60 and SSEA-4 on derived iPSCs colonies. (C) In vitro differentiation ability of iPSCs in the three primary germ cell layers revealed by immunodetection of SMA, MAP2, FOXA2 markers specific for endoderm, ectoderm, and mesoderm, respectively. Hoechst labeling was used for nuclear staining (blue).
iPSCs generated from senescent and centenarian cells with the six-factor-based strategy did not retain marks of cellular aging
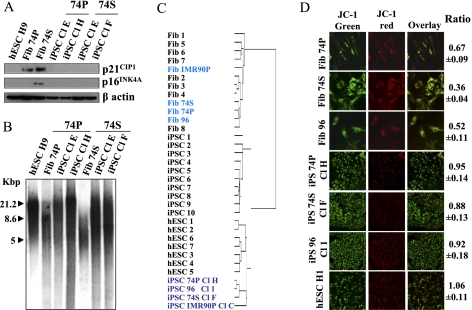
Next we sought to investigate whether iPSCs derived from senescent fibroblasts and aged donors had maintained or lost their aged characteristics. Assuming that increased p16INK4A and p21CIP1 levels are common hallmarks of senescence and aging, we first showed that all iPSCs generated from replicative senescent or proliferative fibroblasts from old donors exhibited down-regulated expression of these proteins, similar to hESC lines (Fig. 3A; Supplemental Fig. 7A,B).
Figure 3.
Disappearance of senescence markers in senescent cell-derived iPSCs. (A) Decrease of p21CIP1 and p16INK4A protein level in iPSCs generated from 74P and 74S cells compared with parental fibroblasts and H9 hESCs, analyzed by Western blotting. β-Actin was used as a loading control. (B) TRF analysis of iPSC clones generated from 74P and 74S cells compared with their parental fibroblasts and H9 hESCs; TRF length is in kilobases (kb). (C) Nonsupervised hierarchical clustering of the global gene expression profiles in fibroblasts, iPSCs, hESCs, and 74- and 96-yr-old fibroblasts and their corresponding iPSCs. (D) Reprogramming of mitochondrial function in iPSCs derived from 74P and 74S and aged 96 cells compared with H1 hESCs, analyzed by JC-1 red/green fluorescence ratio measured by FACS. Red fluorescence indicates a normal membrane potential and green fluorescence indicates a membrane depolarization. A decreasing ratio measures the extent of mitochondrial dysfunction. Experiments were performed in triplicate (±SD for standard deviation).
Resetting telomere function
In humans, progressive telomere shortening is thought to be one of the mechanisms underlying organism aging (Sahin and Depinho 2010). Initial studies of telomere dynamics during reprogramming into iPSCs in mice demonstrated global lengthening of telomeres compared with parental fibroblasts, but also that telomere shortening represents a potent barrier for iPSC generation in engineered telomerase-deficient cells (Marion et al. 2009b). In the case of human cells, there are contradictory reports on the efficiency of telomere lengthening and its maintenance in long-term growth of iPSCs and in their attrition rate in redifferentiation experiments, and contradictory evidence has been amassed previously by the strategy of nuclear transfer for animal cloning (Shiels et al. 1999; Lanza et al. 2000; Alexander et al. 2007), definitely not applicable to humans. Although iPSCs created by current protocols generally exhibit an initial increased telomere size compared with the parental differentiated cells, prematurely aged (shortened) telomeres appear to be a common feature of these iPSCs and their cell progeny (Suhr et al. 2009; Feng et al. 2010; Vaziri et al. 2010).
We measured the telomere length of iPSCs generated from 74S or 74P and found an increased mean size when compared with H9 hESCs (Fig. 3B). Similarly, telomere length was increased in iPSCs derived from senescent or proliferative IMR90 after reprogramming (Supplemental Fig. 8A), like telomeres from centenarians (Supplemental Fig. 8B). Moreover, we were able to maintain all iPSC lines for >110 PDs without either any decrease in telomere length or loss of self-renewal and pluripotency properties, as with H9 hESCs. Interestingly, in some iPSC clones, telomeres were even longer than in H9 hESCs (Supplemental Fig. 9), suggesting possible additional improvements in iPSC generation with OSKMNL for increased proliferation capacity of redifferentiated cells. Collectively, these data demonstrate that our reprogramming protocol leads to erasure of the common marks of senescence and aging in the generated iPSCs, suggesting that it might be especially well suited clinically for deriving stem cells from aged donors.
