Abstract
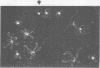
DNA molecules isolated from tailless phage particles (capsids) of bacteriophage P4 virl del10 are known to be knotted. We have found by electron microscopy that 80% of DNA molecules isolated from intact phage particles of P4 virl del10 also contained knots. This observation indicates that the predominant form of P4 virl del10 DNA within the intact phage particle is either knotted or in a configuration that permits knotting upon isolation. In comparison to P4 virl del10 (deleted 1000 basepairs), DNA molecules isolated from intact P4 virl del2 (deleted 650 basepairs) and P4 virl (non-deleted) contained 50% and 15% knots respectively, showing an association of decreased size of deletion of DNA with a decreased fraction of knotted genomes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashikawa I., Furuno T., Kinosita K., Jr, Ikegami A., Takahashi H., Akutsu H. Internal motion of DNA in bacteriophages. J Biol Chem. 1984 Jul 10;259(13):8338–8344. [PubMed] [Google Scholar]
- BERTANI L. E. LYSOGENIC CONVERSION BY BACTERIOPHAGE P2 RESULTING IN AN INCREASED SENSITIVITY OF ESCHERICHIA COLI TO 5-FLUORODEOXYURIDINE. Biochim Biophys Acta. 1964 Aug 12;87:631–640. doi: 10.1016/0926-6550(64)90281-6. [DOI] [PubMed] [Google Scholar]
- Calendar R., Ljungquist E., Deho G., Usher D. C., Goldstein R., Youderian P., Sironi G., Six E. W. Lysogenization by satellite phage P4. Virology. 1981 Aug;113(1):20–38. doi: 10.1016/0042-6822(81)90133-1. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Inman R. B. Location of DNA ends in P2, 186, P4 and lambda bacteriophage heads. J Mol Biol. 1974 Jul 25;87(1):11–22. doi: 10.1016/0022-2836(74)90556-7. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Harrison S. C. DNA arrangement in isometric phage heads. Nature. 1977 Aug 18;268(5621):598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., King J., Harrison S. C., Eiserling F. A. The structural organization of DNA packaged within the heads of T4 wild-type, isometric and giant bacteriophages. Cell. 1978 Jul;14(3):559–568. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- Earnshaw W., Casjens S., Harrison S. C. Assembly of the head of bacteriophage P22: x-ray diffraction from heads, proheads and related structures. J Mol Biol. 1976 Jun 25;104(2):387–410. doi: 10.1016/0022-2836(76)90278-3. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Goldstein R., Sedivy J., Ljungquist E. Propagation of satellite phage P4 as a plasmid. Proc Natl Acad Sci U S A. 1982 Jan;79(2):515–519. doi: 10.1073/pnas.79.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas R., Murphy R. F., Cantor C. R. Testing models of the arrangement of DNA inside bacteriophage lambda by crosslinking the packaged DNA. J Mol Biol. 1982 Jul 25;159(1):71–92. doi: 10.1016/0022-2836(82)90032-8. [DOI] [PubMed] [Google Scholar]
- Hall S. B., Schellman J. A. Flow dichroism of capsid DNA phages. I. Fast and slow T4B. Biopolymers. 1982 Oct;21(10):1991–2010. doi: 10.1002/bip.360211006. [DOI] [PubMed] [Google Scholar]
- Hall S. B., Schellman J. A. Flow dichroism of capsid DNA phages. II. Effect of DNA deletions and intercalating dyes. Biopolymers. 1982 Oct;21(10):2011–2031. doi: 10.1002/bip.360211007. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. Packaging of DNA into bacteriophage heads: a model. J Mol Biol. 1983 Dec 25;171(4):577–580. doi: 10.1016/0022-2836(83)90045-1. [DOI] [PubMed] [Google Scholar]
- Hooper D. C., Wolfson J. S., McHugh G. L., Winters M. B., Swartz M. N. Effects of novobiocin, coumermycin A1, clorobiocin, and their analogs on Escherichia coli DNA gyrase and bacterial growth. Antimicrob Agents Chemother. 1982 Oct;22(4):662–671. doi: 10.1128/aac.22.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman R. B., Schnös M., Simon L. D., Six E. W., Walker D. H., Jr Some morphological properties of P4 bacteriophage and P4 DNA. Virology. 1971 Apr;44(1):67–72. doi: 10.1016/0042-6822(71)90153-x. [DOI] [PubMed] [Google Scholar]
- Kahn M., Hopkins A. Restriction endonuclease cleavage map of bacteriophage P4 DNA. Virology. 1978 Apr;85(2):359–363. doi: 10.1016/0042-6822(78)90444-0. [DOI] [PubMed] [Google Scholar]
- Kahn M., Ow D., Sauer B., Rabinowitz A., Calendar R. Genetic analysis of bacteriophage P4 using P4-plasmid ColE1 hybrids. Mol Gen Genet. 1980 Feb;177(3):399–412. doi: 10.1007/BF00271478. [DOI] [PubMed] [Google Scholar]
