Abstract
Identifying therapeutic targets for cancer treatment relies on consistent changes within particular types or sub-types of malignancy. The ability to define either consistent changes or sub-types of malignancy is often masked by tumor heterogeneity. To elucidate therapeutic targets in cutaneous squamous cell carcinoma (cSCC), the most frequent skin neoplasm with malignant potential, we have developed an integrated approach to gene expression profiling beginning with primary keratinocytes in culture. Candidate drivers of cSCC development were derived by first defining a set of in vitro cancer genes and then comparing their expression in a range of clinical data sets containing normal skin, cSCC and the benign hyper-proliferative condition psoriasis. A small interfering RNA (siRNA) screen of the resulting 21 upregulated genes has yielded targets capable of reducing xenograft tumor volume in vivo. Small-molecule inhibitors for one target, Polo-like kinase-1 (PLK1), are already in clinical trials for other malignancies, and our data show efficacy in cSCC. Another target, C20orf20, is identified as being overexpressed in cSCC, and siRNA-mediated knockdown induces apoptosis in vitro and reduces tumor growth in vivo. Thus, our approach has shown established and uncharacterized drivers of tumorigenesis with potent efficacy as therapeutic targets for the treatment of cSCC.
Keywords: cutaneous squamous cell carcinoma, primary cell culture, gene expression analysis, therapeutic targets, PLK1
Introduction
Keratinocyte skin cancers are the most common neoplasm in Caucasian populations, with an estimated incidence of over 100 000 per year in the United Kingdom (http://info.cancerresearchuk.org/cancerstats/types/skin/incidence/) and a cumulative risk of 70% in a 70y old Australian male (Staples et al., 2006). Cutaneous squamous cell carcinoma (cSCC) is the most common keratinocyte skin cancer with malignant potential and patients presenting with regional metastasis have a poor outcome: 5-year survival in this group is 25–50% (Epstein, 1984; Veness et al., 2007). In the United Kingdom, over 1 in 4 skin cancer deaths can be attributed to non-melanoma skin cancer, principally cSCC (ISD Scotland; http://www.isdscotland.org/isd/183.html). High-risk groups exist where cSCC is a major complication resulting in considerable morbidity and mortality. Organ transplant patients are at a greater than 100-fold increased chance of developing cSCC, leading to a high burden of malignancy (Euvrard et al., 2003), whereas patients with the genetic skin-blistering disease, recessive dystrophic epidermolysis bullosa (RDEB), suffer unprecedented terminal metastasis with over 80% mortality, making cSCC the usual cause of death for this patient group (Fine et al., 2009).
Treatment is always required for cSCC and principally consists of excision and/or radiotherapy for local disease control, with a paucity of options for recurrent and metastatic disease. Pursuit of targeted therapies capable of halting growth and spread of cSCC remains a clear research goal. Recent success with targeted therapies for the treatment of chronic myeloid leukemia, HER2-amplified and BRCA-mutated tumors shows that this goal is achievable and holds great potential (Druker et al., 2001; Piccart-Gebhart et al., 2005; Fong et al., 2009). Screening for cancer targets is hampered by tumor complexity and heterogeneity coupled with difficulties in distinguishing between drivers of tumor characteristics and the characteristics themselves (Merlo et al., 2006). Furthermore, pathways important for cancer are frequently inherent to normal cell function, resulting in clinically limiting side effects when successfully targeted (Cheng and Force, 2010).
In this study, we use the ability to culture primary keratinocytes and use an integrated gene expression profiling approach to derive a small set of tumor-specific genes, which we screen with a small interfering RNA (siRNA), and identify both existing and novel cancer targets capable of specifically inducing tumor cell apoptosis.
Results
Primary cSCC keratinocytes readily form tumors in vivo irrespective of histological grade
In order to model human cSCC without the need for genetic manipulation, we isolated keratinocytes directly from fresh tumor material as described (Rheinwald and Beckett, 1981). To study life-threatening cSCC, we processed tumors that presented with metastasis derived from immunocompetent and immunosuppressed patients, as well as tumors derived from patients with RDEB. We also used cSCC keratinocytes isolated from well-differentiated tumors that did not present with metastasis and non-cSCC primary epidermal keratinocytes (from either routine reduction surgery or normal skin obtained at the time of cSCC surgery). Supplementary Table S1 details the patient donors who provided the tumor samples used in this study. All tumor keratinocytes showed clear genetic alterations as determined by single-nucleotide polymorphism mapping array hybridization and cytogenetic analysis, whereas all peritumoral normal keratinocytes did not (Supplementary Tables S2–S3; Supplementary Figure S1, and data not shown).
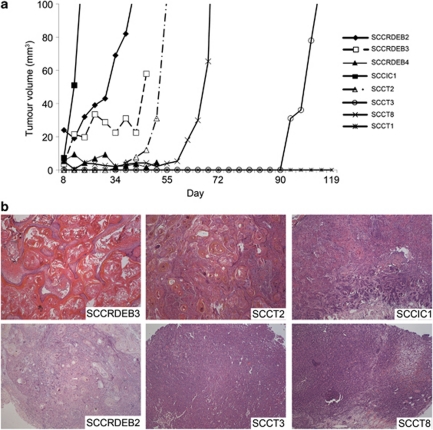
Five of eight cSCC keratinocyte populations readily formed tumors in severe combined immunodeficient (SCID) mice; 1/8 of the populations consistently formed squamous cysts, which failed to reach a volume of 100 mm3 and 2/8 the tumor populations tested did not grow after 120 days (Figure 1a). Xenograft tumors were readily recognized as human cSCC with varied histological grade (Figure 1b). In vivo growth was not restricted to moderately or poorly differentiated tumors, or those derived from patients with RDEB (Figure 1 and Supplementary Table S1).
Figure 1.
cSCC keratinocytes readily form tumors in SCID mice with identical histology to human cSCC. Female SCID Balb/c mice were subcutaneously injected in the right flank with 1–4 × 106 tumor cells mixed with high-concentration Matrigel (Becton Dickinson). Tumor volumes were measured twice a week with calipers and calculated using the formula, V=π4/3((L+W)/4)3, were L is the length and W is the width. (a) Representative growth of a single tumor from eight separate cSCC keratinocyte populations. The average number of days to reach a volume of 100 mm3 from 1–4 separate experiments was as follows: SCCRDEB2 (49 days±5.6 s.d., n=3), SCCIC1 (11.8 days±3.4 s.d., n=12), SCCT2 (59.7 days±8.1 s.d., n=3), SCCT3 (128.3 days±11.7 s.d., n=3) and SCCT8 (65 days±7.1 s.d., n=2). (b) H&E-stained sections of a representative xenograft tumor for each of the six cell populations that showed measurable growth in mice ( × 100 magnification). cSCC, cutaneous squamous cell carcinoma; H&E, hematoxylin and eosin; SCID, severe combined immunodeficient.
