Abstract
Cytosine methylation is the major epigenetic modification of metazoan DNA. Although there is strong evidence that active DNA demethylation occurs in animal cells, the molecular details of this process are unknown. The recent discovery of the TET protein family (TET1–3) 5-methylcytosine hydroxylases has provided a new entry point to reveal the identity of the long-sought DNA demethylase. Here, we review the recent progress in understanding the function of TET proteins and 5-hydroxymethylcytosine (5hmC) through various biochemical and genomic approaches, the current evidence for a role of 5hmC as an early intermediate in active DNA demethylation and the potential functions of TET proteins and 5hmC beyond active DNA demethylation. We also discuss how future studies can extend our knowledge of this novel epigenetic modification.
Key words: TET1, 5-hydroxymethylcytosine, active DNA demethylation, epigenetic, DNA methylation, hippocampus, electroconvulsive stimulation, Gadd45b, BER
History of Active DNA Demethylation
Methylation at the C-5 position of cytosine (C) bases has long been considered the only biologically functional epigenetic covalent modification of the animal genomic DNA. In mammals, 5-methylcytosines (5mCs) are almost exclusively found in CpG dinucleotides, with the exception of non-CpG methylation found in pluripotent stem cells.1,2 CpG methylation plays critical roles in transcriptional silencing of genes and retrotransposons, gene imprinting and X chromosome inactivation.3 Deficiency of DNA methyltransferases (DNMTs), enzymes that add methyl groups onto Cs, leads to profound developmental defects.4 Substantial evidence supports that the CpG methylation pattern across the genome can be replicated across cell division by the maintenance DNMT (DNMT1).5 Indeed, DNMT1 has higher catalytic activity on hemimethylated DNA than on unmethylated DNA, supporting the notion that DNMT1 can replicate the DNA methylation on the parental DNA strand to the newly synthesized strand. It should be noted that inheritability and stability/reversibility of an epigenetic modification are two distinct and separable properties, although they both contribute to the overall duration of the modification.
According to the proposed replication mechanism, DNA methylation can be “diluted” by not replicating the methylated status to the newly synthesized strand. As a result, methylation at this specific locus will be lost in daughter cells upon further division. This process is termed “passive DNA demethylation.”6 On the other hand, much effort has been made to understand the enzymatic removal of this covalent modification (active DNA demethylation) in mammalian cells. Such efforts have been partly encouraged by both the identification of histone demethylases7,8 in mammals and the identification of 5mC glycosylases as DNA demethylases in plants.9 More importantly, it has been shown in many different systems that active DNA methylation does occur in mammalian cells.6 The most well-known example is the rapid loss of 5mC immunoreactivity of the paternal pronucleus in the zygote soon after fertilization.10 In primordial germ cells (PGCs), gene imprinting-related DNA methylation also needs to be re-established.10 In contrast, active DNA methylation occurs in a highly locus-specific manner in differentiated somatic cells. For example, rapid demethylation of interleukin-2 (Il2) promoter in activated T cells has been shown to play a causal role in Il2 expression.11
Despite accumulating evidence for the presence of mammalian DNA demethylase, the identity of such enzymes has been controversial. Methylated DNA binding domain-containing protein MBD2 was reported to efficiently demethylate 5mCs and release methanol with no co-factor requirement.12 However, no independent confirmation from other laboratories has been reported so far. Analogous to plant demethylases, mammalian DNA glycosylase TDG has been shown to possess weak 5mC glycosylase activity,13,14 although it is unclear whether TDG is indeed responsible for active DNA demethylation observed in vivo. Other recent studies have pointed to an alternative mechanism that involves the sequential steps of deamination by AID/APOBEC deaminases15 or DNMTs,16 generation of a T:G mismatch, base excision repair (BER) initiated by T:G mismatch glycosylases, including TDG and MBD4, and refill of unmodified mononucleotides. Multiple lines of evidence have emerged in support of this BER-based mechanism, including extensive presence of DNA break markers, such as γH2A.X and PARP-1, on the paternal pronuclei,17 mechanistic requirements for BER enzymes18 and a modest hypermethylation phenotype in AID-null PGCs.19 Interestingly, demethylation of PGCs was still largely conserved in AID-null PGCs, suggesting that other pathways also play significant roles in the process.