- Lagos R., Goldstein R. Phasmid P4: manipulation of plasmid copy number and induction from the integrated state. J Bacteriol. 1984 Apr;158(1):208–215. doi: 10.1128/jb.158.1.208-215.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. S. Nucleotide sequence of the essential region of bacteriophage P4. Nucleic Acids Res. 1984 Nov 26;12(22):8667–8684. doi: 10.1093/nar/12.22.8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Davis J. L., Calendar R. Novel topologically knotted DNA from bacteriophage P4 capsids: studies with DNA topoisomerases. Nucleic Acids Res. 1981 Aug 25;9(16):3979–3989. doi: 10.1093/nar/9.16.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Perkocha L., Calendar R., Wang J. C. Knotted DNA from bacteriophage capsids. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5498–5502. doi: 10.1073/pnas.78.9.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménissier J., de Murcia G., Lebeurier G., Hirth L. Electron microscopic studies of the different topological forms of the cauliflower mosaic virus DNA: knotted encapsidated DNA and nuclear minichromosome. EMBO J. 1983;2(7):1067–1071. doi: 10.1002/j.1460-2075.1983.tb01547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan R., Wu R., Bode V. C. Arrangement of DNA in lambda bacteriophage heads. 3. Location and number of nucleotides cleaved from lambda-DNA by micrococcal nuclease attack on heads. J Mol Biol. 1972 Aug 21;69(2):201–207. doi: 10.1016/0022-2836(72)90225-2. [DOI] [PubMed] [Google Scholar]
- Raimondi A., Donghi R., Montaguti A., Pessina A., Dehò G. Analysis of spontaneous deletion mutants of satellite bacteriophage P4. J Virol. 1985 Apr;54(1):233–235. doi: 10.1128/jvi.54.1.233-235.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards K. E., Williams R. C., Calendar R. Mode of DNA packing within bacteriophage heads. J Mol Biol. 1973 Aug 5;78(2):255–259. doi: 10.1016/0022-2836(73)90114-9. [DOI] [PubMed] [Google Scholar]
- Saigo K., Uchida H. Connection of the right-hand terminus of DNA to the proximal end of the tail in bacteriophage lambda. Virology. 1974 Oct;61(2):524–536. doi: 10.1016/0042-6822(74)90287-6. [DOI] [PubMed] [Google Scholar]
- Shore D., Dehò G., Tsipis J., Goldstein R. Determination of capsid size by satellite bacteriophage P4. Proc Natl Acad Sci U S A. 1978 Jan;75(1):400–404. doi: 10.1073/pnas.75.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six E. W., Klug C. A. Bacteriophage P4: a satellite virus depending on a helper such as prophage P2. Virology. 1973 Feb;51(2):327–344. doi: 10.1016/0042-6822(73)90432-7. [DOI] [PubMed] [Google Scholar]
- Six E. W. The helper dependence of satellite bacteriophage P4: which gene functions of bacteriophage P2 are needed by P4? Virology. 1975 Sep;67(1):249–263. doi: 10.1016/0042-6822(75)90422-5. [DOI] [PubMed] [Google Scholar]
- Souza L., Geisselsoder J., Hopkins A., Calender R. Physical mapping of the satellite phage P4 genome. Virology. 1978 Apr;85(2):335–342. doi: 10.1016/0042-6822(78)90442-7. [DOI] [PubMed] [Google Scholar]
- Thomas J. O. Chemical linkage of the tail to the right-hand end of bacteriophage lambda DNA. J Mol Biol. 1974 Jul 25;87(1):1–9. doi: 10.1016/0022-2836(74)90555-5. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Sternberg N., Weisberg R. Altered arrangement of the DNA in injection-defective lambda bacteriophage. J Mol Biol. 1978 Aug 5;123(2):149–161. doi: 10.1016/0022-2836(78)90318-2. [DOI] [PubMed] [Google Scholar]
- Volovitch M., Drugeon C., Yot P. Studies on the single-stranded discontinuities of the cauliflower mosaic virus genome. Nucleic Acids Res. 1978 Aug;5(8):2913–2925. doi: 10.1093/nar/5.8.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Martin K. V., Calendar R. On the sequence similarity of the cohesive ends of coliphage P4, P2, and 186 deoxyribonucleic acid. Biochemistry. 1973 May 22;12(11):2119–2123. doi: 10.1021/bi00735a016. [DOI] [PubMed] [Google Scholar]
- Widom J., Baldwin R. L. Tests of spool models for DNA packaging in phage lambda. J Mol Biol. 1983 Dec 25;171(4):419–437. doi: 10.1016/0022-2836(83)90038-4. [DOI] [PubMed] [Google Scholar]
- Wiman M., Bertani G., Kelly B., Sasaki I. Genetic map of Escherichia coli strain C. Mol Gen Genet. 1970;107(1):1–31. doi: 10.1007/BF00433220. [DOI] [PubMed] [Google Scholar]