Primary cSCC keratinocytes are heterogeneous with respect to genetic alteration and cSCC marker expression
In agreement with previous studies, no consistent genetic alterations were observed across all cSCC populations (Supplementary Table S2) (Purdie et al., 2009). Chromosome 8q gain was observed in 7/8 cSCC keratinocyte populations and this correlated with an increase in C-myc expression, an oncogene associated with squamous cell carcinoma (SCC) development (Supplementary Figure S2A) (Pelengaris et al., 1999). To further characterize our cSCC keratinocyte populations, we investigated additional genes, proteins and pathways reported to be important in both human and mouse cSCC, and looked for patterns, which might separate xenograft tumor-forming capability or patient group. p53 and p16 expression varied among cSCC keratinocytes (Supplementary Figure S2B) and we detected TP53 mutations in all eight populations examined (Supplementary Table S1). Phospho-STAT3 expression varied but was consistently increased across all cSCC as compared with primary non-SCC keratinocytes, in agreement with previous reports (Suiqing et al., 2005) (Supplementary Figure S2C). No consistent evidence of activated RAS (determined by antibody detection of GTP-bound RAS) was observed in tumorigenic cultured cSCC keratinocytes as compared with normal primary or non-tumorigenic cSCC human keratinocytes (Supplementary Figures S2D and E).
Comparison of gene expression clearly separates normal keratinocytes from cSCC keratinocytes in quiescent culture
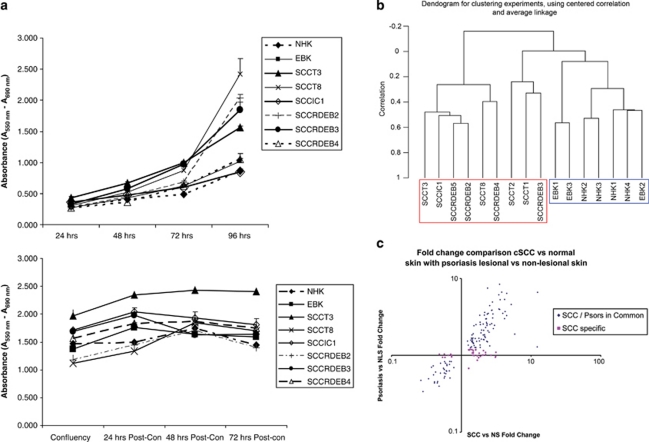
We then performed gene expression analysis using cultures of early-passage primary cells. We chose to use confluent cultures to best mimic the close cell–cell proximity of keratinocytes in vivo and to eliminate changes in gene expression caused by divergent proliferation rates—under the culture conditions used here for RNA harvest, all cells, tumor and normal, showed similar quiescent growth rates (Figure 2a). Un-supervised clustering of normalized array signal intensities clearly segregated normal skin from cSCC (Figure 2b). A comparison between the different sample groups based on normal skin, RDEB skin, cSCC and RDEB cSCC keratinocytes, as well as xenograft tumor histology, or tumor-forming ability, showed that the highest number of differentially expressed genes were identified comparing all cSCC with all non-cSCC cultures (Supplementary Table S4). This analysis defined 435 in vitro differentially expressed cSCC genes (Supplementary Table S5).
Figure 2.
Expression profiling of quiescent cultures of early-passage keratinocytes separates cSCC from normal and identifies potential tumor drivers in vivo. (a) Growth rates of keratinocytes used in this study as assessed by a colorimetric assay of mitochondrial dehydrogenase activity when seeded at low density (upper panel) or at confluence (lower panel). Confluent keratinocytes are quiescent after 48 h of culture. RNA for subsequent array experiment was isolated between 48 and 56 h after confluence. (b) An un-supervised clustering dendrogram of in vitro gene expression data generated by BRB-ArrayTools v3.8.1. cSCC keratinocyte samples (red box) cluster independently of non-cSCC keratinocyte samples (blue box). (c) Average cSCC versus normal skin (NS) fold change for all 154 concordantly expressed in vivo cSCC genes plotted against average psoriatic lesional skin versus psoriatic non-lesional skin (NLS) fold change shows a strong overall correlation (r2=0.84) and identifies genes specifically differentially regulated in cSCC. cSCC, cutaneous squamous cell carcinoma; EBKs, primary non-SCC RDEB keratinocytes; NHKs, primary normal human keratinocytes. A full colour version of this figure is available at the Oncogene journal online
Thirty-five percent of in vitro cSCC genes are expressed concordantly across three independent in vivo data sets
To identify clinically relevant genes from our in vitro cSCC gene set, we analyzed the expression of all probes representing each of the 435 genes in three separate tissue expression data sets containing primary cSCC and normal skin samples. We performed our own experiment comparing RNA isolated from fresh frozen cSCC (n=9) and non-cSCC skin (n=5), and interrogated data from two publicly available experiments (GDS2200 (Nindl et al., 2006) and GSE7553 (Riker et al., 2008)). In agreement with recent observations that little overlap exists between the gene expression profiling of cutaneous or head-and-neck SCC when stringent filtering criteria are applied (Van Haren et al., 2009; Braakhuis et al., 2010), probes representing only six of our 435 gene signature were returned as significantly differentially expressed across all three in vivo data sets based on average fold change >2 and P<0.005 (Supplementary Table S6). However, when using an average fold change of 20%, probes representing 154 of the 435 genes were concordantly expressed across all three separate array platforms, suggesting that a large proportion of the genes identified in culture are relevant to cSCC pathology. This analysis defined a set of 154 in vivo cSCC genes (Supplementary Table S7).
Comparison with the benign hyper-proliferation disorder psoriasis identifies 37 genes as potential drivers of cSCC
Differentially expressed tumor genes can be either a driver or a consequence of disease state. One approach to identify tumor-specific changes is to compare gene expression with comparable benign conditions. Psoriasis offers such a point of comparison as this disease, although completely benign, harbors massive hyper-proliferation, as well as a reactive inflammatory response (Haider et al., 2006). A comparison between fold change of psoriasis lesional versus psoriasis non-lesional skin (average of data sets GSE13355 and GSE14905 (Romanowska et al., 2010)), and fold change of cSCC versus normal skin (average of data sets in Supplementary Table S7), showed a striking relationship between expression (r2=0.84) indicating that the majority of our 154 in vivo cSCC genes were dysregulated similarly in psoriasis (Figure 2c and Supplementary Table S8). Thirty-seven genes did not show similar fold change in psoriasis (Figure 2c and Supplementary Table S9) and we therefore designated these genes as ‘cSCC-specific'.