Indirect/non-catalytic modulators have also been reported to participate in active DNA demethylation. In an overexpression screening, Gadd45a was found to promote global DNA demethylation through DNA repair.20 Similar efforts have not only confirmed the roles of Gadd45 family proteins, but have also identified other proteins that might be involved in this process, including RING finger protein 4.21 The detailed mechanisms by which these factors regulate active DNA demethylation remain to be determined.
Discovery of the TET Proteins and 5hmCs
It was first reported in 1971 that 5hmCs were present in both rodent and frog brain DNA,22 although the reported abundance was significantly higher than recent studies using more accurate methods.23–27 This discovery did not attract significant attention until 2009, when two laboratories independently reported the groundbreaking (re)discovery of 5hmCs in mammalian genomic DNA. In one study, Kriaucionic and Heintz used an elegant genetic labeling approach to purify nuclei from two distinct neuronal subtypes from the mouse cerebellum.28 Purkinje cells have large and generally euchromatic nuclei, whereas granule cells have much smaller and heterochromatic nuclei. When they used the nearest-neighbor assay to determine if the global methylation levels were different between the two subtypes, they observed an uncharacterized mononucleotide signal, more prominent in Purkinje cells than in granule cells. Further chemical characterization identified this unknown signal as 5hmC. They estimated the abundance of 5hmC in Purkinje cells was ∼0.6% of all Cs, which translated to almost 40% of all 5mCs.
In contrast to the serendipity of the Heintz study, Rao and colleagues set out to look for mammalian enzymes that have the potential to modify DNA bases.29 A homology search for the trypanosome thymine hydroxylases JBP1/2 led to three paralogous human TET proteins, with their orthologs found throughout metazoan genomes. An elegant series of experiments showed that TET1 does not modify thymines but, rather, 5mCs both in vitro and in mammalian cells, generating 5hmCs. They further showed that the 5hmC level is higher in mouse embryonic stem cells (mESCs; 4.4% of all CCGGs) than in other cell types they measured, and both Tet1 expression and 5hmC levels are decreased upon differentiation of mESCs, linking this novel modification to pluripotency. Following these two studies, many other laboratories have confirmed and extended these results using various methods.24–27,30,31
5hmC as an Intermediate of Active DNA Demethylation: Indirect Evidence
The discovery of 5hmCs in mammalian DNA immediately led to wide speculation on its biological function, the dominant one being that 5hmCs may represent an intermediate product in the process of active DNA demethylation. The reason is that 5hmC satisfies a possible oxidative demethylation mechanism, analogous to reactions in the thymidine salvage pathway32 and the direct repair of DNA alkylation damages by AlkB oxygenases.33,34 In fact, the first piece of evidence in line with this hypothesis came from the very first paper that identified TET1 as a 5mC hydroxylase,29 where the authors observed a slight but statistically significant increase in unmodified cytosine content upon TET1 overexpression in HEK293 cells.
Studies from other laboratories have provided more evidence supporting the oxidative demethylation hypothesis. Zhang and colleagues showed that Tet1 plays a role in mESC self-renewal and lineage commitment partly by maintaining the promoter of a pluripotency gene Nanog in an unmethylated state.30 Tet1 knockdown caused de novo methylation of the Nanog promoter and gene silencing. Rao and colleagues also reported hypermethylation of a few gene promoters caused by Tet1/2 knockdown in mESCs, but hypomethylation at other promoters was also observed.35 Shi and colleagues reported that TET1 overexpression led to active demethylation of artificially methylated plasmid DNA.31
The TET1 gene was originally identified through its translocation in acute myeloid leukemia (AML).36,37 Later, TET2 was also found to be frequently mutated in various forms of myeloid malignancies.38 A number of studies have provided fascinating links between TET2 malfunction, an oncogenic metabolite 2-hydroxyglutarate (2HG) and myeloid differentiation and malignancies.39–41 One study showed a DNA hypermethylation phenotype (previously known in many AML cases) in TET2mutated AML samples,40 which is consistent with the oxidative demethylation hypothesis. Surprisingly, in another study, Rao and colleagues observed a DNA hypomethylation phenotype in TET2-mutated AML samples.39 While the two studies used different methods for DNA methylation profiling, the exact reasons for the discrepancies remain unclear.
