Abstract
Cell surface lectin staining, examination of Golgi glycosyltransferases stability and localization, and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis were employed to investigate conserved oligomeric Golgi (COG)-dependent glycosylation defects in HeLa cells. Both Griffonia simplicifolia lectin-II and Galanthus nivalus lectins were specifically bound to the plasma membrane glycoconjugates of COG-depleted cells, indicating defects in activity of medial- and trans-Golgi-localized enzymes. In response to siRNA-induced depletion of COG complex subunits, several key components of Golgi glycosylation machinery, including MAN2A1, MGAT1, B4GALT1 and ST6GAL1, were severely mislocalized. MALDI-TOF analysis of total N-linked glycoconjugates indicated a decrease in the relative amount of sialylated glycans in both COG3 KD and COG4 KD cells. In agreement to a proposed role of the COG complex in retrograde membrane trafficking, all types of COG-depleted HeLa cells were deficient in the Brefeldin A- and Sar1 DN-induced redistribution of Golgi resident glycosyltransferases to the endoplasmic reticulum. The retrograde trafficking of medial- and trans-Golgi-localized glycosylation enzymes was affected to a larger extent, strongly indicating that the COG complex regulates the intra-Golgi protein movement. COG complex-deficient cells were not defective in Golgi re-assembly after the Brefeldin A washout, confirming specificity in the retrograde trafficking block. The lobe B COG subcomplex subunits COG6 and COG8 were localized on trafficking intermediates that carry Golgi glycosyltransferases, indicating that the COG complex is directly involved in trafficking and maintenance of Golgi glycosylation machinery.
Keywords: Brefeldin A, COG complex, Golgi, lectin, retrograde trafficking
Introduction
The Golgi apparatus is a hub of intracellular protein trafficking and a major site for the glycosylation of proteins and lipids (Warren and Malhotra 1998). Protein traffic through the Golgi occurs in both anterograde (cis-to-trans) and retrograde (trans-to-cis) directions (Shorter and Warren 2002). The steady-state localization of Golgi glycosylation machinery is maintained through retention and retrieval mechanisms (Opat et al. 2001). Proposed Golgi retention mechanisms (Gleeson et al. 1994; Munro 1998; Nilsson et al. 2009; Tu and Banfield 2010) include enzyme oligomerization/aggregation (Aoki et al. 1992; Nilsson et al. 1993; Opat et al. 2000; Qian et al. 2001) and lipid-based protein partitioning (Tang et al. 1992; Teasdale et al. 1992; Wong et al. 1992; Munro 1995). The motif-based retrieval mechanisms (Schmitz et al. 2008; Tu et al. 2008) include the coat protein (COPI)-dependent (Helms and Rothman 1992; Glick et al. 1997) and COPI-independent (Girod et al. 1999) intra-Golgi and Golgi-endoplasmic reticulum (ER) recycling of components of glycosylation machinery. Both Rab and Arf subfamilies of small GTP-binding proteins and their effectors are required to maintain the Golgi homeostasis and proper localization of Golgi resident glycosyltransferases (Bonifacino and Glick 2004; Ohtsubo and Marth 2006). We, and others, have previously demonstrated that one of the Rab effectors, the conserved oligomeric Golgi (COG) complex, is essential for the proper localization of Golgi-localized glycosyltransferases and several other Golgi resident proteins (Shestakova et al. 2006; Ungar et al. 2006; Smith and Lupashin 2008).
The COG complex, a putative Golgi tethering factor, consists of eight proteins named COG1 through COG8 (Suvorova et al. 2001, 2002; Whyte and Munro 2001; Ungar et al. 2002). Based on yeast genetic studies and observations with electron microscopy (Ungar et al. 2002), the COG subunits have been grouped into two lobes consisting of COG1 to COG4 for lobe A, and COG5 to COG8 for lobe B. Mutations or depletions of the COG complex subunits lead to improper glycosylation of glycoproteins and glycolipids in both yeast and mammalian cells (Bruinsma et al. 2004; Shestakova et al. 2006) and in humans cause multiple organ system pathologies referred to as congenital disorders of glycosylation (CDG) (Wu et al. 2004; Foulquier 2009). Several experimental approaches have been used to characterize the glycosylation and trafficking defects in fibroblasts isolated from COG-deficient human patients, including peanut agglutinin (PNA) lectin-staining of plasma membranes (Foulquier et al. 2006, 2007; Kranz et al. 2007; Morava et al. 2007; Ng et al. 2007), isoelectric focusing of serum glycoproteins (Wu et al. 2004), Brefeldin A (BFA)-induced redistribution of Golgi resident protein to the ER (Steet and Kornfeld 2006), and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis of serum glycoproteins (Foulquier et al. 2007; Kranz et al. 2007; Paesold Burda et al. 2009; Reynders et al. 2009; Zeevaert, Foulquier, Cheillan, et al. 2009, Zeevaert, Foulquier, Dimitrov, et al. 2009). These studies revealed an array of protein glycosylation and trafficking defects in cells that were chronically depleted for COG complex function. However, the systematic characterization of the primary glycosylation/trafficking defects in cells that are acutely depleted for different COG complex subunits was absent. We have previously characterized glycosylation and membrane trafficking defects in both COG3- and COG7-depleted HeLa cells (Shestakova et al. 2006). In this work, we have extended our initial studies to systematically characterize and compare glycosylation and Golgi trafficking defects in HeLa cells acutely depleted for COG2, COG4, COG6 and COG8 subunits.
Results
COG complex depletions alter the N-linked glycosylation of plasma membrane-localized glycoconjugates
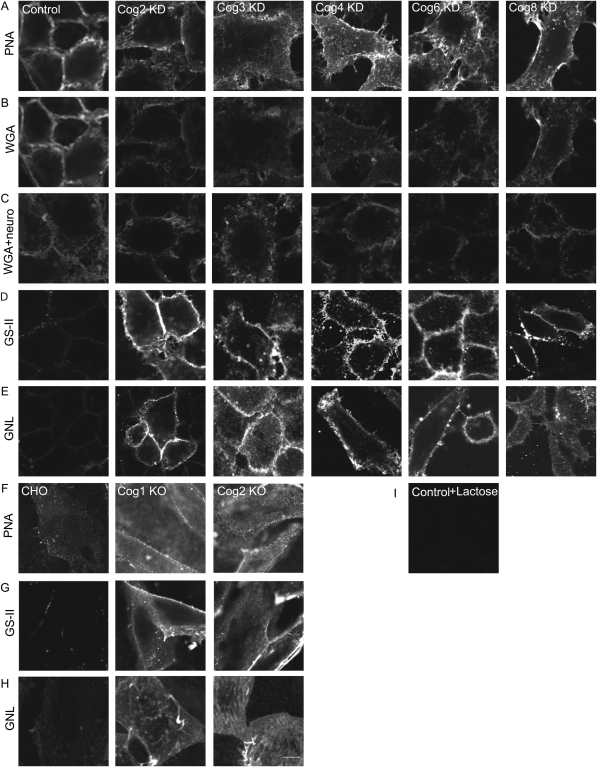
Recently published characterization of human fibroblasts obtained from patients with CDG-II that are deficient for the COG complex subunits demonstrated that the plasma membrane of these mutant cells is positively stained with fluorescently labeled PNA (Foulquier et al. 2006, 2007). The PNA binds specifically to terminal galactosyl residues (Lotan et al. 1975) and therefore PNA-positive staining indicates that there is an increase in the amount of terminal galactoses in plasma membrane O-linked glycoconjugates in COG-deficient cells. We have demonstrated previously that glycosylation of plasma membrane-localized CD44 is significantly altered in both COG3 KD and COG7 KD HeLa cells (Shestakova et al. 2006). To test whether the COG complex-depleted HeLa cells are also prone to accumulate cell surface O-linked glycoconjugates with terminal galactose residues, we have efficiently knocked-down different COG subunits with specific siRNAs (Supplementary data, Figure S1) and then incubated COG-deficient cells with PNA-rhodamine. HeLa cells treated with a scrambled siRNA were used as a negative control. The low-density lipoprotein (ldl) B (COG1 KO) and ldlC (COG2 KO) Chinese hamster ovary (CHO) cells were used as positive controls. Control CHO cells were not capable of binding PNA-rhodamine, while both ldlB and ldlC cells were intensively decorated with the lectin, validating our staining procedure (Figure 1F). All COG knockdown cells were brightly stained with PNA-rhodamine, but, unexpectedly, we found that the control HeLa cells were also capable of binding PNA and were stained to approximately the same intensity as the COG subunit knockdowns (Figure 1A), indicating that PNA staining is not applicable for analyzing glycosylation abnormalities in HeLa cells. Similar results were obtained when cells were incubated with Alexa-488-PNA (data not shown). PNA staining was increased in cells treated with neuraminidase and was abolished in cells incubated in the presence of 100 mM lactose, validating the specificity of the lectin binding to HeLa cells (Figure 1I and Supplementary data, Figure S2).
Fig. 1.