Twenty-two percent of in vivo cSCC genes are differentially regulated in vitro
Of the 37 cSCC-specific genes only 29 were similarly differentially regulated in vitro compared with in vivo; eight genes were differentially expressed (cSCC versus normal) in an opposite manner (Supplementary Table S9). This percentage was identical across all 154 in vivo cSCC genes: 34 (22%) were discordantly regulated in vitro compared with in vivo (Supplementary Tables S5 and S7). Gene ontology analysis showed that these discordantly expressed genes were disproportionately involved in the cytoskeleton or signal transduction (Supplementary Figure S4), suggesting that cellular context is important for their expression. Because of the discordant nature of their expression, we chose not to pursue these eight cSCC-specific genes and focused on the remaining 29 cSCC-specific genes, which were similarly regulated (relative to normal skin) in vitro and in vivo. Supplementary Figure S3 provides a flow diagram detailing how we derived this gene set.
RNA interference screen identifies PLK1 and C20orf20 as genes critical for tumor cell survival
As 21 of the remaining 29 cSCC-specific genes were upregulated, we screened two cSCC keratinocyte populations, SCCIC1 and SCCRDEB2, by siRNA knockdown of each gene individually and assessing cell viability by a colorimetric assay of mitochondrial dehydrogenase activity. All three duplexes targeting Polo-like kinase-1 (PLK1) and C20orf20, either individually or pooled, consistently reduced cell viability in a high-throughput format (Supplementary Figure S5). Three further potential targets showed ‘hits' consistently with 2/3 siRNAs (Supplementary Figure S5).
PLK1 knockdown and inhibition induces significant G2/M arrest and apoptosis in cSCC keratinocytes compared with normal-proliferating keratinocytes
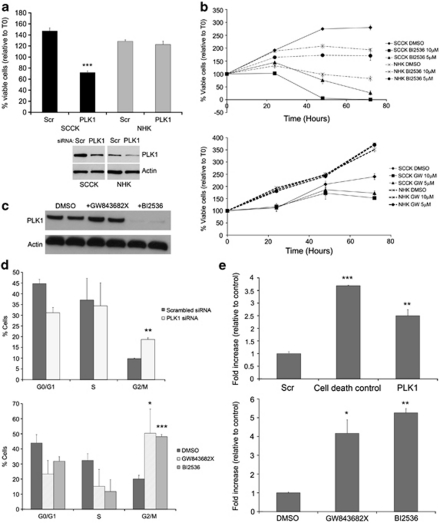
Both RNA interference-mediated depletion of PLK1 and activity inhibition using the small-molecule inhibitors BI 2536 and GW843682X resulted in a potent reduction of cell viability in cSCC cells, with little effect on the growth of normal primary keratinocytes at comparable doses (Figures 3a and b). siRNA knockdown of PLK1 was performed more than 10 times in cSCC cells (SCCRDEB2 and SCCIC1) and five times in normal primary keratinocytes. On each separate occasion, a significant reduction in viability when compared with scrambled non-targeting control was seen in cSCC cells, whereas no significant change was observed in normal primary keratinocytes using the MTS assay as described (P<0.05; Figure 3a and data not shown). Knockdown of PLK1 at the level of protein expression relative to actin was assessed by western blotting on three separate occasions and the extent of knockdown ranged from 48.4 to 80.8% in cSCC and 43 to 80.2% in normal primary keratinocytes (Figure 3a, and data not shown). GW843682X did not affect normal primary keratinocyte growth, whereas only very high doses of the inhibitor BI 2536 reduced the viability of proliferating normal primary keratinocytes (Figure 3b). In agreement with separate studies evaluating BI 2536 in chronic myeloid leukemia (Gleixner et al., 2010) and GW843682X in melanoma (Schmit et al., 2009b), western analysis showed that, unlike exposure to GW843682X, BI 2536 significantly reduced the amount of PLK1 protein after 24 h (Figure 3c). Cell-cycle analysis showed an accumulation of cells at the G2/M phase following PLK1 inhibition and depletion (Figure 3d), and a cell death detection ELISA showed substantial induction of apoptosis, through an increase in cleaved nucleosomes in the cytoplasm, following both PLK1 inhibition and depletion in cSCC cells (Figure 3e). The concentration of the inhibitor BI 2536 needed for half-maximal growth inhibition (EC50) of SCCRDEB2, SCCIC1 and normal primary keratinocytes in the presence of media supplements required for normal primary keratinocyte growth was found to be 190 n, 440 n and 2.98 μ respectively.
Figure 3.
PLK1 knockdown and inhibition induces significant G2/M arrest and apoptosis in cSCC keratinocytes compared with that in normal-proliferating keratinocytes. (a) SCCRDEB2 (SCCK) keratinocytes and primary normal human keratinocytes (NHKs) were transfected with a pool of three separate siRNAs targeting PLK1 and both cell viability and PLK1 protein expression were determined. The percentage of viable cells relative to time 0 (T0) is shown. Whole-cell lysate was extracted from the same SCCRDEB2 keratinocytes or NHKs treated with the same transfection reagent mix at the same time as those used for the viability assessment shown, and PLK1 protein levels were determined by western blotting (lower panel). All results shown represent the mean±s.d.; ***P<0.001 compared with the scrambled control siRNA (SCR) (n=3). (b) The SCCIC1 (SCCK) keratinocytes and NHKs cell viability time course over 72 h of treatment with the PLK1 inhibitors BI 2536 (top panel) and GW843682X (bottom panel). The percentage viable cell number is shown relative to values at T0 (<100% represents a net decline in cell number, >100% represents a net increase in cell number). Representatives of a minimum of three experiments are shown. The results shown represent the mean±s.d., n=3. The NHKs used in each experiment were isolated from different donors. (c) Whole-cell lysate was extracted from SCCIC1 keratinocytes and subjected to western blot analysis of PLK1 protein levels 24 h after exposure to 10 μ GW843682X or 5 μ BI 2536 as indicated. (d) Cell-cycle analysis in SCCIC1 keratinocytes treated either with a pool of three separate PLK1 targeting siRNA (top panel) or with the PLK1 inhibitors GW843682X (10 μ) and BI 2536 (5 μ) (bottom panel), and stained for BrdU and propidium iodide. Results expressed as percentage of cells at the G0/G1, S and G2/M phases of cell cycle 20 or 16 h following treatment, respectively. The results shown are the mean±s.d. of three independent experiments. (e) SCCIC1 keratinocytes transfected with a pool of three separate siRNAs targeting PLK1 or a cell death-positive control siRNA (upper panel), or treated with GW843682X (10 μ) and BI 2536 (5 μ) (lower panel), were assayed for induction of apoptosis using a Cell Death Detection ELISA 24 or 20 h after treatment. The results show the fold increase in absorbance associated with increased cytoplasmic nucleosomes relative to scrambled control non-targeting siRNA or drug vehicle control, respectively. Shown is a representative experiment, with each experiment performed a minimum of three times. The results are the mean±s.d., n=3; *P<0.05, **P<0.01, ***P<0.001 compared with the control. BrdU, 5-Bromodeoxyuridine; cSCC, cutaneous squamous cell carcinoma; PLK, Polo-like kinase; siRNA, small interfering RNA.