Multiple groups have asked the same question, whether 5hmC plays any role in zygotic paternal DNA demethylation.18,42,43 Strikingly, 5hmC immunoreactive signals were found exclusively on the paternal pronuclei, highly correlated with the disappearance of its 5mC immunoreactivity. The responsible hydroxylase appeared to be TET3. This observation resolved the apparent discrepancy between the global loss of 5mC immunoreactivity and the modest decrease in bisulfite sequencing-measured methylation levels of individual loci. It has been further shown that, although modest demethylation occurs, 5hmC signals on the paternal pronuclei persist upon multiple rounds of cell division, demarcating the paternally originated chromosomes.42,43 These observations raise fascinating questions about how the imbalance of 5hmC levels between the two sets of chromosomes affects early development.
Direct Evidence for 5hmC Demethylase Activity in Mammalian Cells
Although the abovementioned studies have significantly strengthened the oxidative demethylation hypothesis, it was still unproven whether 5hmCs generated by TET proteins are indeed converted to unmodified Cs. Changes in the methylation status of specific genomic loci could very well be indirect consequences of gain- or loss-of-function of TET proteins. This possibility may explain some of the differences in the methylation phenotypes in TET1/2 loss-of-function models. To directly test the hypothesis, an artificial DNA probe was designed consisting of a human Ubiquitin C promoter driving a GFP open reading frame (UbC-GFP).44 This design could potentially allow one to measure any transcriptional regulatory effect of cytosine modifications. It was fortunate that most Taq DNA polymerases could incorporate pre-modified 5m-dCTP (5-methyl-2-deoxycytidine-5-triphosphate) and 5hm-dCTP (5-hydroxymethyl2-deoxycytidine-5-triphosphate) just as efficiently as dCTP (2-deoxycytosine-5-triphosphate), making it possible to generate fully pre-modified probes by PCR. Fully hydroxymethylated UbC-GFP probes (5hmC-GFP) exhibit modest yet significant sensitivity to a methylation-sensitive enzyme HpaII at 48 h after transfection into HEK293 cells. More importantly, bisulfite sequencing, which also does not distinguish 5mC and 5hmC, shows the presence of unmodified Cs in the transfected 5hmC-GFP probes but not in untransfected 5hmC-GFP probes or transfected 5mC-GFP probes. These results provide the first direct evidence for a robust 5hmC-specific DNA demethylase activity in mammalian cells. Notably, such demethylation activity for 5hmCs is not limited to a CpG context.
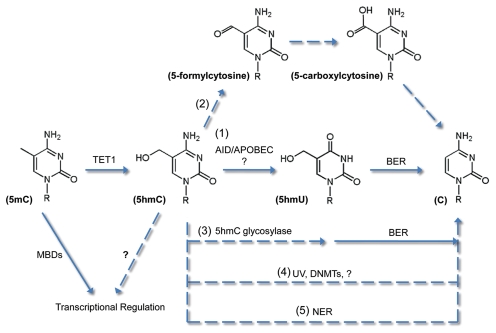
This simple assay using an easily accessible cell line, such as HEK293 cells, greatly facilitated the understanding of its molecular details (Fig. 1).44 First, pharmacological inhibition of key BER enzymes could partially block 5hmC demethylation, consistent with a requirement for these enzymes in zygotic demethylation.18 Second, overexpression of each of the 12 known human DNA glycosylases does not have significant impact on global 5hmC levels or demethylation efficiency of 5hmC-GFP, suggesting that BER might be initiated by an indirect mechanism. Finally, over-expression of a few members of the AID/APOBEC deaminase family could enhance demethylation of 5hmC-GFP probes, suggesting that these enzymes might have a direct or indirect role in this process. It will be important for future studies to determine directly the capacity of these deaminases to use 5hmC as their substrates to generate 5-hydroxymethyluracil (5hmU).
Figure 1.
Potential mechanisms for active DNA demethylation through 5hmC. (1) Evidence suggests that 5hmC may be converted to 5hmU by AID/APOBEC deaminases and repaired by the base excision repair (BER) pathway. (2) 5hmC may also be further oxidized to 5-carboxylcytosine and decarboxylated/repaired to C. (3) A 5hmC glycosylases may excise 5hmC and directly initiate BER. (4) Direct conversion from 5hmC to C may also occur. (5) Nuclear excision repair (NER) pathway may excise 5hmC-containing DNA strands. Besides promoting DNA demethylation, 5hmC may regulate transcription through unidentified 5hmC-binding proteins. Solid arrows indicate catalytic steps that are supported by experimental evidence, whereas dashed arrows indicate speculative processes.