Lectin staining of COG complex-depleted HeLa cells reveals altered glycosylations of plasma membrane glycoconjugates. HeLa cells were mock-transfected or transfected with siRNA to COG2, COG3, COG4, COG6 and COG8. Seventy two hours after transfection, the cells were chilled to 4°C, stained with PNA-rhodamine, WGA-fluorescein, GS-II-Alexa 594 or GNL-Alexa 647 for 30 min at 4°C, fixed with 4% paraformaldehyde, and visualized by fluorescent microscopy (A–B, D–E, respectively). (C) Cells were treated with neuraminidase before WGA-fluorescein binding. CHO, ldlB (COG1 KO) and ldlC (COG2 KO) cells were chilled to 4°C, stained with PNA-rhodamine, GS-II-Alexa 594 or GNL-Alexa 647 conjugate for 30 min at 4°C, fixed in 4% paraformaldehyde and visualized by fluorescent microscopy (F–H, respectively). (I) PNA-rhodamine binding to control HeLa cells was performed in the presence of 10 mM lactose. Samples were viewed by dual-laser confocal microscopy (63× oil objective). Bar, 10 μm.
In order to find the reagents that will specifically probe for glycosylation abnormalities in HeLa cells, we tested four lectins with different sugar specificity: Galanthus nivalis agglutinin (GNL; binds to glycoconjugates with terminal mannose) (Van Damme and Peumans 1988; Fouquaert et al. 2007), Griffonia simplicifolia lectin-II (GS-II; binds to GlcNAc and agalactosylated N-glycans) (Lamb et al. 1983; Tateno et al. 2007), Lotus tetragonolobus lectin (LTL; binds to Fucα1-3GlcNAc) (Pereira and Kabat 1974; Tateno et al. 2007) and wheat germ agglutinin (WGA; binds to GlcNAcβ1-4GlcNAcβ1-4GlcNAc and Neu5Ac) (Nagata and Burger 1974; Tateno et al. 2007). For WGA staining, a slight decrease in lectin binding to the surface of all tested COG-deficient cells was detected (Figure 1B), indicating a defect in protein sialylation. This result was in a good agreement with protein sialylation defects previously observed in fibroblasts isolated from COG-related CDG patients (Wu et al. 2004; Kranz et al. 2007; Zeevaert et al. 2008). Cell treatment with neuraminidase prior to incubation with WGA partially reduced the lectin binding to control HeLa cells (Figure 1C and Supplementary data, Figure S3). For LTL staining, there was no difference between the control and the COG subunit knockdown cells, indicating that COG-deficient cells are not significantly altered in expressing cell surface glycoconjugates with terminal fucoses (data not shown). The plasma membrane of all tested COG KD cells was, however, specifically stained with GS-II (Figure 1D) and GNL (Figure 1E). Likewise, the plasma membrane of both ldlB and ldlC cells was also distinctly stained with GS-II (Figure 1G) and GNL (Figure 1H). No staining was observed for either control HeLa or CHO cells. This indicated that the COG subunit knockdown cells express plasma membrane-localized galactosylated N-glycans and glycoconjugates with an increased amount of terminal mannoses. Importantly, lectin staining demonstrated no difference between the knockdowns of COGs 2–4 (lobe A) or COGs 6–8 (lobe B), indicating that the whole functioning COG complex is essential for the correct processing of plasma membrane glycoconjugates.
Golgi glycosyltransferases are mostly stable but severely mislocalized in COG-depleted cells
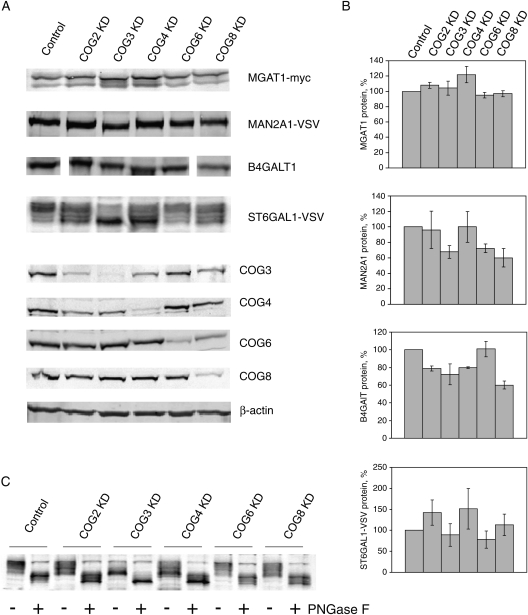
What is the primary cause of the alterations in N-linked glycosylation of plasma membrane-localized glycoconjugates found in COG-depleted cells? Our previous investigation of glycosylation defects in COG3- and COG7-depleted HeLa cells indicated that in these cells, myc-tagged medial-Golgi enzyme MGAT1 (α-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase/GlcNAcT1-myc) was extensively mislocalized to vesicle-like membrane compartment (Shestakova et al. 2006). Three-day knockdown of COG subunits did not compromise the stability of the MGAT1 or MAN2A1 (α-mannosidase II-VSV) proteins, while the longer depletion had a detrimental effect on MAN2A1. We therefore investigated both protein levels and intracellular localization of medial- (MGAT1 and MAN2A1) and trans-Golgi (B4GALT1 and ST6GAL1)-localized glycosyltransferases in COG2-, COG4-, COG6- and COG8-depleted cells using cell lines that stably express tagged versions of Golgi enzymes (Figures 2 and 3). We found that MGAT1-myc was mostly stable in all COG-depleted cells. The protein level of MAN2A1-VSV was unchanged in COG2 and COG4 KD cells and was found slightly lower in COG3, COG6 and COG8 KD cells. The endogenous trans-Golgi-localized B4GALT1 (UDP-Gal:betaGlcNAc-beta-1,4-galactosyltransferase) was mostly stable in COG6-depleted cells and slightly depleted (∼80% of wt level) in COG2 and COG4 KD cells. Intriguingly, B4GALT1 expression in COG3 and COG8 KD cells was reduced to 60–70% of the wild-type level. Although no significant depletion was found for the protein levels of ST6GAL1-VSV (β-galactoside α-2,6-sialyltransferase 1) in all investigated COG-deficient cells, the sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) mobility of VSV-positive material was altered, indicating COG-dependent defects in ST6GAL1 protein modifications. ST6GAL1 protein sequence has two N-glycosylation sites (Chen et al. 2009) and therefore its own glycosylation could be altered in COG-depleted cells. Indeed, sample deglycosylation with PNGase F resulted in fast-migrating ST6GAL1 protein species in both control and COG-depleted cells (Figure 1C). Further defects in O-glycosylation of ST6GAL1 protein in COG-depleted cells may contribute to an additional increase in ST6GAL1 protein mobility.
Fig. 2.
Stability of Golgi enzymes in COG KD cells. HeLa cells that stably express VSV or myc-tagged Golgi enzymes were mock-transfected or transfected with siRNA to COG2, COG3, COG4, COG6 and COG8. Seventy two hours after transfection, cells were collected and lysed in hot 2% SDS. Proteins from cell lysates were separated on SDS–PAGE and western blotted with antibody as indicated (A). Experiment was repeated at least three times and protein expression data were quantified. Actin expression was used as a standard (B). (C) Lysates of HeLa cells that stably express ST6GAL1-VSV were incubated with buffer or treated with PNGase F (New England Biolab) overnight before the WB with anti-VSV antibodies.
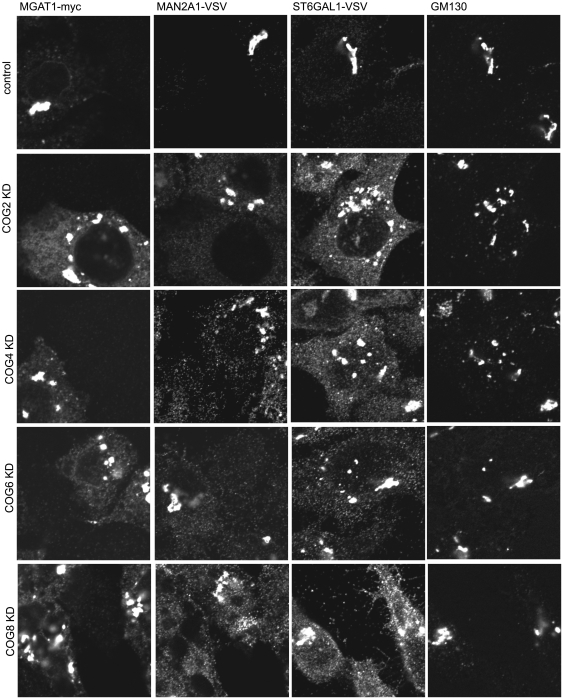
Fig. 3.
Localization of Golgi enzymes in COG KD cells. HeLa cells that stably express VSV or myc-tagged Golgi enzymes were mock-transfected or transfected with siRNA to COG2, COG4, COG6 and COG8. Seventy two hours after transfection, cells were fixed, stained with antibodies as indicated and analyzed by laser confocal fluorescent microscopy.
While the stability of Golgi enzymes was not significantly compromised in HeLa cells after 3 days of COG subunit depletion, the localization of all tested enzymes was dramatically altered (Figure 3). In all analyzed COG KD cells, MGAT1, MAN2A1 and ST6GAL1 proteins were found in multiple vesicle-like structures, as well as in large fragmented Golgi mini-stacks that were positive for the Golgi matrix protein GM130. Importantly, no difference was observed between cells depleted for either lobe A or lobe B COG subcomplexes.