C20orf20 knockdown induces apoptosis in cSCC cells in the absence of cell-cycle arrest and has no measurable effect on normal keratinocyte growth
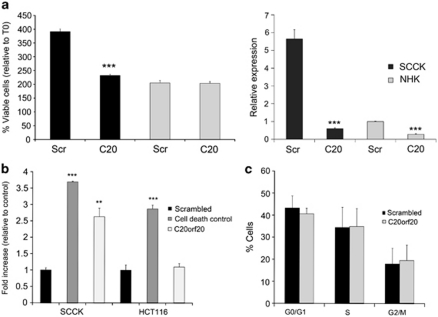
The chromosomal segment harboring C20orf20 has been identified as being frequently amplified in colorectal cancer (Carvalho et al., 2009) and cervical cancer (Scotto et al., 2008), and most recently, in parallel to our work, C20orf20 was identified as being overexpressed in colorectal cancer (Yamaguchi et al., 2010). This colorectal cancer study showed that a reduction of C20orf20 expression through stable short-hairpin RNA inhibited proliferation in the colon carcinoma lines HCT116 and SW480 (shown by a 10% decrease in S-phase replicating cells), without evidence of apoptosis (Yamaguchi et al., 2010). We investigated whether C20orf20 knockdown yielded similar results in cSCC and whether any effect on normal keratinocytes was observed. Knockdown in cSCC resulted in reduced cell viability, with no measurable effect on normal keratinocytes under similar conditions (Figure 4a). siRNA knockdown of C20orf20 was performed more than 20 times in cSCC cells (SCCRDEB2 and SCCIC1) and eight times in normal primary keratinocytes. On each separate occasion, a significant reduction in viability when compared with scrambled non-targeting control was seen in cSCC cells, whereas no significant change was observed in normal primary keratinocytes using the MTS assay as described (P<0.05; Figure 4a, and data not shown). Knockdown of C20orf20 relative to actin was assessed by reverse transcription–real-time quantitative PCR on two separate occasions and the extent of knockdown was found to be 64.3 and 89.4% in cSCC, and 51.9 and 72.3% in normal primary keratinocytes (Figure 4a, and data not shown). Depletion in cSCC induced a significant apoptotic response in the absence of any change in cell-cycle parameters (Figure 4b and c), the opposite to that seen in colorectal cancer cell lines. In agreement with the previous study, knockdown of C20orf20 in the HCT116 cells did not show an increase in apoptosis (Yamaguchi et al., 2010) (Figure 4b), indicating either a more potent effect in cSCC or a different mode of action.
Figure 4.
C20orf20 knockdown induces apoptosis in cSCC cells without cell-cycle arrest, with no measurable effect on normal keratinocyte growth. (a) SCCIC1 keratinocytes and primary normal human keratinocytes (NHKs) were transfected with a pool of three separate siRNAs targeting C20orf20 (C20) or a scrambled non-targeting control siRNA (Scr), and cell viability was assessed by MTS assay (left panel). The results shown are 48 h after seeding of transfected cells and expressed relative to T0. The relative expression of C20orf20 following siRNA transfection was assessed by reverse transcription–real-time quantitative PCR on the same cells treated with the same transfection reagent mix at the same time as those used for the viability assessment shown, with all data shown relative to the NHK scrambled non-targeting control (right panel). (b) SCCIC1 keratinocytes and HCT116 colorectal cancer cells were transfected with a scrambled non-targeting control, a cell death-positive control or a pool of three separate C20orf20 targeting siRNA, and apoptosis was assessed 24 h later by a Cell Death Detection ELISA. The results show the fold increase in absorbance associated with increased cytoplasmic nucleosomes relative to non-targeting siRNA. Shown is a representative experiment, with each experiment performed a minimum of three times. The results are the mean±s.d., n=3; **P<0.01, ***P<0.001 compared with the control. (c) Cell-cycle analysis in SCCIC1 keratinocytes treated with C20orf20 siRNA. Cells were incubated for 20 h after transfection and labeled with BrdU and propidium iodide. Results expressed as percentage of cells at the G0/G1, S and G2/M phases. The results shown are the mean ±s.d. of three independent experiments. BrdU, 5-Bromodeoxyuridine; cSCC, cutaneous squamous cell carcinoma; siRNA, small interfering RNA.
PLK1 inhibition and C20orf20 siRNA knockdown target cSCC in vivo
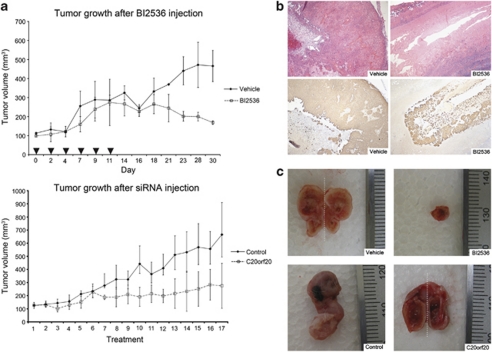
To assess the in vivo action of PLK1 inhibition and C20orf20 depletion we injected either the PLK1 inhibitor BI 2536 or a C20orf20-targeting siRNA into established SCCIC1 xenograft tumors. In each case we saw direct evidence of effective tumor targeting (Figure 5a). In as little as 2 weeks, tumors harvested from animals treated with BI 2536 showed marked reduction in the presence of tumor keratinocytes compared with vehicle controls (Figure 5b). Treatment with C20orf20 siRNA reduced tumor volume over time compared with a non-targeting siRNA control (Figure 5a). The largest C20orf20 siRNA-treated tumors showed marked reduction in the number of tumor keratinocytes present and were hollow in appearance (Figure 5c).
Figure 5.