To further investigate the ability of our six-factor-based reprogramming strategy to erase cellular marks of aging, we analyzed three iPSC clones in detail (iPSC 74P Cl H, iPSC 74S Cl F, and iPSC 96 Cl 1). First, we demonstrated that these clones, whose identities were confirmed by DNA fingerprinting analyses (Supplemental Table 1), exhibited perfectly normal karyotypes (Supplemental Fig. 10). Then, we showed that they acquired the capacity to progress into terminal differentiation, forming organized organ-like structures characteristic of the three embryonic lineages in teratoma (Supplemental Fig. 11), and that the six transgenes used for reprogramming were almost completely down-regulated (Supplemental Fig. 12).
Then, we performed transcriptome analysis of the three iPSCs and their parental counterparts, compared them with an iPSC IMR90 generated with six factors (iPSC IMR90P Cl C) from proliferative IMR90 fibroblasts (Fib IMR90P), and a built set of publicly available data for hESCs and iPSCs generated with four factors from different laboratories (Supplemental Table 2; Assou et al. 2007). We first confirmed that the specific pluripotency genes were expressed in our iPSCs at a level similar to hESCs and iPSCs (Supplemental Fig. 13; Guenther et al. 2010) and then performed a nonsupervised hierarchical clustering. Strikingly, gene expression profiles of proliferative and senescent aged fibroblasts clustered together compared with embryonic and postnatal fibroblasts, indicating that they share a general common aging signature, and our derived iPSCs (obtained using six factors) clustered with hESCs and separated them from previously described iPSCs derived with only four factors (Fig. 3C; Supplemental Fig. 14). Furthermore, selected genes involved in telomere metabolism and maintenance (Vaziri et al. 2010) were similarly reset in our iPSCs, which clustered with hESCs and not with iPSCs generated by four factors (Supplemental Fig. 15). Finally, we found that those iPSCs expressed higher levels of telomerase transcripts compared with hESCs (Supplemental Fig. 16) as well as longer telomeres measured by FISH (Supplemental Fig. 17), demonstrating that our six-factor-based strategy was more efficient for telomere function resetting.
Resetting mitochondria metabolism
Since oxidative stress and mitochondrial dysfunction are well described common features of senescence and aging (Passos et al. 2007; Moiseeva et al. 2009), we wondered whether these functions were also specifically reprogrammed from senescent and aged cells to acquire the anaerobic-like metabolism observed in hESCs. Clustering of transcriptomes using a specific subset of genes previously described as involved in these regulatory pathways (Armstrong et al. 2010; Prigione et al. 2010) confirmed that our iPSCs have reset these functions to an embryonic-like state (Supplemental Fig. 18). The overall mitochondrial activity was measured by evaluating the (ΔΨm) trans-membrane potential calculated by the fluorescence intensity ratio of the JC-1 dye with confocal microscopy and flow cytometry analysis. As previously shown, the red/green ratio decreases in fibroblasts with senescence and aging (Fig. 3D; Passos et al. 2007; Moiseeva et al. 2009). Strikingly, we found an increased membrane potential in iPSCs to a level similar to that in hESCs, confirming the reprogramming of the altered mitochondrial activity from old and senescent fibroblasts to an hESC-like metabolism. Similar results were obtained with iPSCs from IMR90P and IMR90S (Supplemental Fig. 19). Furthermore, we did not observe differences in the reset number, distribution, or morphology of mitochondria in iPSCs when compared with hESCs (Supplemental Figs. 20, 21). Analysis of mitochondrial properties in iPSCs illustrates how nuclear reprogramming in resetting gene expression programs to pluripotency might also reprogram cellular organelle function.
Collectively, our results indicate that senescent and aged cellular physiology is not a barrier to reprogramming and that our six-factor-based reprogramming strategy generates iPSCs indistinguishable from hESCs with some optimized properties.
