Abstract
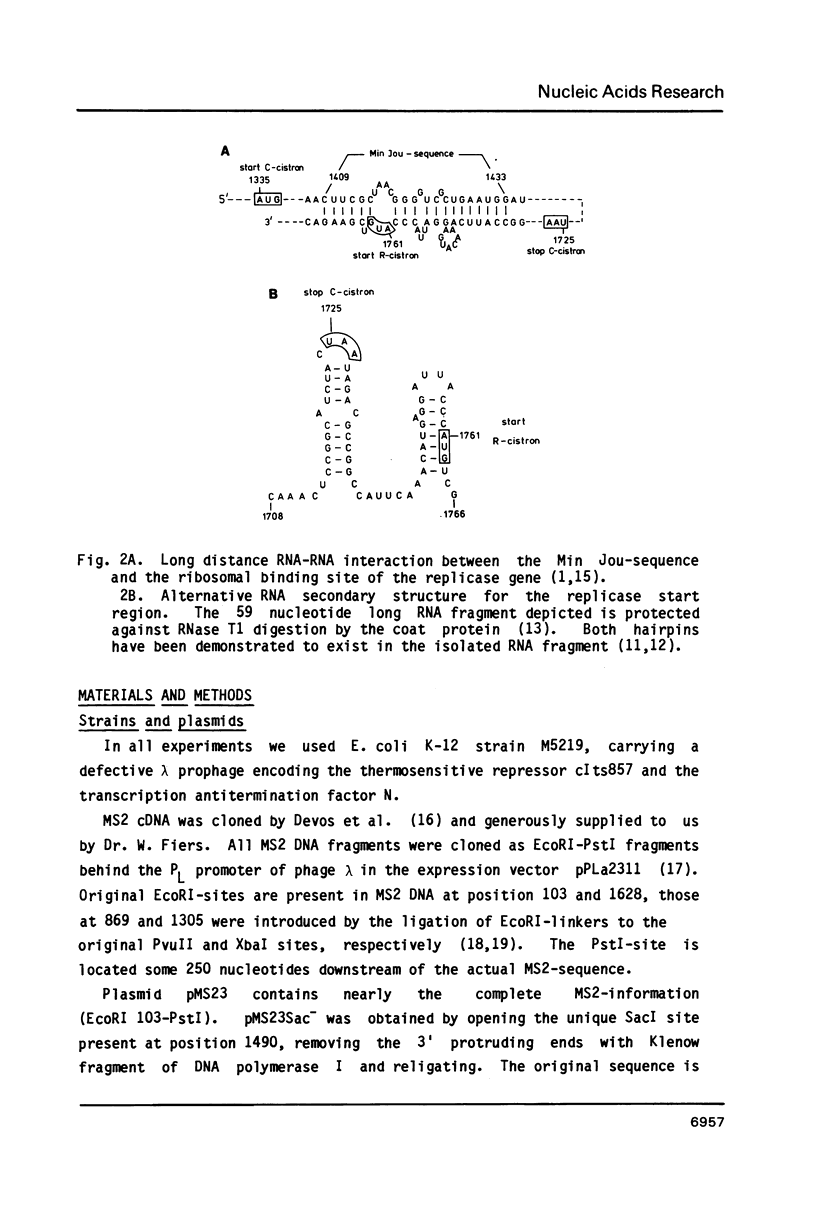
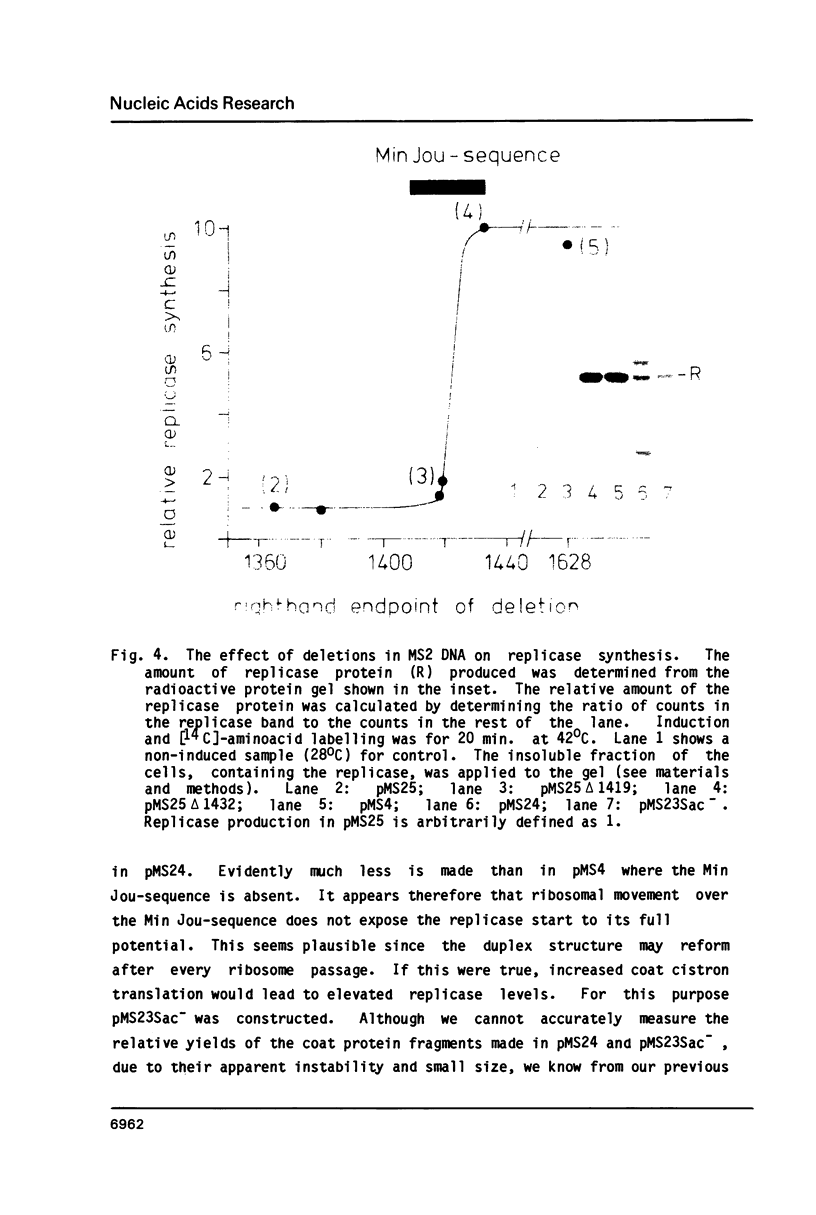
We have analyzed the molecular mechanism that makes translation of the MS2 replicase cistron dependent on the translation of the upstream coat cistron. Deletion mapping on cloned cDNA of the phage shows that the ribosomal binding site of the replicase cistron is masked by a long distance basepairing to an internal coat cistron region. Removal of the internal coat cistron region leads to uncoupled replicase synthesis. Our results confirm the model as originally proposed by Min Jou et al. (1). Activation of the replicase start is sensitive to the frequency of upstream translation, but never reaches the level of uncoupled replicase synthesis.
Full text
PDF










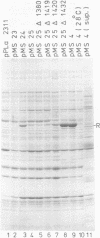
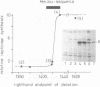
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksoy S., Squires C. L., Squires C. Translational coupling of the trpB and trpA genes in the Escherichia coli tryptophan operon. J Bacteriol. 1984 Feb;157(2):363–367. doi: 10.1128/jb.157.2.363-367.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Urbanowski J., Weissbach H., Nestor J., Yanofsky C. In vitro synthesis of the tryptophan operon leader peptides of Escherichia coli, Serratia marcescens, and Salmonella typhimurium. Proc Natl Acad Sci U S A. 1983 May;80(10):2879–2883. doi: 10.1073/pnas.80.10.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Yanofsky C. A ribosome binding site sequence is necessary for efficient expression of the distal gene of a translationally-coupled gene pair. Nucleic Acids Res. 1984 Jun 11;12(11):4757–4768. doi: 10.1093/nar/12.11.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos R., van Emmelo J., Contreras R., Fiers W. Construction and characterization of a plasmid containing a nearly full-size DNA copy of bacteriophage MS2 RNA. J Mol Biol. 1979 Mar 15;128(4):595–619. doi: 10.1016/0022-2836(79)90295-x. [DOI] [PubMed] [Google Scholar]
- Eggen K., Nathans D. Regulation of protein synthesis directed by coliphage MS2 RNA. II. In vitro repression by phage coat protein. J Mol Biol. 1969 Jan;39(2):293–305. doi: 10.1016/0022-2836(69)90318-0. [DOI] [PubMed] [Google Scholar]
- Fedoroff N. V., Zinder N. D. Structure of the poly(G) polymerase component of the bacteriophage f2 replicase. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1838–1843. doi: 10.1073/pnas.68.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976 Apr 8;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Gesteland R. F., Spahr P. F. Translation of R-17 RNA fragments. Cold Spring Harb Symp Quant Biol. 1969;34:707–716. doi: 10.1101/sqb.1969.034.01.080. [DOI] [PubMed] [Google Scholar]