The single-strand, single-nucleotide resolution of bisulfite sequencing revealed more mechanistic properties of the mammalian 5hmC demethylase activity.44 First, 5hmC demethylation appears to be highly processive, with a few DNA strands being extensively demethylated, while others barely showed any demethylated 5hmCs. Within each cluster of demethylated 5hmCs, very few 5hmCs are skipped (unpublished observations), implying that a “hopping” mechanism is unlikely. Second, transcription plays a critical role in the process. Analysis of promoter-truncated transcriptionally inactive 5hmC-GFP probes showed much less demethylation. It is possible that the 5hmC demethylase targets single-strand DNA, or that the 5hmC demethylase is recruited by specific components of the transcription initiation/elongation machineries. Finally, of the two DNA strands in the transcribed region, 5hmC demethylation is much more efficient on the non-transcribed (sense) strand than on the transcribed (antisense) strand. All these mechanistic aspects resemble some of the known properties of AID and somatic hypermutation,45,46 making deamination a more promising candidate step for 5hmC demethylation.
Other possibilities exist for the bona fide 5hmC demethylase (Fig. 1). First, it is not known whether TET proteins or other enzymes can further oxidize the hydroxymethyl group to generate formyl and carboxyl groups, which may serve as better leaving groups. Second, 5hmC glycosylase activity was found in calf thymus extract over 20 y ago,47 although the identity of this glycosylase still remains unknown. Third, direct reversal of 5hmCs to unmodified Cs has been reported to occur under certain conditions or to be performed by bacterial methyltransferase HhaI.48 Finally, similar to its potential role in 5mC demethylation,20,49 nucleotide excision repair (NER), which does not involve modi-fication-specific recognition, may be utilized to re-synthesize the DNA strand that contains 5hmCs. On the other hand, 5hmC in certain genomic contexts may remain stable and resistant to the 5hmC demethylase. It may reverse the silencing effect of 5mCs mediated by MBD proteins, which do not seem to recognize 5hmC-containing DNA,50 or it may have its own protein “readers” to achieve transcriptional regulations.
Active DNA Demethylation in the Mammalian Brain
An advantageous experimental system to study active DNA demethylation would be non-dividing cells. Post-mitotic neurons in the mammalian brain, in particular, have attracted much attention because of their remarkable ability to alter gene expression profiles in response to external stimuli.51–53 Active DNA demethylation has been shown to occur both in cultured neurons54 and in various brain regions in vivo.55–57 Genetic58,59 and pharmacological57,60 studies have further indicated important roles for DNA methylation/demethylation dynamics in regulating neuronal plasticity and animal behaviors.
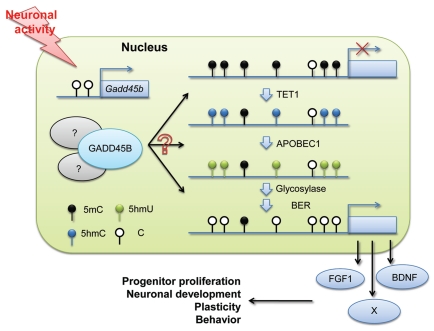
The adult dentate gyrus provides a relative homogenous population of post-mitotic neurons that can be activated synchronously in vivo and made readily accessible in large quantities.54 Such properties are particularly important for epigenetic analysis, because each diploid cell displays only two locus-specific modifications. In a search for epigenetic factors that may mediate neuronal activation-induced changes in gene expression, Gadd45b (growth arrest and DNA damage induced gene 45 β) was found to be dramatically induced by eletroconvulsive stimulation (ECS), a treatment currently used in clinics for depression disorders.54 As discussed above, Gadd45 family proteins have been shown to be enhancing factors in DNA demethylation.15,20,21,49,61 Importantly, Gadd45b plays a critical role in ECS-induced demethylation of Bdnf (brain-derived neurotrophic factor) and Fgf1 (fibroblast growth factor 1) promoters in the adult mouse dentate gyrus (Fig. 2). Activity-induced expression of these neurogenic niche factors and activity-enhanced neurogenesis in the dentate gyrus are both greatly attenuated in Gadd45bdeficient mice, suggesting an important role for DNA demethylation in these processes.51,62 Moreover, endogenously expressed Tet1 and AID homolog Apobec1 also appear to be involved in neuronal activity-induced demethylation of Bdnf and Fgf1 promoters.44 Consistent with this dynamic DNA methylation regulation in neurons, cultured mouse hippocampal neurons can also demethylate the fully modified 5hmC-GFP probes with high processivity.44 It remains to be determined whether or not and how Gadd45b and TET1 may interact to regulate active DNA demethylation in neurons in vivo (Fig. 2). The impact of Gadd45b or TET1 deletion in mature neurons on neuronal function and animal behaviors also needs to be determined. It is interesting to note that Gadd45b has been shown to be abnormally expressed in mental retardation patients carrying mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex,63 and in brain tissue of autistic patients,64,65 supporting a critical role of epigenetic neuronal DNA demethylation in normal brain function.