The acute COG complex depletions have a minor effect on the structure of total cellular N-linked glycoconjugates
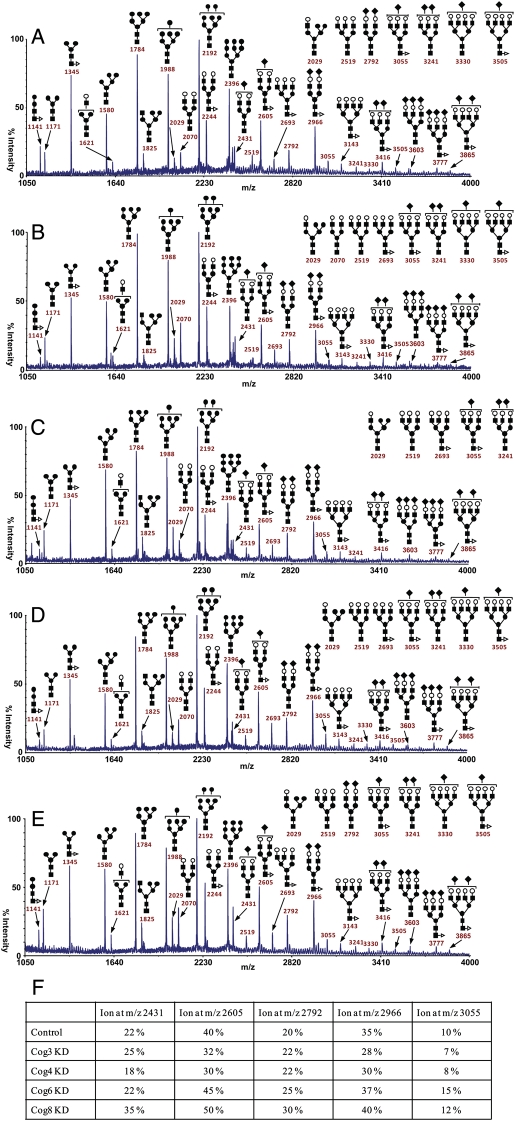
To get a better understanding of the nature of glycosylation defects that occur in cells with the acute COG subunit knockdown (i.e., to get a picture of primary COG-dependent glycosylation defects), we performed a mass spectroscopy analysis of total N-linked glycoconjugates isolated from the whole lysates of control HeLa cells and siRNA-treated HeLa cells. MALDI-TOF-MS of permethylated oligosaccharides can be used for relative quantitation of oligosaccharides and is as reliable as chromatographic methods for elucidating glycan profiles. After detergent extraction of cells, glycoproteins were reduced, carboxyamidomethylated and digested with trypsin. N-glycans were then released with PNGase F before permethylation and purification on Sep-Pak prior to MALDI-TOF mass profiling (Figure 4A–F) (Morelle and Michalski 2007). The structural assignments were made based on the MALDI mass and MS-MS experiments. High-mannose-type structures having five to nine mannose residues and no core fucosylation (Man5–9GlcNAc2) (m/z 1580, 1784, 1988, 2192 and 2396), minor hybrid structures [m/z 1825 (Hex5HexNAc3) and 2029 (Hex6HexNAc3)] and complex-type structures of compositions (Fuc0–1-NeuAc0–3Hex5–7HexNAc4–6) were observed in control HeLa cells and were also found in COG-deficient cells. High-mannose-type structures were more abundant than complex-type structures. Comparison between the N-glycans from control HeLa cells and COG-deficient cells revealed no major differences for high-mannose-type structures. However, the comparison of the MALDI-TOF-MS spectra of the N-glycans purified from control HeLa cells and COG3- and COG4-deficient cells indicates a decrease in sialylation, as illustrated by the major sialylated biantennary glycans (m/z 2605 and 2966). Surprisingly, the data also show a minor increase in sialylation in the N-glycans of COG6- and COG8-deficient cells. Thus, these results suggest that there are differences between the sialylated N-glycans from COG3- and COG4-deficient cells (lobe A) and COG6- and COG8-deficient cells (lobe B).
Fig. 4.
MALDI-TOF analysis of N-linked glycoconjugates isolated from COG-deficient HeLa cells. Mass spectrometry results of total cellular N-linked glycoconjugates of control (A), COG3 KD (B), COG4 KD (C), COG6 KD (D) and COG8 KD (E). Analysis was repeated at least two times for each knockdown. (F) Quantification of these results shown in panels A–E. The structural assignments were made based on the MALDI mass and MS-MS experiments. A minor portion of the monofucosylated glycans carries fucose on an antenna rather than the core. Symbols: galactose, open circles; mannose, closed circles; GlcNAc, closed squares; fucose, open triangles; NeuAc, closed diamonds.
COG complex-deficient HeLa cells are defective in the BFA-induced redistribution of Golgi resident glycosyltransferases to the ER
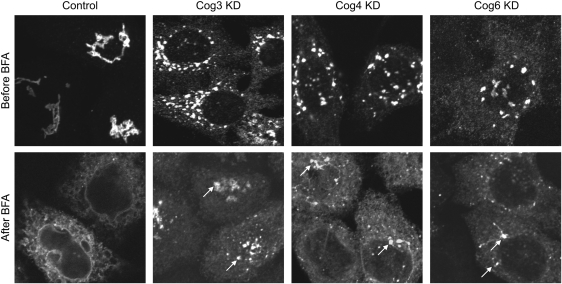
It has been demonstrated previously that the BFA-induced retrograde Golgi-to-ER trafficking is markedly delayed in fibroblasts isolated from COG-deficient human patients (Steet and Kornfeld 2006; Foulquier et al. 2007; Kranz et al. 2007; Ng et al. 2007; Paesold Burda et al. 2009). To determine whether BFA-induced retrograde traffic of Golgi glycosyltransferases is affected in the COG complex-depleted HeLa cells, BFA, at a relatively low (0.25 µg/mL) concentration (Steet and Kornfeld 2006), was added to control, COG2 KD, COG3 KD, COG4 KD, COG6 KD and COG8 KD GALNT2-GFP (polypeptide N-acetylgalactosaminyltransferase 2/GalNAcT2-GFP) HeLa cells for 1 h, after which the subcellular localization of GALNT2-GFP was detected using fluorescent microscopy. We found that all types of COG-depleted cells are defective in the BFA-induced Golgi redistribution of GALNT2-GFP to the ER (Figure 5). Similar inhibition of Golgi disassembly was found in cells treated with a higher concentration (5.0 µg/mL) of BFA (data not shown).
Fig. 5.
COG complex-depleted cells are defective in the BFA-induced Golgi redistribution to the ER. GALNT2-GFP HeLa cells were mock-transfected or transfected with siRNA to COG3, COG4 and COG6. Seventy two hours after transfection, cells were fixed (before BFA panel) or were exposed to 0.25 µg/mL BFA for 1 h, fixed and analyzed by fluorescent microscopy (after BFA panel). White arrows show remnants of the Golgi apparatus for the COG subunit knockdown cells (63× oil objective). Bar, 10 μm.
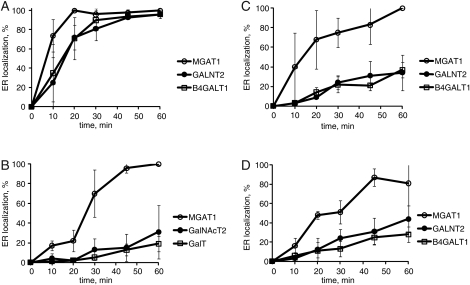
To quantify the BFA resistance in COG subunit knockdown cells and to investigate this effect further, we applied BFA on control and COG complex-depleted (COG3 KD, COG4 KD and COG6 KD) HeLa cells for an hour-long assay with cells being fixed at 0, 10, 20, 30, 45 and 60 min and then analyzed the redistribution of medial-Golgi-localized MGAT1-myc (Hull et al. 1991), medial-/trans-Golgi GALNT2-GFP and trans-Golgi enzyme ST6GAL1 (Teasdale et al. 1994) as described in Materials and Methods. Three assays were completed and the average number of cells with ER-localized Golgi enzymes was calculated. For control cells (Figure 5A), MGAT1-myc, GALNT2-GFP and ST6GAL1 were redistributed to the ER within 20 min of application of BFA. The medial-Golgi resident protein (MGAT1) was distributed to the ER first followed by the medial-/trans- (GALNT2) and trans-Golgi proteins (ST6GAL1); however, this difference was not statistically significant in control cells. Retrograde delivery of all three enzymes was significantly delayed in COG-deficient cells (Figure 5B–D). We found a 20-min delay in the redistribution of the medial-Golgi enzyme MGAT1-myc, while the relocalization of more distal markers (medial-/trans- and trans-Golgi markers) was dramatically inhibited. This suggests that trans-Golgi enzymes are moving to the ER through the cis-Golgi compartments and that the COG complex is primarily responsible for the retrograde trafficking of medial-/trans-Golgi enzymes. Furthermore, we found a very similar BFA resistance in cells that were depleted for either lobe A (COG3 KD and COG4 KD) or lobe B (COG6 KD) COG complex subunits. This suggests again that the entire COG complex is essential for the retrograde trafficking of the different Golgi-localized glycosylation enzymes.