PLK1 inhibition and C20orf20 knockdown reduces tumor growth in vivo. (a) Top panel: SCCIC1 tumors treated with the PLK1 inhibitor BI 2536. Treatment group tumors (n=3) were injected with BI 2536 at a dose of 25 mg/kg, six times over 2 weeks (arrows). The control group (n=2) was injected with the vehicle following the same schedule. Bottom panel: SCCIC1 tumors treated with C20orf20 siRNA. Treatment group (n=3) tumors were injected with 580 pmol of a pool of three separate C20orf20 targeting siRNA duplexes in 100 μl of phosphate-buffered saline. The control group (n=3) was injected with 580 pmol of a scrambled non-targeting control siRNA duplex in 100 μl of phosphate-buffered saline. The animals were killed 2 days after the last treatment. (b) A representative H&E-stained section (top) and a keratin-immunostained section (bottom) of control (left) and BI 2536-treated (right) SCCIC1 tumors after 2 weeks of treatment. Although no difference in tumor volume between the control and the treated group was seen at this time point, histology shows that the BI 2536-treated tumors consisted of mainly inert material (keratin) without discernable SCCIC1 tumor cells, whereas control tumors contained numerous SCCIC1 cells ( × 100 magnification). (c) Photographs from the experiment shown in panel a of vehicle- (top left, tumor bisected showing both halves delineated by the dashed line) and BI 2536-treated (top right) SCCIC1 tumors. The largest SCCIC1 tumor treated with C20orf20 siRNA in the experiment shown in panel a (bottom right, tumor bisected, showing both halves delineated by a dashed line) is hollow in appearance compared with the scrambled non-targeting control siRNA-treated tumor (bottom left). H&E, hematoxylin and eosin; PLK, Polo-like kinase; siRNA, small interfering RNA.
Discussion
Our approach to target identification has yielded definite therapeutic targets for cSCC in the form of PLK1 and C20orf20 (Figure 5). The discovery that PLK1 is overexpressed and required for survival in cSCC cells mirrors observations in a number of different tumor types (Takai et al., 2005). In agreement with our findings, a recent study has shown PLK1 overexpression in cSCC using immunohistochemical staining of tissue arrays (Schmit et al., 2009a). Here, we show that cSCC keratinocytes undergo established hallmarks of PLK1 inhibition and depletion; mitotic arrest, inhibition of proliferation and apoptosis, and as reported previously, cell death occurs, preferentially in cancer cells compared with normal cells, thus providing a therapeutic window (Liu et al., 2006; Schmit and Ahmad, 2007; Schmit et al., 2009b) (Figure 3). Normal cells require knockdown of p53 in addition to PLK1 to invoke cell death (Liu et al., 2006), and various reports suggest increased sensitivity to PLK1 inhibition when p53 is defective (Guan et al., 2005; Degenhardt and Lampkin, 2010). This is especially pertinent in the case of cSCC as both our data and that of others show the majority of cSCC to harbor TP53 mutation (Supplementary Table S1) (Giglia-Mari and Sarasin, 2003). The potential of PLK1 as a therapeutic target that could be fast-tracked into human trials for cSCC is enhanced by the fact that a number of small-molecule inhibitors are already in clinical development (Schoffski, 2009; Degenhardt and Lampkin, 2010). Among these, the inhibitor BI 2536 has progressed to phase-II trials for both hematological and solid-tumor malignancies (http://www.clinicaltrials.gov), and is shown here to have dramatic efficacy in treating cSCC in vivo (Figure 5). We show that in vitro cSCC keratinocytes are up to 16 times more sensitive to the inhibitor BI 2536 than normal primary keratinocytes. Together, these data suggest that targeting PLK1 has great promise for effective cSCC therapy.
Our observation that knockdown of C20orf20 can induce apoptosis and reduce tumor growth in cSCC highlights this gene, and the histone acetyltransferase complex (TIP60 HAT) with which it associates, as potential targets for therapeutic development. C20orf20 depletion in cSCC resulted in reduced cell viability attributable to induction of apoptosis at levels comparable with PLK1 knockdown, and without effect on cell cycle (Figure 4). This is in contrast to recent data on colorectal cancer cells, which we confirm here showing a reduction in proliferation without engagement of apoptosis (Yamaguchi et al., 2010). In addition, normal keratinocytes, which express this gene at very low levels, remain unaffected by C20orf20 knockdown (Figure 4a).
C20orf20 was initially identified as a protein capable of binding to two components of the TIP60 HAT complex, MRG15 and MRGX (Bertram and Pereira-Smith, 2001; Cai et al., 2003). MRG15 and MRGX are stable components of both HAT and histone deacetylase complexes, and overexpression of C20orf20, which specifically associates with TIP60 HAT, has been shown to increase their protein levels, indicating regulation of stability and/or synthesis (Hayakawa et al., 2007). It has been suggested that C20orf20 influences the acetylation of histones and potentially transcription factors such as p53 (Gu and Roeder, 1997), by controlling the balance of MRG proteins associated with either TIP60 HAT or histone deacetylase complexes (Hayakawa et al., 2007). In the study by Yamaguchi et al. (2010), knockdown of another TIP60 HAT C20orf20-binding partner, BRD8, produced effects similar to C20orf20 depletion. It is possible that the effects of C20orf20 are mediated through interaction with other HAT complex proteins, and it will be necessary to systematically knockdown these components to identify functional partners specific to cSCC. Because of the increased potency seen in cSCC compared with that in colon cancer it is tempting to speculate that C20orf20 is ‘wired' differently in different tumor types, leading to varied responses upon reduced expression. It should be noted that expression at the mRNA level was around two-fold higher in cSCC cells compared with that in HCT116 cells (data not shown), perhaps indicating a greater C20orf20-dependent pro-survival drive in cSCC than in colon carcinoma. It will be important to investigate the mode of action for C20orf20-specific apoptotic induction to clarify its potential as a cancer target.