Figure 2.
Neuronal activity-induced DNA demethylation in the adult brain. Neuronal activation in the dentate gyrus induces expression of Gadd45b, which plays an essential role in demethylation of Bdnf and Fgf1 promoters. This process also involves Tet1 and Apobec1. Gadd45b-dependent DNA demethylation leads to expression of important genes, such as Bdnf and Fgf1, and can potentially have a broad impact in the mature nervous system.
Genomic Distribution of TET Proteins and 5hmCs
The past decade has witnessed a revolution of genomics incited by massively parallel DNA sequencing technologies. Soon after the discovery of TET proteins and 5hmCs, significant efforts have been made to characterize TET protein binding and 5hmC deposition across the mammalian genome, resulting in a flurry of papers recently published from a number of laboratories.66–74 The first published 5hmC profile was that of mouse cerebellum,71 where 5hmCs were present in high abundance. He, Jin and colleagues ingeniously leveraged on the known biology of 5hmC in T-even bacteriophage and used T4 β-glucosyltransferase to tag 5hmCs with glucose-like moieties that could be further biotinylated and affinity-enriched.71 Unlike 5mC, which is abundant all over the genome, 5hmC signals appeared to be enriched around genes. Furthermore, 5hmC levels in gene bodies positively correlated with gene expression. Similar findings were reported later using an hMeDIP (hydroxymethylated DNA immunoprecipitation) method.72
Several studies have focused on mESCs and have used high quality TET1 antibodies to generate Tet1 ChIP-seq profiles.67,69,73 Surprisingly, Tet1 shows strong binding to predominantly CpG islands, which are known to be largely resistant to DNA methylation. Tet1-bound promoters show significantly lower 5mC levels, reflecting the enzymatic action of Tet1. Bisulfite sequencing further showed that aberrant DNA methylation occurs on these normally unmethylated CpG islands upon Tet1 knockdown,69 establishing a role for Tet1 in maintaining the unmethylated states of these critical regulatory regions. This finding has been confirmed on a genome-wide scale by Shi and colleagues.73 It will be interesting to determine whether this function of Tet1 is mediated by its CXXC domain, which usually interacts with unmethylated CpGs. Surprisingly, Tet1 binds to both active and repressed (bivalent) genes in mESCs. Tet1 knockdown led to a loss of H3K27me3 modification and transcriptional derepression of many bivalent genes, placing Tet1 in an upstream position in Polycomb repressive complex-mediated gene silencing in mESCs.69
Unlike 5mCs, 5hmCs are enriched at gene transcription start sites (TSS).71 Loss-of-function experiments show that Tet1 binds to 30–50% of genes marked by 5hmC,67,68,70 suggesting that other Tet proteins may function in parallel. Multiple studies have made the unexpected observation that Tet1 knockdown leads to more de-repressed genes than deactivated genes. Helin and colleagues showed an even more surprising finding that such transcriptional effects were conserved in DNMT triple-knockout mESCs,67 suggesting that Tet1 can regulates gene expression independently of its 5mC hydroxylase activity. They further reported the physical interaction and similar genome distribution between Tet1 and SIN3A repressive complex, linking this well-studied transcriptional regulator to the novel functions of Tet1.