Reprogramming senescent and centenarian fibroblasts with a six-factor-based strategy leads to redifferentiated cells with rejuvenated physiology
The last important and as-yet-unanswered question is whether reprogramming through pluripotency from senescent cells and cells from very aged patients allows the production of young redifferentiated cells.
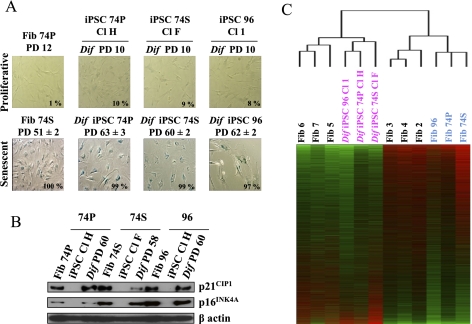
First, we demonstrated that fibroblasts derived from 74P, 74S, and 96 iPSCs did not enter prematurely into senescence, contrary to previous results for redifferentiated iPSCs generated from different healthy proliferating cells or from premature aging syndromes (Feng et al. 2010; Liu et al. 2011). Indeed, our iPSCs had no SA-β-Gal activity after 10 PDs (Fig. 4A, top panel) and proliferated at a rate equivalent to young proliferative fibroblasts. To exclude the possibility that our reprogramming strategy was associated with any mutations in the senescence induction pathways, we demonstrated the ability of redifferentiated fibroblasts to finally re-enter in replicative senescence after extensive culture, as shown by increased SA-β-Gal activity associated with cell cycle arrest (Fig. 4A, bottom panel); the reincreased expression of p16INK4A and p21CIP1 (Fig. 4B); and a reshortened telomere size (Supplemental Fig. 22). But importantly, the number of PDs to reach replicative senescence arrest has been increased after reprogramming through the pluripotent state. Indeed, the 74P parental fibroblasts were infected at PD 12, which without infection would last only a further 39 PDs before replicative senescence led to an iPSC 74P Cl H, which reached replicative senescence after 63 PDs when redifferentiated into fibroblasts, gaining >50% of additional proliferation capacity compared with their parental fibroblasts. Similarly, iPSC 74S Cl F produced from senescent 74S fibroblasts, which had totally exhausted their replicative life span, reentered into replicative senescence after 60 PDs when redifferentiated into fibroblasts. A similar resetting of proliferation capacity was observed in the aged 96 parental fibroblasts initially close to replicative senescence. This demonstrates a rejuvenated proliferation potential of aged and senescent cells induced by our six-factor-based strategy.
Figure 4.
Rejuvenated features of fibroblasts redifferentiated from iPSCs generated from senescent and aged fibroblasts with a six-factor-based reprogramming strategy. (A) SA-β-gal staining of proliferative (PD 10) and replicative senescence (PD 60–63) fibroblasts redifferentiated from iPSCs (PD 90). The percentage of positive cells is indicated in the inset. Experiments were performed in triplicate (±SD for standard deviation). (B) Restoration of p21CIP1 and p16INK4A protein expression in iPSC-derived fibroblasts is triggered during replicative senescence. Western blot analysis using β-actin as the loading control. (C) Nonsupervised hierarchical clustering with gene expression profiles of iPSCs redifferentiated into fibroblasts compared with their 74P-, 74S-, and 96-yr-old parental fibroblasts, postnatal fibroblasts, and embryonic fibroblasts differentiated from H1 hESCs.
In addition to short telomeres, senescent or aged cells closely share profound and numerous transcriptional modifications indicative of their cellular aging, with major impacts on their physiology. To confirm and assess rejuvenation of the global cellular physiology, we analyzed their transcriptomes, and this showed that the global gene expression profiles of redifferentiated fibroblasts from 74P, 74S, and 96 iPSCs actually cluster with young proliferative embryonic fibroblasts derived from the H1 hESCs, whereas their parental counterparts share a common aging signature, separating them from postnatal fibroblasts (Fig. 4C). This result was also confirmed by gene expression profile clustering of genes associated with oxidative stress (Supplemental Fig. 23) and mitochondrial activity (Supplemental Table 3). These results thus demonstrate a rejuvenated physiology of senescent and aged cells after the six-factor-based reprogramming through the pluripotent state.