By identifying both PLK1 and C20orf20 as demonstrable cancer targets we re-enforce the notion that although cultured tumor cells fall short of faithfully replicating the complex nature of human cancers, they are nevertheless invaluable in our goal to understand and ultimately treat this disease (Masters, 2000; Sharma et al., 2010). The majority of arguments against the use of cultured cells to investigate tumor biology are based on the marked differences seen in the expression profiling of cultured cancer cells compared directly with tumor tissue (Perou et al., 1999; Ross et al., 2000; Welsh et al., 2001; Dairkee et al., 2004). Few examples exist where normal cells are included in such analysis and in these cases, expression profiles in culture, as would be expected from disparate environments, cluster separately from tissue (Perou et al., 1999). The use of quiescent, confluent cultures represents a departure from traditional in vitro mRNA expression experiments, which use cultures growing in the log phase, typically considered ‘healthy' (Perou et al., 1999; Welsh et al., 2001). This was prompted by observations that junction complexes can take 48 h to mature in cultured keratinocytes (Wallis et al., 2000; South et al., 2003) and that varying cell–cell adhesion can modulate numerous signaling cascades (Wu and Bonavida, 2009). By using cultured material we have been able to assess gene expression in the absence of a surrounding microenvironment and supported by the cells own matrix. Although it is well documented that this does not reflect the situation in vivo (Weaver et al., 1997; Creighton et al., 2003), it has enabled us to compare tumor with normal in the absence of variation resulting from tumor heterogeneity. In doing so we make the following observations: RDEB cSCC keratinocytes possess expression profiles similar to other cSCC, indicating common initiation and maintenance pathways, and, even after using a quiescent in vitro model, many of the ‘cSCC-specific' genes identified are involved in the cell cycle and proliferation (BUB1, PLK1, CDC25C, WDHD1; Supplementary Table S9), in keeping with features common to all cancers (Hanahan and Weinberg, 2000) and suggesting that dysregulation of the cell cycle is apparent even in the absence of marked proliferation differences. We believe that the strategy described here will be applicable to epithelial cancers where culture of primary cells is well established (Brattain et al., 1981; Rheinwald and Beckett, 1981; Smith et al., 1981; Burns et al., 1993).
In summary, we have used an integrative approach including expression profiling and in vivo assays to identify novel targets in cSCC. We hope this work will lead to the use of PLK1 inhibitors in the treatment of cSCC and to the development of targeted therapies based around the biology of C20orf20.
Materials and methods
All human samples were collected after informed, written consent and in accordance with the Helsinki guidelines. All animals were used in accordance with the UK Home Office regulations.
Keratinocyte isolation and growth
Primary keratinocytes were isolated from fresh tumor or normal specimens and initially grown in the presence of a mitotically inactivated 3T3 feeder layer as described (Rheinwald and Beckett, 1981). Tumor populations were verified by single-nucleotide polymorphism mapping (Purdie et al., 2007) or cytogenetic analysis (Cunningham et al., 2002) as described. Normal keratinocytes were routinely expanded and passaged no more than twice prior to experimental assays in the presence of 3T3 cells. Tumor keratinocytes were initially established with a 3T3 feeder layer and subsequently cultured without. With the exception of cultures used for expression analysis (below), all keratinocytes used in the assays described here were cultured in the absence of 3T3 cells. All keratinocytes were cultured in ‘keratinocyte media' containing 10% serum and growth factors as described (Rheinwald and Beckett, 1981). For expression analysis keratinocytes were seeded in the presence of a 3T3 feeder layer and grown to confluence. One day prior to confluence any remaining adherent 3T3 cells were removed using versene (Invitrogen, Paisley, UK) or a weak solution of trypsin/versene (Invitrogen).
In vivo tumor growth and treatment
For tumorigenicity assays a suspension of 1–4 × 106 tumor cells was mixed with high-concentration Matrigel (Becton Dickinson, Oxford, UK) and injected subcutaneously into the flanks of SCID Balb/c mice. For tumor treatment 4 × 106 SCCIC1 cells were used. Tumors were measured by a caliper and treatment began when volume reached 100 mm3.
Proliferation and cell viability assays
Colorimetric assays of mitochondrial dehydrogenase activity were used to measure growth curves and cell viability. The initial proliferation rates of primary cultures were determined using Cell Proliferation Kit-I (MTT) (Roche, Burgess Hill, UK) according to the manufacturer's instructions. All subsequent proliferation/viability assays were performed using the MTS CellTitre 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) according to the manufacturer's instructions. In each case all cells were seeded into replica 96-well plates and readings were taken at intervals up to a maximum of 96 h. An increase in mitochondrial dehydrogenase activity using this assay directly correlated with cell number determined using a CASY Model TT cell counter (Roche Diagnostics Ltd, West Sussex, UK) (data not shown).
Gene expression and analysis
According to the manufacturer's instructions, total RNA was extracted from cells (passage <7) or frozen tissue sections using the RNeasy kit (Qiagen, Crawley, UK) and hybridized to Human Hybridize 6-Sample BeadChips (whole-genome gene expression for BeadStation) (Illumina, San Diego, CA, USA). RNA was isolated from keratinocyte cultures 2 days after confluence. V1 arrays were used for the cell culture analysis and V2 arrays were used for the tissue analysis. Cubic-spline-normalized signal intensities for each probe were determined using Illumina's BeadStudio Data Analysis Software. Data were analyzed by Student's t-test. In our initial in vitro experiment, probes were scored that met the following three criteria: (a) P<0.001, average fold change (FC) >2.5, with expression above the signal intensity detection threshold of 10; (b) P<0.001, FC>1.5, with intermediate expression (signal intensity >50); (C) P<0.005, FC>2.5, with high expression (signal intensity >100).
For all other array analysis, average FC and Student's t-test P-values were generated for all probes representing each of the 435 in vitro cSCC genes where present on each of the arrays described using normalized signal intensities either generated in BeadStudio (our own experiment) or available at the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/ (Edgar et al., 2002)).
RNA interference screen
We used the top three siRNA oligonucleotides per gene as ranked by Sigma-Aldrich (Dorset, UK). In each experiment a negative control (MISSION siRNA Universal Negative Control #1; Sigma-Aldrich) and a positive control (AllStars Hs Cell Death Control siRNA; Qiagen) were used. Cells were seeded in 96-well plates at 5000 cells/well in 100 μl of keratinocyte media and transfected 24 h later with siRNA (40 n final concentration) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) diluted in Opti-MEM (Invitrogen) according to the manufacturer's instructions. Cell viability was assessed at 48, 72 and 96 h after transfection, and a reading was taken at time 0 (before transfection), using the MTS CellTitre 96 AQueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer's instructions.
PLK1 and C20orf20 siRNA and PLK1 inhibition
For RNA interference transfection, cells were seeded in six-well plates at 2.5 × 105 cells/well and 24 h later transfected as described above. For each experiment a pool of three individual siRNA oligonucleotides was used. Cells were left for 16 h then trypsinized, counted on a CASYModel TT counter (Roche Diagnostics Ltd) and seeded in 96-well plates at 3000 cells/well in 100 μl of keratinocyte media. Cell viability was determined using the MTS assay as described. For small-molecule inhibitor treatment, 50 μl of cells were seeded in 96-well plates at 3000 cells/well, incubated at 37 °C for 2 h and then 50 μl of media containing two-times-concentrated drug added to produce the stated final concentrations. The effective concentrations at which cellular growth was inhibited by 50% (EC50) after 48 h of treatment were extrapolated by the five-parameter logistic model using the curve fitting program Masterplex ReaderFit 2010 (MiraiBio Group, Hitachi Solutions America, Ltd, San Francisco, CA, USA).