Concluding Remarks
DNA methylation has been traditionally perceived as a binary switch in the genomic DNA. As a majority of CpGs in the mammalian genome are methylated, its impact on genome functions has been a major topic in epigenetics. With the discovery of TET proteins and 5hmC, we now know that Cs in the genome can exist in more than just two forms. Technological barriers need to be overcome to adapt to such new knowledge. Future methods that can simultaneously distinguish C, 5mC and 5hmC,75,76 preferably compatible with high-throughput sequencing, will certainly be the new driving force for the field.
5hmC, recently regarded as “the sixth base” in the genome, greatly expands the epigenetic plasticity of the genome and requires researchers to revisit virtually all known biology of DNA methylation. Our current understanding of this novel epigenetic modification is only the tip of the iceberg.
Acknowledgments
We thank K. Christian for comments. Supported by Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, NIH (NS048271, HD069184) to G.L.M., NIH (AG024984, NS047344) to H.J.S., The FARMS Fellowship to J.U.G., and by postdoctoral fellowship from MSCRF to Y.S. and C.Z.
Abbreviations
- TET
ten-eleven translocated oncogene
- C
cytosine
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- Gadd45
growth arrest and DNA damage induced gene 45
- BER
base excision repair
- NER
nucleotide excision repair
- AID
activation-induced cytidine deaminase
- APOBEC
apolipoprotein B mRNA editing enzyme catalytic polypeptide
- DNMT
DNA methyltransferase
- MBD
methylated DNA binding domain-containing protein
References
- 1.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 4.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 9.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surani MA, Hajkova P. Epigenetic Reprogramming of Mouse Germ Cells toward Totipotency. Cold Spring Harb Symp Quant Biol. 75:211–218. doi: 10.1101/sqb.2010.75.010. [DOI] [PubMed] [Google Scholar]
- 11.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 12.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 13.Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, et al. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardeland U, Bentele M, Jiricny J, Schar P. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Métivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 17.Wossidlo M, Arand J, Sebastiano V, Lepikhov K, Boiani M, Reinhardt R, et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 21.Hu XV, Rodrigues TM, Tao H, Baker RK, Miraglia L, Orth AP, et al. Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc Natl Acad Sci USA. 2010;107:15087–15092. doi: 10.1073/pnas.1009025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinney SM, Chin HG, Vaisvila R, Bitinaite J, Zheng Y, Esteve PO, et al. Tissue specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genome. J Biol Chem. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krais AM, Park YJ, Plass C, Schmeiser HH. Determination of genomic 5-hydroxymethyl-2′-deoxycytidine in human DNA by capillary electrophoresis with laser induced fluorescence. Epigenetics. 2011;6:560–565. doi: 10.4161/epi.6.5.15678. [DOI] [PubMed] [Google Scholar]
- 25.Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS ONE. 2010;5:15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Münzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, et al. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.20100203. [DOI] [PubMed] [Google Scholar]
- 27.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 32.Warn-Cramer BJ, Macrander LA, Abbott MT. Markedly different ascorbate dependencies of the sequential alpha-ketoglutarate dioxygenase reactions catalyzed by an essentially homogeneous thymine 7-hydroxylase from Rhodotorula glutinis. J Biol Chem. 1983;258:10551–10557. [PubMed] [Google Scholar]
- 33.Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 34.Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 35.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 37.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 38.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/jccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 43.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 46.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 47.Cannon SV, Cummings A, Teebor GW. 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem Biophys Res Commun. 1988;151:1173–1179. doi: 10.1016/S0006291X(88)80489-3. [DOI] [PubMed] [Google Scholar]
- 48.Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nat Chem Biol. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 49.Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/jmolcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meaney MJ, Ferguson-Smith AC. Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci. 2010;13:1313–1318. doi: 10.1038/nn1110-1313. [DOI] [PubMed] [Google Scholar]
- 54.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 55.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13:1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 57.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 58.LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 61.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–567. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garbett K, Ebert PJ, Mitchell A, Lintas C, Manzi B, Mirnics K, et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol Dis. 2008;30:303–311. doi: 10.1016/j.nbd.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011 doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 69.Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin SG, Wu X, Li AX, Pfeifer GP. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-wide Regulation of 5hmC, 5mC and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wallace EV, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun (Camb) 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]