- Gralla J., Steitz J. A., Crothers D. M. Direct physical evidence for secondary structure in an isolated fragment of R17 bacteriophage mRNA. Nature. 1974 Mar 15;248(445):204–208. doi: 10.1038/248204a0. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Shulman R. G., Yamane T., Steitz J. A. High resolution proton NMR study of an isolated fragment of R17 bacteriophage mRNA. Nature. 1974 Mar 15;248(445):225–226. doi: 10.1038/248225a0. [DOI] [PubMed] [Google Scholar]
- Jacobson A. B., Kumar H., Zuker M. Effect of spermidine on the conformation of bacteriophage MS2 RNA. Electron microscopy and computer modeling. J Mol Biol. 1985 Feb 20;181(4):517–531. doi: 10.1016/0022-2836(85)90424-3. [DOI] [PubMed] [Google Scholar]
- Jacobson A. B., Spahr P. F. Studies on the secondary structure of single-stranded RNA from the bacteriophage MS2. II Analysis of the RNase IV cleavage products. J Mol Biol. 1977 Sep 25;115(3):279–294. doi: 10.1016/0022-2836(77)90155-3. [DOI] [PubMed] [Google Scholar]
- Jansone I., Berzin V., Gribanov V., Gren E. J. The regulatory region of MS2 phage RNA replicase cistron. III. Characterization of fragments resulting from S1 nuclease digestion. Nucleic Acids Res. 1979;6(5):1747–1760. doi: 10.1093/nar/6.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastelein R. A., Berkhout B., Overbeek G. P., van Duin J. Effect of the sequences upstream from the ribosome-binding site on the yield of protein from the cloned gene for phage MS2 coat protein. Gene. 1983 Sep;23(3):245–254. doi: 10.1016/0378-1119(83)90015-x. [DOI] [PubMed] [Google Scholar]
- Keller W. The RNA lariat: a new ring to the splicing of mRNA precursors. Cell. 1984 Dec;39(3 Pt 2):423–425. doi: 10.1016/0092-8674(84)90449-5. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D., Weissmann C. Possible mechanism for transition of viral RNA from polysome to replication complex. Nat New Biol. 1971 May 12;231(19):42–46. doi: 10.1038/newbio231042a0. [DOI] [PubMed] [Google Scholar]
- Kuroda M. I., Yanofsky C. Evidence for the transcript secondary structures predicted to regulate transcription attenuation in the trp operon. J Biol Chem. 1984 Oct 25;259(20):12838–12843. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lodish H. F. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein biosynthesis. J Mol Biol. 1970 Jun 28;50(3):689–702. doi: 10.1016/0022-2836(70)90093-8. [DOI] [PubMed] [Google Scholar]