Apoptosis detection
Apoptosis was detected using the Cell Death Detection ELISAPLUS (Roche Diagnostics Ltd). Cells were seeded in 24-well plates at 0.5 × 105 cells/well for 24 h and either transfected with siRNA (three individual oligonucleotides pooled) or treated with small-molecule inhibitors for 24 or 16 h, respectively, as described, before collecting lysates and performing ELISA according to the manufacturer's instructions.
Antibodies and chemicals
The antibodies and chemicals used in this study were the following: β-Actin (mAbcam 8226; Abcam, Cambridge, UK); PLK1 (#208G4; Cell Signaling Technology Inc, Danvers, MA, USA); BI 2536 (Selleck Chemicals LLC, Houston, TX, USA), GW843682X (Sigma-Aldrich); anti-BrdU antibody (Becton Dickinson).
Protein quantification
The amount of PLK1 protein was quantified from western blot experiments using Image J (http://rsb.info.nih.gov/ij/) by calculating an integrated density value through multiplication of band area and gray intensity. This value was divided by the corresponding value for the actin loading control to produce a relative intensity.
Cell-cycle analysis
5-Bromodeoxyuridine (BrdU; Sigma-Aldrich) was added at 30 μ final concentration for 20 min. Cells were collected and fixed by dropping a 1-ml cell suspension in phosphate-buffered saline into 3 ml ice-cold ethanol while vortexing. Pepsin (Sigma) was added at 1 mg/ml in 30 m HCL for 30 min and DNA was denatured with 2 N HCL for 20 min. An anti-BrdU antibody diluted in phosphate-buffered saline/0.5% Tween/0.5% bovine serum albumin was added for 1 h followed by 30-min incubation with a fluorescein isothiocyanate-sheep anti-mouse IgG (Sigma). Propidium iodide (Sigma) was added in the final wash step at a concentration of 25 μg/ml and samples were analyzed using a FACScan flow cytometer and the CellQuest software (Becton Dickinson).
Real-time quantitative PCR
A 5-μg weight of RNA was incubated with random primers and M-MLV reverse transcriptase (Promega) to generate cDNA. For quantitative measurement of C20orf20 mRNA, the SYBR Green Master Mix (Qiagen) was used with the following primers: 5′-ATTCTTCCATTCCCGAATCC-3′ and 5′-CCCAAACTCCCTGAAGATGA-3′. Actin primers (5′-CCTTGCACATGCCGGAG-3′ and 5′-GCACAGAGCCTCGCCTT-3′) were used as internal control. The QIAgility (Qiagen) automated PCR workstation was used to set up PCR samples; reactions were performed using the Rotor-Gene Q (Qiagen) and expression was calculated by the ΔΔCT method (Livak and Schmittgen, 2001).
Acknowledgments
We thank David Kelsell, Edel O'Toole, Ian MacKenzie and Alan Storey for help in securing funding. We thank David Meek for critically reading the manuscript. Clustering dendrogram analyses were performed using BRB-ArrayTools developed by Dr Richard Simon and the BRB-ArrayTools Development Team. We thank Dr Andreas Volz for assistance with cell culture and Professor Bryan Young and Tracy Chaplin for help with single-nucleotide polymorphism analysis. We thank the Tayside Tissue Bank for RNA isolation.
Funding: This work was funded by DebRA, the dystrophic epidermolysis bullosa research association (http://www.debra.org.uk/).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Supplementary Material
References
- Bertram MJ, Pereira-Smith OM. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–121. doi: 10.1016/s0378-1119(01)00372-9. [DOI] [PubMed] [Google Scholar]
- Braakhuis BJ, Brakenhoff RH, Leemans CR. Gene expression profiling in head and neck squamous cell carcinoma. Curr Opin Otolaryngol Head Neck Surg. 2010;18:67–71. doi: 10.1097/MOO.0b013e32833693ce. [DOI] [PubMed] [Google Scholar]
- Brattain MG, Brattain DE, Fine WD, Khaled FM, Marks ME, Kimball PM, et al. Initiation and characterization of cultures of human colonic carcinoma with different biological characteristics utilizing feeder layers of confluent fibroblasts. Oncodev Biol Med. 1981;2:355–366. [PubMed] [Google Scholar]
- Burns JE, Baird MC, Clark LJ, Burns PA, Edington K, Chapman C, et al. Gene mutations and increased levels of p53 protein in human squamous cell carcinomas and their cell lines. Br J Cancer. 1993;67:1274–1284. doi: 10.1038/bjc.1993.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, et al. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- Cheng H, Force T. Molecular mechanisms of cardiovascular toxicity of targeted cancer therapeutics. Circ Res. 2010;106:21–34. doi: 10.1161/CIRCRESAHA.109.206920. [DOI] [PubMed] [Google Scholar]
- Creighton C, Kuick R, Misek DE, Rickman DS, Brichory FM, Rouillard JM, et al. Profiling of pathway-specific changes in gene expression following growth of human cancer cell lines transplanted into mice. Genome Biol. 2003;4:R46. doi: 10.1186/gb-2003-4-7-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham J, Sales M, Pearce A, Howard J, Stallings R, Telford N, et al. Does isochromosome 7q mandate bone marrow transplant in children with Shwachman–Diamond syndrome. Br J Haematol. 2002;119:1062–1069. doi: 10.1046/j.1365-2141.2002.03940.x. [DOI] [PubMed] [Google Scholar]
- Dairkee SH, Ji Y, Ben Y, Moore DH, Meng Z, Jeffrey SS. A molecular ‘signature' of primary breast cancer cultures; patterns resembling tumor tissue. BMC Genomics. 2004;5:47. doi: 10.1186/1471-2164-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt Y, Lampkin T. Targeting Polo-like kinase in cancer therapy. Clin Cancer Res. 2010;16:384–389. doi: 10.1158/1078-0432.CCR-09-1380. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E., Sr Metastases of sun-induced SCC. J Dermatol Surg Oncol. 1984;10:418. [PubMed] [Google Scholar]
- Euvrard S, Kanitakis J, Claudy A. Skin cancers after organ transplantation. N Engl J Med. 2003;348:1681–1691. doi: 10.1056/NEJMra022137. [DOI] [PubMed] [Google Scholar]
- Fine JD, Johnson LB, Weiner M, Li KP, Suchindran C. Epidermolysis bullosa and the risk of life-threatening cancers: the National EB Registry experience, 1986–2006. J Am Acad Dermatol. 2009;60:203–211. doi: 10.1016/j.jaad.2008.09.035. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G, Sarasin A. TP53 mutations in human skin cancers. Hum Mutat. 2003;21:217–228. doi: 10.1002/humu.10179. [DOI] [PubMed] [Google Scholar]