Taken together, our results show that it is possible, using an adequate reprogramming strategy, to efficiently reprogram senescent cells and cells derived from very old individuals into iPSCs and that cellular senescence and aging are not barriers to reprogramming toward pluripotency. Crucially, we also demonstrated for the first time that, when redifferentiated back into fibroblasts, our cells have rejuvenated extended life spans and characteristics of young proliferative embryonic fibroblasts; thus, these cells have been completely rid of their former aged cellular phenotype. Our demonstration of the reversibility of major aspects of the cellular aging physiology provides a totally unexpected insight into the perceived importance of epigenetic modifications in aging and provides a new paradigm for cell rejuvenation.
Because iPSCs can bypass the ethical concerns related to hESC derivation and potential issues of allogenic rejection, an evident ultimate goal for iPSC research is their use for therapy, as they may represent an ideal source of immune system-tolerated adult cells for future clinical applications. In this context, we believe that our six-factor-based reprogramming, in reversing the aged altered cellular physiology, may constitute an optimal strategy for developing cell-based therapies for aged patients.
Materials and methods
SA-β-Gal
SA-β-Gal activity was detected as previously described (Matsuura et al. 2007).
Generation of human iPSCs
iPSCs were generated using lentiviral vectors containing cDNAs of human OCT4, SOX2, NANOG, LIN28, KLF4, and C-MYC genes in pSin vectors, as described (Takahashi et al. 2007; Yu et al. 2007).
In vitro differentiation assays
Embryoid bodies were generated from iPSCs as previously described, plated onto gelatin-coated tissue culture dishes, and grown for an additional 2 wk into the differentiating medium. For differentiation into fibroblast-like cells from iPSCs, they were cultured in differentiating conditions for 1 wk, selected, and subcultured according to regular fibroblast cell culture protocols.
Teratoma formation assay
For teratoma formation assays, undifferentiated cells were injected into rear leg muscles of NOD/SCID mice. Tumors were resected 2–4 mo after injection and fixed before paraffin embedding. Sections were subjected to hematoxylin and eosin staining before analysis under microscope.
Karyotypes
At least 25 metaphases were analyzed for each cell line using a conventional microscope and IKAROS software (Metasystems).
Bisulphite sequencing
Genomic DNA was treated with EZ-DNA Methylation kit (Zymo Research). The promoter regions of the human Oct3/4 and Nanog genes were amplified by PCR and subcloned into pGEM-T easy vector system (Promega). Ten random clones were picked and checked by sequencing.
Microarray analysis
Total RNA from each sample was prepared, and hybridization with Affymetrix HG-U133 Plus 2.00 GeneChip was performed according to the manufacturer's protocol. Microarrays were processed in the Microarray Core Facility of the Institute for Research in Biotherapy of Montpellier (http://irb.chu-montpellier.fr). A gene expression profile of each cell line was established using the Cluster and TreeView programs.
Mitochondrial membrane potential
Mitochondrial membrane potential was measured with the JC-1 dye (Molecular Probes/Invitrogen).
Telomere length analysis
Telomere length analysis was measured using TeloTAGGG telomere length kit (Roche).
Acknowledgments
We thank Dr. M. Coué, Dr. C. Pfarr, Dr. D. Fisher, and Dr. J. Venables for critical reading and comments of the manuscript. We thank Dr. F. Moreau-Gaudry (University Bordeaux II) for subcloning c-Myc and Klf4 in pSin vectors. We also thank Dr. O. Feraud from the Stem Cell Core Facility of Villejuif for advice, technical assistance, and teratoma formation, and Dr. C. Crozet for gifts of MEF feeder. We also thank Q. Bai for help in transcriptome data analysis, and Dr. C. Cazevieille and C. Sanchez for technical assistance and interpreting of the ME ultrastructural data. Affymetrix microarrays were processed in IRB the Core Facility, CHRU-INSERM-UMI Montpellier. We acknowledge Montpellier RIO Imaging (MRI) for the imaging analysis and FACS facility. This work was supported by an AVENIR INSERM Program/INCa (Convention 2007/3D1616/InsermAvenir-22-1/NG-NC), la Fondation pour la Recherche Médicale (FRM: projet DCR20091217183), l'Association pour la Recherche contre le Cancer (ARC) for the Lemaitre Laboratory; and by grants from the Région Languedoc-Roussillon (Chercheur d'Avenir 09-13198 01) and the Agence Nationale de la Recherche (ANR-07-BLAN-0076-01) for the DeVos Laboratory.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.173922.111.