COG complex-deficient HeLa cells are normal in Golgi reassembly after the BFA washout
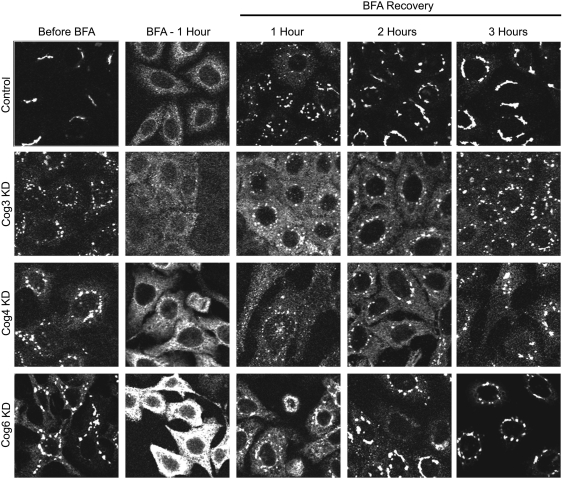
To test whether the COG KD cells are specifically deficient for the retrograde movement of Golgi enzymes, we performed a BFA washout experiment and examined the reassembly of the Golgi. Control, COG3 KD, COG4 KD and COG6 KD cells that carry GALNT2-GFP were exposed to BFA to redistribute Golgi proteins to the ER. After that, the medium was exchanged and the dynamics of reassembly of GALNT2-GFP was captured using fluorescent microscopy (Figure 6). We found that the control and COG-depleted cells behave similarly in restoring their pre-BFA phenotypes, indicating that the COG complex is dispensable for the anterograde trafficking of Golgi enzymes (Figure 6).
Fig. 6.
BFA-induced relocalization of trans-Golgi enzymes is specifically blocked in COG complex-deficient cells. GALNT2-GFP and MGAT1-myc HeLa cells were mock-transfected or transfected with siRNA to COG3, COG4 and COG6. Seventy two hours after transfection, cells were exposed to 0.25 µg/mL BFA for 0-, 10-, 20-, 30-, 45- and 60-min, fixed, stained for MGAT1-myc and GalT, and analyzed by fluorescent microscopy. Images were quantified by counting cells that show a complete redistribution of Golgi enzymes to the ER. Four images were captured with the 63× oil objective on a laser confocal microscope for each time point. The mean result of three independent experiments using control cells (A), COG3 KD cells (B), COG4 KD cells (C) and COG6 KD cells (D) was quantified. Error bars represent standard deviations between three independent experiments.
The COG complex is required for the Sar1p-DN-induced relocalization of the Golgi enzymes to the ER
To further investigate the COG complex requirement for the retrograde trafficking, we employed a dominant-negative (DN) mutant of Sar1p GTPase that interferes with the formation of ER-derived COPII vesicles, causing a dis-balance in ER-Golgi bidirectional membrane trafficking and subsequent redistribution of Golgi resident enzymes to the ER (Stroud et al. 2003).
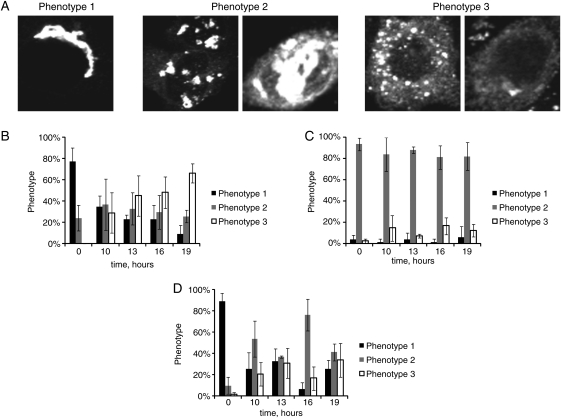
Both control and COG complex-deficient GALNT2-GFP HeLa cells were transfected with a plasmid expressing Sar1 DN (pSar1p-T39N) to induce retrograde trafficking of the medial-/trans-Golgi protein GALNT2-GFP (see Materials and Methods). We have found that even in control cells, relocalization of GALNT2-GFP was not complete. Three distinct Sar1 DN-induced GALNT2-GFP phenotypes were observed (Figure 7A): group 1 phenotype with cells which have a normal juxtanuclear Golgi ribbon, group 2 phenotype with cells which have an appearance of Golgi mini-cisterns (similar to a COG3 KD phenotype) and group 3 phenotype with Golgi enzyme localized to a vesicle-like haze and the ER.
Fig. 7.
COG complex knockdown cells are not deficient in Golgi reassembly assay. MGAT1-myc HeLa cells were mock-transfected or transfected with siRNA to COG3, COG4 and COG6. Seventy two hours after transfection, cells were exposed to BFA (0.25 µg/mL) for 1 h and then allowed to recover in fresh 10% FBS medium for times indicated. The cells were fixed before BFA application, 60 min after BFA and 0-, 1-, 2- and 3-h for recovery, permeabilized with Triton X-100, stained with anti-myc antibodies and analyzed by IF microscopy (63× oil objective). Bar, 10 μm.
Our analysis revealed that most of the control cells had been converted from phenotype of group 1 to phenotype of group 3 after 19 h of Sar1 DN expression (Figure 7B), indicating that the Golgi enzymes had been efficiently redistributed to the ER. For the COG3 KD (Figure 7C) and COG6 KD (Figure 7D) cells, we found a significant defect in Sar1 DN-induced GalNAc-T2-GFP redistribution (group 2 phenotype was maintained in COG3 KD cells and group 1 phenotype was persistent for COG6 KD cells). This result is in a good agreement with the results obtained in the experiment with BFA, supporting our conclusion that the whole COG complex is specifically required for the efficient retrograde trafficking of Golgi glycosylation enzymes.
Recycling Golgi enzymes and COG complex subunits are partially colocalize on MGAT1-positive vesicular structures
To test whether the COG complex is directly involved in vesicle-mediated recycling of Golgi enzymes, MGAT1-myc cells were transfected with fluorescently labeled lobe B COG subunits RFP-COG6, COG6-GFP and COG8-GFP. Forty eight hours after the transfection, the cells were viewed with confocal microscopy to determine the colocalization of the COG subunits with MGAT1 (Figure 8). All three COG subunit constructs were colocalized to each other and with MGAT1-myc on perinuclear Golgi membranes, indicating that the presence of the N- or C-terminally attached fluorescent proteins does not interfere with the function of the tagged proteins. Importantly, MGAT1 was also colocalized to a population of the COG6- and COG8-positive vesicular structures, directly implicating the COG complex in vesicle-mediated trafficking of Golgi glycosylation machinery (Figure 8B and C). Interestingly, the endogenous COG3 was mostly absent from these peripheral vesicle structures (Figure 8E–F), implicating that lobe A of the COG complex is primarily associated with large Golgi membranes and that the tethering of lobe B/MGAT1-positive vesicles to the Golgi could be regulated through the formation of the lobe A/B octameric COG complex.
Fig. 8.
Sar1p T39N-induced disassembly of the Golgi is delayed in COG complex-deficient cells. GALNT2-GFP HeLa cells were mock-transfected or transfected with siRNA to COG3 and COG6. Seventy two hours after transfection, the cells were transfected with plasmid expressing Sar1p T39N. Transfected cells were fixed 19 h later and analyzed by IF microscopy using 63× oil objective. Three phenotypes of localization of GALNT2-GFP were observed: phenotype 1 has a normal juxtanuclear or dilated Golgi localization. Phenotype 2 has a punctate distribution of the enzyme present in both the Golgi and ER localization. Phenotype 3 has small vesicles or ER localization of the enzyme (A). Bar, 10 μm. Graphs showing redistribution of GALNT2-GFP from the Golgi apparatus to the ER in control cells (B), COG3 KD cells (C) and COG6 KD cells (D). For statistical analysis, three individuals blindly classified the phenotypes. Error bars represent standard deviations between each individual's phenotype classifications (n > 50).
Discussion
Lectin staining is an essential tool for the elucidation of defects in Golgi-localized glycosylation machinery. Wu et al. (2004) had employed a PNA-Alexa Fluor 488 conjugate stain on COG7-mutant fibroblasts showing hyposialylation of the plasma membrane glycoconjugates. Similar plasma membrane PNA staining has been done on other COG-deficient fibroblasts (Foulquier et al. 2006, 2007; Kranz et al. 2007). While the characterization of a primary cell culture is obviously important, the use of model cell lines, such as HeLa and CHO cells, provides a unique opportunity to investigate the function of a particular gene product in a controlled and reproducible manner. PNA staining was not informative in HeLa cell line, since we detected a significant PNA binding to cell surface of control HeLa cells, most likely indicating the capacity of these cancer cells to express a significant amount of galactose-terminated O-linked glycoconjugates on the plasma membrane.
WGA staining of plasma membrane glycoconjugates revealed a slight decrease in lectin binding, indicating a defect in protein sialylation, previously observed in human COG-deficient patients. For the LTL, SNA and RCA staining, there was no difference between the control and the COG subunit knockdown cells, indicating that COG-deficient cells are not significantly altered in expressing cell surface glycoconjugates with terminal galactoses or fucoses (Supplementary data, Figure S4 and data not shown).
Our search for COG-specific tools yielded two lectins, GS-II and GNL, which efficiently decorated the cell surfaces of all tested COG-deficient cells. In a good agreement with this new result, we have recently demonstrated that HeLa cells expressing COG4 human-mutant alleles are positive in a GNL binding assay (Richardson et al. 2009), and Linstedt lab found that GS-II is capable of binding cells deficient for GM130 (Puthenveedu et al. 2006). Staining of plasma membrane glycoconjugates with GNL and GS-II lectins in the COG-deficient cells indicated that the COG complex is important for the proper localization of both medial-Golgi enzymes MGAT1 and MAN2A1. We previously demonstrated that these enzymes are mislocalized to CCD vesicles in COG3 KD cells (Zolov and Lupashin 2005). We have now expanded that initial observation and demonstrated that all tested Golgi enzymes are extensively mislocalized in both COG lobe A and COG lobe B-depleted cells, indicating that the entire COG complex is involved in vesicular trafficking of both MGAT1 and MAN2A1. In agreement with this idea, we now show that lobe B of the COG subcomplex is partially localized on vesicles that carry MGAT1.