- Gleixner KV, Ferenc V, Peter B, Gruze A, Meyer RA, Hadzijusufovic E, et al. Polo-like kinase 1 (Plk1) as a novel drug target in chronic myeloid leukemia: overriding imatinib resistance with the Plk1 inhibitor BI 2536. Cancer Res. 2010;70:1513–1523. doi: 10.1158/0008-5472.CAN-09-2181. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Guan R, Tapang P, Leverson JD, Albert D, Giranda VL, Luo Y. Small interfering RNA-mediated Polo-like kinase 1 depletion preferentially reduces the survival of p53-defective, oncogenic transformed cells and inhibits tumor growth in animals. Cancer Res. 2005;65:2698–2704. doi: 10.1158/0008-5472.CAN-04-2131. [DOI] [PubMed] [Google Scholar]
- Haider AS, Peters SB, Kaporis H, Cardinale I, Fei J, Ott J, et al. Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J Invest Dermatol. 2006;126:869–881. doi: 10.1038/sj.jid.5700157. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, et al. RBP2 is an MRG15 complex component and downregulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Lei M, Erikson RL. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol Cell Biol. 2006;26:2093–2108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Masters JR. Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol. 2000;1:233–236. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- Nindl I, Dang C, Forschner T, Kuban RJ, Meyer T, Sterry W, et al. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol Cancer. 2006;5:30. doi: 10.1186/1476-4598-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- Purdie KJ, Harwood CA, Gulati A, Chaplin T, Lambert SR, Cerio R, et al. Single nucleotide polymorphism array analysis defines a specific genetic fingerprint for well-differentiated cutaneous SCCs. J Invest Dermatol. 2009;129:1562–1568. doi: 10.1038/jid.2008.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdie KJ, Lambert SR, Teh MT, Chaplin T, Molloy G, Raghavan M, et al. Allelic imbalances and microdeletions affecting the PTPRD gene in cutaneous squamous cell carcinomas detected using single nucleotide polymorphism microarray analysis. Genes Chromosomes Cancer. 2007;46:661–669. doi: 10.1002/gcc.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowska M, Reilly L, Palmer CN, Gustafsson MC, Foerster J. Activation of PPARbeta/delta causes a psoriasis-like skin disease in vivo. PLoS One. 2010;5:e9701. doi: 10.1371/journal.pone.0009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Schmit TL, Ahmad N. Regulation of mitosis via mitotic kinases: new opportunities for cancer management. Mol Cancer Ther. 2007;6:1920–1931. doi: 10.1158/1535-7163.MCT-06-0781. [DOI] [PubMed] [Google Scholar]
- Schmit TL, Zhong W, Nihal M, Ahmad N. Polo-like kinase 1 (Plk1) in non-melanoma skin cancers. Cell Cycle. 2009a;8:2697–2702. doi: 10.4161/cc.8.17.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit TL, Zhong W, Setaluri V, Spiegelman VS, Ahmad N. Targeted depletion of Polo-like kinase (Plk) 1 through lentiviral shRNA or a small-molecule inhibitor causes mitotic catastrophe and induction of apoptosis in human melanoma cells. J Invest Dermatol. 2009b;129:2843–2853. doi: 10.1038/jid.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffski P. Polo-like kinase (PLK) inhibitors in preclinical and early clinical development in oncology. Oncologist. 2009;14:559–570. doi: 10.1634/theoncologist.2009-0010. [DOI] [PubMed] [Google Scholar]
- Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- Smith HS, Lan S, Ceriani R, Hackett AJ, Stampfer MR. Clonal proliferation of cultured nonmalignant and malignant human breast epithelia. Cancer Res. 1981;41:4637–4643. [PubMed] [Google Scholar]
- South AP, Wan H, Stone MG, Dopping-Hepenstal PJ, Purkis PE, Marshall JF, et al. Lack of plakophilin 1 increases keratinocyte migration and reduces desmosome stability. J Cell Sci. 2003;116:3303–3314. doi: 10.1242/jcs.00636. [DOI] [PubMed] [Google Scholar]
- Staples MP, Elwood M, Burton RC, Williams JL, Marks R, Giles GG. Non-melanoma skin cancer in Australia: the 2002 national survey and trends since 1985. Med J Aust. 2006;184:6–10. doi: 10.5694/j.1326-5377.2006.tb00086.x. [DOI] [PubMed] [Google Scholar]
- Suiqing C, Min Z, Lirong C. Overexpression of phosphorylated-STAT3 correlated with the invasion and metastasis of cutaneous squamous cell carcinoma. J Dermatol. 2005;32:354–360. doi: 10.1111/j.1346-8138.2005.tb00906.x. [DOI] [PubMed] [Google Scholar]
- Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24:287–291. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- Van Haren R, Feldman D, Sinha AA. Systematic comparison of nonmelanoma skin cancer microarray datasets reveals lack of consensus genes. Br J Dermatol. 2009;161:1278–1287. doi: 10.1111/j.1365-2133.2009.09338.x. [DOI] [PubMed] [Google Scholar]
- Veness MJ, Porceddu S, Palme CE, Morgan GJ. Cutaneous head and neck squamous cell carcinoma metastatic to parotid and cervical lymph nodes. Head Neck. 2007;29:621–631. doi: 10.1002/hed.20576. [DOI] [PubMed] [Google Scholar]
- Wallis S, Lloyd S, Wise I, Ireland G, Fleming TP, Garrod D. The alpha isoform of protein kinase C is involved in signaling the response of desmosomes to wounding in cultured epithelial cells. Mol Biol Cell. 2000;11:1077–1092. doi: 10.1091/mbc.11.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, et al. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- Wu K, Bonavida B. The activated NF-kappaB–Snail–RKIP circuitry in cancer regulates both the metastatic cascade and resistance to apoptosis by cytotoxic drugs. Crit Rev Immunol. 2009;29:241–254. doi: 10.1615/critrevimmunol.v29.i3.40. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Sakai M, Shimokawa T, Yamada Y, Nakamura Y, Furukawa Y. C20orf20 (MRG-binding protein) as a potential therapeutic target for colorectal cancer. Br J Cancer. 2010;102:325–331. doi: 10.1038/sj.bjc.6605500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.