- Lodish H. F., Zinder N. D. Mutants of the bacteriophage f2. 8. Control mechanisms for phage-specific syntheses. J Mol Biol. 1966 Aug;19(2):333–348. doi: 10.1016/s0022-2836(66)80008-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Min Jou W., Haegeman G., Ysebaert M., Fiers W. Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein. Nature. 1972 May 12;237(5350):82–88. doi: 10.1038/237082a0. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Chou M. Y., Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U S A. 1984 Apr;81(7):1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Oppenheim D. S., Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980 Aug;95(4):785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Remaut E., Waele P. D., Marmenout A., Stanssens P., Fiers W. Functional expression of individual plasmid-coded RNA bacteriophage MS2 genes. EMBO J. 1982;1(2):205–209. doi: 10.1002/j.1460-2075.1982.tb01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Pleij C. W., Bosch L. Three-dimensional models of the tRNA-like 3' termini of some plant viral RNAs. EMBO J. 1983;2(7):1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D., Lodish H. F. Messenger characteristics of nascent bacteriophage RNA. Proc Natl Acad Sci U S A. 1970 Oct;67(2):710–716. doi: 10.1073/pnas.67.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H., Webster R. E., Zinder N. D. Bacteriophage coat protein as repressor. Nature. 1968 May 11;218(5141):533–536. doi: 10.1038/218533a0. [DOI] [PubMed] [Google Scholar]
- Saito H., Richardson C. C. Processing of mRNA by ribonuclease III regulates expression of gene 1.2 of bacteriophage T7. Cell. 1981 Dec;27(3 Pt 2):533–542. doi: 10.1016/0092-8674(81)90395-0. [DOI] [PubMed] [Google Scholar]
- Schümperli D., McKenney K., Sobieski D. A., Rosenberg M. Translational coupling at an intercistronic boundary of the Escherichia coli galactose operon. Cell. 1982 Oct;30(3):865–871. doi: 10.1016/0092-8674(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Translational control of IS10 transposition. Cell. 1983 Sep;34(2):683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Nakada D. Control of translation of MS2 RNA cistrons by MS2 coat protein. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1744–1750. doi: 10.1073/pnas.57.6.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Weber K. Isolation and characterization of amber mutants of bacteriophage R17. J Mol Biol. 1967 Sep 14;28(2):311–330. doi: 10.1016/s0022-2836(67)80012-3. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Nomura M. Feedback regulation of ribosomal protein synthesis in E. coli: localization of the mRNA target sites for repressor action of ribosomal protein L1. Cell. 1981 Apr;24(1):243–249. doi: 10.1016/0092-8674(81)90520-1. [DOI] [PubMed] [Google Scholar]