Our results indicated that the acute depletion of COG complex subunits caused not only the mislocalization of trans-Golgi enzymes ST6GAL1 and B4GALT1, but also altered the stability and/or modifications of these proteins. We found a 30–40% reduction in the cellular level of the endogenous ST6GAL1 in both COG3 and COG8 KD cells. Even more striking, all COG-deficient cells expressed underglycosylated species of ST6GAL1-VSV. Underglycosylation of ST6GAL1 reflected on the activity of this enzyme; as a consequence, the terminal sialylation of secreted and plasma membrane-exposed glycoconjugates diminished in COG-deficient cells resulting in reduced binding of WGA. The glycosylation of ST6GAL1 protein in HeLa cells was most extremely distorted upon COG3 and COG4 depletion and, as a result, MALDI-TOF of total N-glycans purified from COG3- and COG4-deficient cells indicates a decrease of sialylation.
While this paper was under review, Dr. F. Foulquier's laboratory published their work (Peanne et al. 2011) describing differential effects of COG3 and COG7 depletion on the stability of GFP-tagged B4GALT1 and ST6GAL1 in HeLa cells. They found that both proteins were stable in COG3 KD cells, but extremely unstable in COG7 KD cells. We have not specifically analyzed COG7 depletion in this work, partially because it has been done before by us and other laboratories (Shestakova et al. 2006; Sohda et al. 2010), and partially because we found that the double knockdown with COG7 siRNA was toxic for the cell lines used in this work. Since Dr. Foulquier's conclusions were not in agreement with our results, we have repeated knockdowns of COG3, COG4, COG6, COG7 and COG8 in cells obtained from Dr. Foulquier laboratory. The depletion of the COG complex subunits was very efficient in every case (Supplementary data, Figure S5) and indeed, in agreement with Dr. Foulquier's publication, we observed that B4GALT1-GFP was less stable in COG7 KD cells when compare with COG3 KD cells (Supplementary data, Figure S6). Surprisingly, this construct was stable in COG8 KD cells and its level was similar in COG4 and COG6 KD cells. Even more striking, the cellular level of the endogenous B4GALT1 was very similar in all analyzed COG KD cells. Interestingly, the endogenous ST6GAL1 in this cell line was very sensitive to COG complex depletion, but again no major differences were detected between lobe A and lobe B-depleted cells, supporting our conclusion that the entire COG complex is involved in localization of Golgi enzymes. We also observed that the stability and/or glycosylation of both ST6GAL1 and cis-Golgi-localized GPP130 were severely altered in both COG4 (lobe A) and COG7 (lobe B) cells, which may indicate the central role of these subunits in COG subcomplexes.
MALDI-TOF of total N-linked glycoproteins provides valuable information on the array of glycosylation defects associated with COG complex deficiencies. This methodology was used before only for secreted glycoproteins (Foulquier et al. 2007; Kranz et al. 2007; Reynders et al. 2009). Our data show that the glycosylation defects found for the entire population of cellular N-linked glycoproteins are rather minor.
To directly test the effect of COG complex deficiencies on the retrograde trafficking of Golgi glycosylation machinery, we induced Golgi retrograde membrane trafficking by either BFA or Sar1 DN treatments and compared the dynamics of the redistribution of different Golgi enzymes in control and COG-deficient cells. Our results indicated that cells lacking either lobe A or lobe B of the COG complex are delayed in their retrograde Golgi-to-ER delivery of the tested components of the Golgi glycosylation machinery, implying that the entire COG complex works as a global regulator of recycling/retrieval of Golgi resident glycosyltransferases. This is in a good agreement with results obtained with lectin staining. Again, COG complex-deficient cells were not defective in Golgi reassembly after BFA washout, indicating specificity of the COG complex in retrograde trafficking.
The strongest inhibition of the retrograde trafficking was observed for trans-Golgi-localized enzymes GALNT2 and B4GALT1, indicating that in BFA-treated cells, such as in control cells, trans-Golgi-localized glycosyltransferases are recycled through the early Golgi compartments and that the COG complex specifically regulates this leg of recycling of Golgi resident proteins. The COG-dependent delay in Golgi redistribution to the ER, in response to BFA treatment observed in this study, is well correlated with similar defects observed in fibroblasts isolated from human patients carrying mutations in different COG complex subunits (Wu et al. 2004). The reason for this delay is not clear, but our experiments indicate that the dynamics of BFA-induced COPI dissociation is altered in COG-deficient HeLa cells (data not shown). Arf-GEF proteins GBF1 and BIG1 are important for the BFA-induced Golgi disassembly (Zhao et al. 2002), and our preliminary results indicated that these proteins are mislocalized in the COG-deficient HeLa cells (Smith and Lupashin, in preparation). Oka et al. (2004), Ram et al. (2002) and Suvorova et al. (2002) have previously demonstrated that the COG complex functionally, and physically, interacts with the COPI complex and its intracellular dynamic is likely to be altered in COG-deficient cells.
The colocalization of the lobe B COG subunits with vesicles that carry the MGAT1 demonstrates the COG complex's direct role in vesicular trafficking of the resident Golgi enzymes.
The Warren lab (Malsam et al. 2005) demonstrated the existence of at least two distinct classes of Golgi-derived COPI vesicles that carry different populations of Golgi resident proteins. One vesicle population was regulated by p115/Giantin, a pair of coiled-coil tethering factors, while the other one was regulated by the coiled-coil tethering factors golgin-84/CASP. Sohda et al. (2007) has recently demonstrated that COG2 interacts with p115, implying that the COG complex regulates the p115/Giantin class of COPI vesicles. On the other hand, both golgin-84 and CASP were previously described as GEARS (proteins dependent on a proper COG complex function) (Oka et al. 2004), so it is likely that the COG complex also regulates trafficking of the golgin-84/CASP class of COPI vesicles.
Our results demonstrate that cell surface lectin staining and mass spectrometry analysis of glycosylation-deficient cells cannot substitute each other. A combination of these two methods is a valuable approach for detecting a complete set of COG complex-related glycosylation deficiencies.
In this work, we have detected a relatively small number of COG-positive vesicle-like structures carrying a Golgi enzyme. It is likely that at steady state, the vesicle recycling process is very efficient and the number of “free” transport vesicles is limited. COG depletion leads to a dramatic increase in the number of nontethered CCD vesicles (Zolov and Lupashin 2005; Shestakova et al. 2006). These vesicles likely represent a mixed population of COG complex-dependent intra-Golgi membrane carriers that serve to recycle/retrieve different components of Golgi glycosylation machinery. Further investigation of CCD vesicles should uncover the other components of vesicle targeting machinery that directs the efficient intra-Golgi vesicular trafficking and Golgi maintenance.
Materials and Methods
Reagents and antibodies
Lectins (with their concentrations) used were as follows: G. simplicifolia lectin-II (GS-II)-Alexa 594 (100 µg/mL, Invitrogen, Carlsbad, CA), Alexa 488-Peanut Agglutinin (PNA) (20 µg/mL, Invitrogen), PNA-rhodamine (20 µg/mL, Vector Laboratories, Burlingame, CA), Galanthus nivalus lectin (GNL)-Alexa 647 (20 µg/mL, Vector), L. tetragonolobus lectin (LTL)-Alexa 647 (20 µg/mL, Vector) and WGA-rhodamine and WGA-fluorescein (both 5 µg/mL; Vector). GNL and LTL were labeled with an Alexa 647 protein labeling kit from Invitrogen (Carlsbad, CA).
Antibodies used for immunofluorescent (IF) microscopy or western blotting (WB) were purchased through commercial sources, gifts from generous individual investigators or generated by us via affinity purification. Antibodies (and their dilutions) were as follows. Mouse monoclonal antibodies: GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA) WB 1:1000, β-actin (Sigma, St. Louis, MO) WB 1:10,000, β-COP (Sigma) IF 1:200, GalT (a gift from Dr. Brian Storrie, UAMS) IF 1:50, Cog3 (this lab) WB 1:1000 (Smith et al. 2009); rabbit polyclonal antibodies: myc (Bethyl Laboratories, Montgomery, TX) IF 1:3000, affi-pure Cog3 (this lab) WB 1:10,000 (Suvorova et al. 2001), affi-pure Cog4 (this lab) WB 1:1000, affi-pure Cog6 (this lab) WB 1:1000, affi-pure Cog8 (this lab) WB 1:1000 (Smith et al. 2009); goat polyclonal antibodies: B4GALT1 (R&D Systems, Minneapolis, MN) WB 1:1000, ST6GAL1 (R&D Systems) WB 1:1000.
Secondary anti-goat-horseradish peroxidase (HRP), anti-mouse-HRP and anti-rabbit-HRP for WB were obtained from Jackson ImmunoResearch Laboratories (Baltimore, MD). IRDye 680 goat anti-rabbit, IRDye 700 goat anti-mouse and IRDye 800 donkey anti-goat for WB were obtained from LI-COR Biosciences (Lincoln, NE). Anti-rabbit HiLyte 488, anti-rabbit HiLyte 555 and anti-mouse HiLyte 647 for IF were obtained from AnaSpec, Inc (San Jose, CA).