References
- Alexander B, Coppola G, Perrault SD, Peura TT, Betts DH, King WA 2007. Telomere length status of somatic cell sheep clones and their offspring. Mol Reprod Dev 74: 1525–1537 [DOI] [PubMed] [Google Scholar]
- Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, Przyborski S, Lako M 2010. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells 28: 661–673 [DOI] [PubMed] [Google Scholar]
- Assou S, Le Carrour T, Tondeur S, Strom S, Gabelle A, Marty S, Nadal L, Pantesco V, Reme T, Hugnot JP, et al. 2007. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells 25: 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. 2009. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J 2011. Cellular senescence: putting the paradoxes in perspective. Curr Opin Genet Dev 21: 107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F 2007. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740 [DOI] [PubMed] [Google Scholar]
- Feng B, Ng JH, Heng JC, Ng HH 2009. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4: 301–312 [DOI] [PubMed] [Google Scholar]
- Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim KS, Lanza R 2010. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 28: 704–712 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA 2010. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R 2009. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462: 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R 2010. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell 143: 508–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza RP, Cibelli JB, Blackwell C, Cristofalo VJ, Francis MK, Baerlocher GM, Mak J, Schertzer M, Chavez EA, Sawyer N, et al. 2000. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science 288: 665–669 [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Wu Z, Wang Y, Cheng L, Cui C, Gao Y, Chen T, Rao L, Chen S, Jia N, et al. 2008. Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Res 18: 600–603 [DOI] [PubMed] [Google Scholar]
- Liu GH, Barkho BZ, Ruiz S, Diep D, Qu J, Yang SL, Panopoulos AD, Suzuki K, Kurian L, Walsh C, et al. 2011. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature 472: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009a. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA 2009b. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4: 141–154 [DOI] [PubMed] [Google Scholar]
- Matsuura F, Hirano K, Ikegami C, Sandoval JC, Oku H, Yuasa-Kawase M, Tsubakio-Yamamoto K, Koseki M, Masuda D, Tsujii K, et al. 2007. Senescent phenotypes of skin fibroblasts from patients with Tangier disease. Biochem Biophys Res Commun 357: 493–498 [DOI] [PubMed] [Google Scholar]
- Moiseeva O, Bourdeau V, Roux A, Deschenes-Simard X, Ferbeyre G 2009. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol Cell Biol 29: 4495–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113: 703–716 [DOI] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, et al. 2007. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol 5: e110 doi: 10.1371/journal.pbio.0050110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J 2010. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 28: 721–733 [DOI] [PubMed] [Google Scholar]
- Sahin E, Depinho RA 2010. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature 464: 520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels PG, Kind AJ, Campbell KH, Wilmut I, Waddington D, Colman A, Schnieke AE 1999. Analysis of telomere length in Dolly, a sheep derived by nuclear transfer. Cloning 1: 119–125 [DOI] [PubMed] [Google Scholar]
- Suhr ST, Chang EA, Rodriguez RM, Wang K, Ross PJ, Beyhan Z, Murthy S, Cibelli JB 2009. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS ONE 4: e8124 doi: 10.1371/journal.pone.0008124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Chapman KB, Guigova A, Teichroeb J, Lacher MD, Sternberg H, Singec I, Briggs L, Wheeler J, Sampathkumar J, et al. 2010. Spontaneous reversal of the developmental aging of normal human cells following transcriptional reprogramming. Regen Med 5: 345–363 [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA 2009. Human induced pluripotent stem cells free of vector and transgene sequences. Science 314: 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]