Cell culture
HeLa cells stably expressing GALNT2-GFP and MGAT1-myc were obtained from Dr. Brian Storrie (UAMS) (Nilsson et al. 1994; Rottger et al. 1998). These cells were cultured in Dulbecco's modified Eagle medium (DMEM)/F-12 50/50 medium supplemented with 15 mM HEPES, 2.5 mM l-glutamine, 10% fetal bovine serum (FBS) and 0.4 mg/mL G418 sulfate. Cells were grown at 37°C and 5% CO2 in a 90% humidified incubator. HeLa cells stably expressing GFP-tagged B4GALT1 (aa 1–50) (GalT1-GFP) were obtained from Dr. Francois Foulquier (Universite des Sciences et Technologies de Lille, France). Cells were grown in DMEM supplemented with 10% FBS and 2.5 mM l-glutamine at 37°C and 5% CO2 in a 90% humidified incubator.
CHO cells, along with the COG1 knockout (ldlB/COG1 KO) and the COG2 knockout (ldlC/COG2 KO) mutant CHO cells, were obtained from Dr. Monty Krieger (MIT). CHO cells were grown in DMEM/F-12 50/50 with 15 mM HEPES, 2.5 mM l-glutamine, 5% FBS and 1% antibiotic/antimycotic (100 U/mL penicillin G, 100 µg/mL streptomycin and 0.25 µg/mL amphotericin B). Cells were grown at 37°C and 5% CO2 in a 90% humidified incubator. All cell culture media and sera were obtained from Invitrogen (Carlsbad, CA).
siRNA-induced knockdowns
siRNA duplexes for COG2, COG3, COG4, COG6 and COG8 (Zolov and Lupashin 2005; Smith et al. 2009) were obtained from Dharmacon (Chicago, IL). Transfection was performed using either oligofectamine or lipofectamine RNAiMAX siRNA Transfection Reagent (Invitrogen), following a protocol recommended by Invitrogen. Two cycles of transfections (100 nm siRNA each) were performed and cells were analyzed 72 h after the second cycle.
Plasmid preparation and transfection
The plasmid for pSar1p-T39N was described previously (Stroud et al. 2003). The RFP-COG6, COG6-GFP and COG8-GFP plasmids were created by subcloning COG6 or COG8 (Shestakova et al. 2007) into pRFP-C1 and pGFP-N1 plasmids (Clonetech, Palo Alto, CA), respectively. Plasmids were isolated from bacterial cells using the QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). Plasmid DNA transfections into tissue culture cells were performed with Lipofectamine 2000 (Invitrogen) or with FuGene (Roche Applied Science, Indianapolis, IN), according to the manufacturer's protocol.
Fig. 9.
Vesicles that carry MGAT1 contain lobe B COG complex subunits. RFP-COG6 and COG6-GFP plasmids (A, B), RFP-COG6 and COG8-GFP (C, D) or RFP-COG6 (E, F) were cotransfected in MGAT1-myc-myc HeLa cells. After 48 h, cells were fixed, stained with anti-myc and/or anti-COG3 antibodies and developed using HyLite-647-conjugated anti-rabbit antibodies, HyLite-647-conjugated anti-mouse antibodies and/or HyLite-488-conjugated anti-mouse antibodies. Cells were viewed by dual-laser confocal microscopy (63× oil objective). RFP-COG6, COG6-GFP and COG8-GFP positively colocalize with the MGAT1-myc on large Golgi cisternae as well as small vesicle-like membranes (red arrows). Staining with endogenous COG3 reveal no colocalization with the RFP-COG6/MGAT1-myc-positive vesicle structures.
IF microscopy
For IF, cells were grown on 12 mm glass coverslips (#1, 0.17 mm thickness) 1 day before transfection. Seventy two hours after siRNA transfection, cells are fixed and stained with antibodies as previously described (Jiang and Storrie 2005; Shestakova et al. 2006).
In short, cells were fixed in 4% paraformaldehyde (16% stock solution; Electron Microscopy Sciences, Port Washington, PA) and then treated with 1% Triton X-100 for 1 min. After following incubation with 50 mM ammonium chloride for 5 min, cells were washed with phosphate-buffered saline (PBS) and blocked twice for 10 min with 1% bovine serum albumin (BSA) and 0.1% saponin in PBS. Cells were then incubated for 30–60 min at room temperature with primary antibody diluted in the 1% cold fish gelatin, 0.1% saponin in PBS, washed four times with PBS and incubated for 30 min with fluorescently tagged secondary antibody (1:400 Hilyte; AnaSpec) in antibody buffer at room temperature. After that, coverslips were washed four times with PBS, rinsed with ddH2O and mounted on glass microscope slides using Prolong® Gold antifade reagent (Invitrogen). Cells were imaged with the 63× oil 1.4 numerical aperture (NA) objective of an LSM510 Zeiss Laser inverted microscope outfitted with confocal optics. Image acquisition was controlled with LSM510 software (Release Version 4.0 SP1).
Lectin staining of intact cells
Lectin staining was performed with both fixed and unfixed cells. In the first protocol, control and knocked-down HeLa cells (generally, 96 h after the siRNA-induced transfection) were rinsed in 1× DPBS and fixed with 1% paraformaldehyde for 10 min at room temperature. After incubation in 50 mM ammonium chloride for 5 min at room temperature, cells were washed in PBS and blocked with 0.1% BSA in PBS. Finally, cells were incubated with PNA-rhodamine and WGA-fluorescein for 1 h at room temperature, washed with PBS and visualized using confocal microscopy.
For control experiments such as desialylation of cell monolayers with Vibrio cholerae neuraminidase (Roche), cells were incubated with neuraminidase, 50 mU/mL for 1 h at 37°C with the following with lecting staining.
To confirm that the cell surface binding of lectins is a carbohydrate-specific phenomenon, control staining in the presence of 10 mM lactose was performed.
For lectin staining of unfixed cells, control and knocked-down (generally, 96 h after the siRNA-induced transfection) GALNT2-GFP HeLa cells were rinsed in 1× DPBS at 4°C and placed in cold DPBS and allowed to chill at 4°C for 15 min to reduce endocytosis. PNA-rhodamine, GS-II-Alexa 594, GNL-Alexa 647, LTL-Alexa 647 and WGA-fluorescein were all diluted in cold 10% FBS DMEM/F-12 (50:50) media without antibiotics and allowed to incubate for 20 min at 4°C. Cells were washed with DPBS at 4°C four times and fixed in 4% paraformaldehyde for confocal microscopy.
BFA-induced Golgi fragmentation and recovery
BFA was used in a final concentration of 0.25 µg/mL (Steet and Kornfeld 2006). BFA in 10% FBS in DMEM/F-12 medium without antibiotics/antimycotics was applied over the 60 min time course on stably expressing MGAT1-myc and GALNT2-GFP HeLa cells grown on glass coverslips. Cells were fixed and stained as indicated and visualized using confocal microscopy. The resulting images were quantified by manual counting to estimate the percentage of cells containing no Golgi remnants (complete ER distribution; n > 50).
For BFA recovery experiment, cells after 60 min of incubation with BFA were washed three times with 1× DPBS, transferred to a preheated fresh media with 10% FBS and incubated at 37°C and 5% CO2 in a 90% humidified incubator. Cells were fixed at 1-, 2-, and 3-h and analyzed on the laser confocal microscope.
N-glycan preparation for mass spectrometry
Control, COG3 KD, COG4 KD and COG6 KD HeLa cells were grown on 10 cm culture plates to 90% confluency. Cells were lysed and homogenized by Dounce homogenizer in Tris buffer (50 mM, pH 7.4) with 1% (w/v) SDS. The homogenate was dialysed in 12–14 kDa cut-off dialysis tubing in an ammonium hydrogen carbonate solution (50 mM, pH 7.4) for 48 h at 4°C and lyophilized. The proteins/glycoproteins were reduced and carboxyamidomethylated followed by sequential tryptic and peptide N-glycosidase F digestion and Sep-Pak purification. Permethylation of the freeze-dried glycans, MALDI-TOF-MS and electrospray MS/MS analyses of permethylated glycans were performed as described elsewhere (Morelle and Michalski 2007).
SDS–PAGE and WB
SDS–PAGE and WB were performed as described previously (Suvorova et al. 2002). The blots were incubated first with primary antibodies and then with a secondary immunoglobulin G (IgG) antibody conjugated with IRDye 680 or IRDye 800 dyes. The blots were scanned and analyzed with an Odyssey Infrared Imaging System (LI-COR). Alternatively, a signal was detected using an enhanced chemiluminescent reagent for HRP from Pierce (Rockford, IL) and quantified using imagej software (http://rsb.info.nih.gov/ij/).
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by the National Science Foundation (MCB-0645163) (V.L.), the National Institute of Health (1R01GM083144-01A1) (V.L.), the Centre National de la Recherche (Unité Mixte de Recherche CNRS/USTL 8576) (W.M.) and the Ministère de la Recherche et de l'Enseignement Supérieur (W.M.). The mass spectrometry facility used in this study was funded by the European Community (FEDER), the Région Nord-Pas de Calais (France) and the Université des Sciences et Technologies de Lille.
Conflict of interest statement
None declared.
Abbreviations
BFA, Brefeldin A; BSA, bovine serum albumin; CCD, COG complex dependent; CDG, congenital disorders of glycosylation; CHO, Chinese hamster ovary; COG, conserved oligomeric Golgi; COPI, coat protein; DMEM, Dulbecco's modified Eagle medium; ER, endoplasmic reticulum; FBS, fetal bovine serum; GNL, Galanthus nivalis lectin; GS-II, Griffonia simplicifolia lectin-II; HRP, horseradish peroxidase; IF, immunofluorescent; ldl, low-density lipoprotein; LTL, Lotus tetragonolobus lectin; MALDI-TOF, matrix-assisted laser desorption ionization-time of flight; PBS, phosphate-buffered saline; PNA, peanut agglutinin; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; WB, western blotting; WGA, wheat germ agglutinin.
Supplementary Material
Acknowledgements
We are thankful to R. Duden, F. Foulquier, M. Krieger, B. Storrie and others who provided reagents and critical reading of the manuscript. We also would like to thank Oleksandra Pavliv and Jackob Szwedo for technical support.
References
- Aoki D, Lee N, Yamaguchi N, Dubois C, Fukuda MN. Golgi retention of a trans-Golgi membrane protein, galactosyltransferase, requires cysteine and histidine residues within the membrane-anchoring domain. Proc Natl Acad Sci USA. 1992;89:4319–4323. doi: 10.1073/pnas.89.10.4319. doi:10.1073/pnas.89.10.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. doi:10.1016/S0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bruinsma P, Spelbrink RG, Nothwehr SF. Retrograde transport of the mannosyltransferase Och1p to the early Golgi requires a component of the COG transport complex. J Biol Chem. 2004;279:39814–39823. doi: 10.1074/jbc.M405500200. doi:10.1074/jbc.M405500200. [DOI] [PubMed] [Google Scholar]
- Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J Proteome Res. 2009;8:651–661. doi: 10.1021/pr8008012. doi:10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- Foulquier F. COG defects, birth and rise! Biochim Biophys Acta. 2009;1792:896–902. doi: 10.1016/j.bbadis.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Foulquier F, Ungar D, Reynders E, Zeevaert R, Mills P, Maria Teresa García-Silva MT, Briones P, Winchester B, Morelle W, Krieger M, et al. A new inborn error of glycosylation due to a Cog8 deficiency reveals a critical role for the Cog1–Cog8 interaction in COG complex formation. Hum Mol Genet. 2007;16:717–730. doi: 10.1093/hmg/ddl476. doi:10.1093/hmg/ddl476. [DOI] [PubMed] [Google Scholar]
- Foulquier F, Vasile E, Schollen E, Callewaert N, Raemaekers T, Quelhas D, Jaeken J, Mills P, Winchester B, Krieger M, et al. Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc Natl Acad Sci USA. 2006;103:3764–3769. doi: 10.1073/pnas.0507685103. doi:10.1073/pnas.0507685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquaert E, Hanton SL, Brandizzi F, Peumans WJ, Van Damme EJM. Localization and topogenesis studies of cytoplasmic and vacuolar homologs of the Galanthus nivalis agglutinin. Plant Cell Physiol. 2007;48:1010–1021. doi: 10.1093/pcp/pcm071. doi:10.1093/pcp/pcm071. [DOI] [PubMed] [Google Scholar]
- Girod A, Storrie B, Simpson JC, Johannes L, Goud B, Roberts LM, Michael Lord J, Nilsson J, Pepperkok R. Evidence for a COP-I-independent transport route from the Golgi complex to the endoplasmic reticulum. Nat Cell Biol. 1999;1:423–430. doi: 10.1038/15658. doi:10.1038/15658. [DOI] [PubMed] [Google Scholar]
- Gleeson PA, Teasdale RD, Burke J. Targeting of proteins to the Golgi apparatus. Glycoconj J. 1994;11:381–394. doi: 10.1007/BF00731273. doi:10.1007/BF00731273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Elston T, Oster G. A cisternal maturation mechanism can explain the asymmetry of the Golgi stack. FEBS Lett. 1997;414:177–181. doi: 10.1016/s0014-5793(97)00984-8. doi:10.1016/S0014-5793(97)00984-8. [DOI] [PubMed] [Google Scholar]
- Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature. 1992;360:352–354. doi: 10.1038/360352a0. doi:10.1038/360352a0. [DOI] [PubMed] [Google Scholar]
- Hull E, Sarkar M, Spruijt MPN, Höppener JWM, Dunn R, Schachter H. Organization and localization to chromosome 5 of the human UDP-N-acetylglucosamine:alpha-3-d-mannoside beta-1,2-N-acetylglucosaminyltransferase I gene. Biochem Biophys Res Commun. 1991;176:608–615. doi: 10.1016/s0006-291x(05)80227-x. doi:10.1016/S0006-291X(05)80227-X. [DOI] [PubMed] [Google Scholar]
- Jiang S, Storrie B. Cisternal rab proteins regulate Golgi apparatus redistribution in response to hypotonic stress. Mol Biol Cell. 2005;16:2586–2596. doi: 10.1091/mbc.E04-10-0861. doi:10.1091/mbc.E04-10-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz C, Ng BG, Sun L, Sharma V, Eklund EA, Miura Y, Ungar D, Lupashin V, Dennis Winkel R, Cipollo JF, et al. COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum Mol Genet. 2007;16:731–741. doi: 10.1093/hmg/ddm028. doi:10.1093/hmg/ddm028. [DOI] [PubMed] [Google Scholar]
- Lamb JE, Shibata S, Goldstein IJ. Purification and characterization of Griffonia simplicifolia leaf lectins. Plant Physiol. 1983;71:879–887. doi: 10.1104/pp.71.4.879. doi:10.1104/pp.71.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R, Skutelsky E, Danon D, Sharon N. The purification, composition, and specificity of the anti-T lectin from peanut (Arachis hypogaea) J Biol Chem. 1975;250:8518–8523. [PubMed] [Google Scholar]
- Malsam J, Satoh A, Pelletier L, Warren G. Golgin tethers define subpopulations of COPI vesicles. Science. 2005;307:1095–1098. doi: 10.1126/science.1108061. doi:10.1126/science.1108061. [DOI] [PubMed] [Google Scholar]
- Morava E, Zeevaert R, Korsch E, Huijben K, Wopereis S, Matthijs G, Keymolen K, Lefeber DJ, De Meirleir L, Wevers RA. A common mutation in the COG7 gene with a consistent phenotype including microcephaly, adducted thumbs, growth retardation, VSD and episodes of hyperthermia. Eur J Hum Genet. 2007;15:638–645. doi: 10.1038/sj.ejhg.5201813. doi:10.1038/sj.ejhg.5201813. [DOI] [PubMed] [Google Scholar]
- Morelle W, Michalski JC. Analysis of protein glycosylation by mass spectrometry. Nat Protoc. 2007;2:1585–1602. doi: 10.1038/nprot.2007.227. doi:10.1038/nprot.2007.227. [DOI] [PubMed] [Google Scholar]
- Munro S. A comparison of the transmembrane domains of Golgi and plasma membrane proteins. Biochem Soc Trans. 1995;23:527–530. doi: 10.1042/bst0230527. [DOI] [PubMed] [Google Scholar]
- Munro S. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. doi:10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata Y, Burger MM. Wheat germ agglutinin: Molecular characteristics and specificity for sugar binding. J Biol Chem. 1974;249:3116–3122. [PubMed] [Google Scholar]
- Ng BG, Kranz C, Hagebeuk EEO, Duran M, Abeling NGGM, Wuyts B, Ungar D, Lupashin V, Hartdorff CM, Poll-The BT, et al. Molecular and clinical characterization of a Moroccan Cog7 deficient patient. Mol Genet Metab. 2007;91:201–204. doi: 10.1016/j.ymgme.2007.02.011. doi:10.1016/j.ymgme.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Au CE, Bergeron J. Sorting out glycosylation enzymes in the Golgi apparatus. FEBS Lett. 2009;583:3764–3769. doi: 10.1016/j.febslet.2009.10.064. doi:10.1016/j.febslet.2009.10.064. [DOI] [PubMed] [Google Scholar]
- Nilsson T, Hoe MH, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger EG, Warren G. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G. Kin recognition: A model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. doi:10.1016/0014-5793(93)80906-B. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. doi:10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Oka T, Ungar D, Hughson FM, Krieger M. The COG and COPI complexes interact to control the abundance of GEARs, a subset of Golgi integral membrane proteins. Mol Biol Cell. 2004;15:2423–2435. doi: 10.1091/mbc.E03-09-0699. doi:10.1091/mbc.E03-09-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opat AS, Houghton F, Gleeson PA. Medial Golgi but not late Golgi glycosyltransferases exist as high molecular weight complexes: Role of luminal domain in complex formation and localization. J Biol Chem. 2000;275:11836–11845. doi: 10.1074/jbc.275.16.11836. doi:10.1074/jbc.275.16.11836. [DOI] [PubMed] [Google Scholar]
- Opat AS, Houghton F, Gleeson PA. Steady-state localization of a medial-Golgi glycosyltransferase involves transit through the trans-Golgi network. Biochem J. 2001;358:33–40. doi: 10.1042/0264-6021:3580033. doi:10.1042/0264-6021:3580033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paesold Burda P, Maag C, Troxler H, Foulquier F, Kleinert P, Schnabel S, Baumgartner M, Hennet T. Deficiency in COG5 causes a moderate form of congenital disorders of glycosylation. Hum Mol Genet. 2009;18(22):4350–4356. doi: 10.1093/hmg/ddp389. [DOI] [PubMed] [Google Scholar]
- Peanne R, Legrand D, Duvet S, Mir A-M, Matthijs G, Rorher J, Foulquier F. Differential effects of lobe a and lobe B of the Cog complex on the stability of B4galt1 and St6gal1. Glycobiology. 2011;7:864–876. doi: 10.1093/glycob/cwq176. [DOI] [PubMed] [Google Scholar]
- Pereira ME, Kabat EA. Specificity of purified hemagglutinin (lectin) from Lotus tetragonolobus. Biochemistry. 1974;13:3184–3192. doi: 10.1021/bi00712a029. doi:10.1021/bi00712a029. [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol. 2006;8:238–248. doi: 10.1038/ncb1366. doi:10.1038/ncb1366. [DOI] [PubMed] [Google Scholar]
- Qian R, Chen C, Colley KJ. Location and mechanism of alpha 2,6-sialyltransferase dimer formation: Role of cysteine residues in enzyme dimerization, localization, activity, and processing. J Biol Chem. 2001;276:28641–28649. doi: 10.1074/jbc.M103664200. doi:10.1074/jbc.M103664200. [DOI] [PubMed] [Google Scholar]
- Ram RJ, Li B, Kaiser CA. Identification of sec36p, sec37p, and sec38p: Components of yeast complex that contains sec34p and sec35p. Mol Biol Cell. 2002;13:1484–1500. doi: 10.1091/mbc.01-10-0495. doi:10.1091/mbc.01-10-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders E, Foulquier F, Teles EL, Quelhas D, Morelle W, Rabouille C, Annaert W, Matthijs G. Golgi function and dysfunction in the first COG4-deficient CDG type II patient. Hum Mol Genet. 2009;18:3244–3256. doi: 10.1093/hmg/ddp262. doi:10.1093/hmg/ddp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BC, Smith RD, Ungar D, Nakamura A, Jeffrey PD, Lupashin VV, Hughson FM. Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene. Proc Natl Acad Sci USA. 2009;106:13329–13334. doi: 10.1073/pnas.0901966106. doi:10.1073/pnas.0901966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottger S, White J, Wandall HH, Olivo JC, Stark A, Bennett EP, Whitehouse C, Berger EG, Clausen H, Nilsson T. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J Cell Sci. 1998;111(Pt 1):45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- Schmitz KR, Liu J, Li S, Setty TG, Wood CS, Burd CG, Ferguson KM. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev Cell. 2008;14:523–534. doi: 10.1016/j.devcel.2008.02.016. doi:10.1016/j.devcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A, Suvorova E, Pavliv O, Khaidakova G, Lupashin V. Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J Cell Biol. 2007;179:1179–1192. doi: 10.1083/jcb.200705145. doi:10.1083/jcb.200705145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A, Zolov S, Lupashin V. COG complex-mediated recycling of Golgi glycosyltransferases is essential for normal protein glycosylation. Traffic. 2006;7:191–204. doi: 10.1111/j.1600-0854.2005.00376.x. doi:10.1111/j.1600-0854.2005.00376.x. [DOI] [PubMed] [Google Scholar]
- Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18:379–420. doi: 10.1146/annurev.cellbio.18.030602.133733. doi:10.1146/annurev.cellbio.18.030602.133733. [DOI] [PubMed] [Google Scholar]
- Smith RD, Lupashin VV. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr Res. 2008;343:2024–2031. doi: 10.1016/j.carres.2008.01.034. doi:10.1016/j.carres.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Willett R, Kudlyk T, Pokrovskaya I, Paton AW, Paton JC, Lupashin VV. The COG complex, Rab6 and COPI define a novel Golgi retrograde trafficking pathway that is exploited by SubAB toxin. Traffic. 2009;10:1502–1517. doi: 10.1111/j.1600-0854.2009.00965.x. doi:10.1111/j.1600-0854.2009.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yamamoto A, Nakamura N, Ogata S, Sakisaka S, Hirose S, Ikehara Y, Oda K. Interaction of Golgin-84 with the COG complex mediates the intra-Golgi retrograde transport. Traffic. 2010;11:1552–1566. doi: 10.1111/j.1600-0854.2010.01123.x. doi:10.1111/j.1600-0854.2010.01123.x. [DOI] [PubMed] [Google Scholar]
- Sohda M, Misumi Y, Yoshimura S-I, Nakamura N, Fusano T, Ogata S, Sakisaka S, Ikehara Y. The interaction of two tethering factors, p115 and COG complex, is required for Golgi integrity. Traffic. 2007;8:270–284. doi: 10.1111/j.1600-0854.2006.00530.x. doi:10.1111/j.1600-0854.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- Steet R, Kornfeld S. COG-7-deficient human fibroblasts exhibit altered recycling of Golgi proteins. Mol Biol Cell. 2006;17:2312–2321. doi: 10.1091/mbc.E05-08-0822. doi:10.1091/mbc.E05-08-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud WJ, Jiang S, Jack G, Storrie B. Persistence of Golgi matrix distribution exhibits the same dependence on Sar1p activity as a Golgi glycosyltransferase. Traffic. 2003;4:631–641. doi: 10.1034/j.1600-0854.2003.00122.x. doi:10.1034/j.1600-0854.2003.00122.x. [DOI] [PubMed] [Google Scholar]
- Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157:631–643. doi: 10.1083/jcb.200111081. doi:10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova ES, Kurten RC, Lupashin VV. Identification of a human orthologue of Sec34p as a component of the cis-Golgi vesicle tethering machinery. J Biol Chem. 2001;276:22810–22818. doi: 10.1074/jbc.M011624200. doi:10.1074/jbc.M011624200. [DOI] [PubMed] [Google Scholar]
- Tang BL, Wong SH, Low SH, Hong W. The transmembrane domain of N-glucosaminyltransferase I contains a Golgi retention signal. J Biol Chem. 1992;267:10122–10126. [PubMed] [Google Scholar]
- Tateno H, Uchiyama N, Kuno A, Togayachi A, Sato T, Narimatsu H, Hirabayashi J. A novel strategy for mammalian cell surface glycome profiling using lectin microarray. Glycobiology. 2007;17:1138–1146. doi: 10.1093/glycob/cwm084. doi:10.1093/glycob/cwm084. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, D'Agostaro G, Gleeson PA. The signal for Golgi retention of bovine beta-1,4-galactosyltransferase is in the transmembrane domain [erratum appears in 1992 J. Biol. Chem. 267:13113] J Biol Chem. 1992;267:4084–4096. [PubMed] [Google Scholar]
- Teasdale RD, Matheson F, Gleeson PA. Post-translational modifications distinguish cell surface from Golgi-retained beta 1,4 galactosyltransferase molecules: Golgi localization involves active retention. Glycobiology. 1994;4:917–928. doi: 10.1093/glycob/4.6.917. doi:10.1093/glycob/4.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Banfield DK. Localization of Golgi-resident glycosyltransferases. Cell Mol Life Sci. 2010;67:29–41. doi: 10.1007/s00018-009-0126-z. doi:10.1007/s00018-009-0126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Tai WC, Chen L, Banfield DK. Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science. 2008;321:404–407. doi: 10.1126/science.1159411. doi:10.1126/science.1159411. [DOI] [PubMed] [Google Scholar]
- Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Gerard Waters M. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. doi:10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar D, Oka T, Krieger M, Hughson FM. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. doi:10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Van Damme EJ, Peumans WJ. Biosynthesis of the snowdrop (Galanthus nivalis) lectin in ripening ovaries. Plant Physiol. 1988;86:922–926. doi: 10.1104/pp.86.3.922. doi:10.1104/pp.86.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Malhotra V. The organisation of the Golgi apparatus. Curr Opin Cell Biol. 1998;10:493–498. doi: 10.1016/s0955-0674(98)80064-1. doi:10.1016/S0955-0674(98)80064-1. [DOI] [PubMed] [Google Scholar]
- Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the Exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. doi:10.1016/S1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- Wong SH, Low SH, Hong W. The 17-residue transmembrane domain of beta-galactoside alpha-2,6-sialyltransferase is sufficient for Golgi retention. J Cell Biol. 1992;117:245–258. doi: 10.1083/jcb.117.2.245. doi:10.1083/jcb.117.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Steet RA, Bohorov O, Bakker J, Newell J, Krieger M, Spaapen L, Kornfeld S, Freeze HH. Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat Med. 2004;10:518–523. doi: 10.1038/nm1041. doi:10.1038/nm1041. [DOI] [PubMed] [Google Scholar]
- Zeevaert R, Foulquier F, Cheillan D, Cloix I, Guffon N, Sturiale L, Garozzo D, Matthijs G, Jaeken J. A new mutation in COG7 extends the spectrum of COG subunit deficiencies. Eur J Med Genet. 2009;52:303–305. doi: 10.1016/j.ejmg.2009.06.006. doi:10.1016/j.ejmg.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Zeevaert R, Foulquier F, Dimitrov B, Reynders E, Van Damme-Lombaerts R, Simeonov E, Annaert W, Matthijs G, Jaeken J. Cerebrocostomandibular-like syndrome and a mutation in the conserved oligomeric Golgi complex, subunit 1. Hum Mol Genet. 2009;18:517–524. doi: 10.1093/hmg/ddn379. doi:10.1093/hmg/ddn379. [DOI] [PubMed] [Google Scholar]
- Zeevaert R, Foulquier F, Jaeken J, Matthijs G. Deficiencies in subunits of the conserved oligomeric Golgi (COG) complex define a novel group of congenital disorders of glycosylation. Mol Genet Metab. 2008;93:15–21. doi: 10.1016/j.ymgme.2007.08.118. doi:10.1016/j.ymgme.2007.08.118. [DOI] [PubMed] [Google Scholar]
- Zhao X, Lasell TK, Melançon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: Evidence for distinct functions in protein traffic. Mol Biol Cell. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. doi:10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolov SN, Lupashin VV. Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J Cell Biol. 2005;168:747–759. doi: 10.1083/jcb.200412003. doi:10.1083/jcb.200412003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.