Abstract
Animal fertilization is governed by the interaction (binding) of proteins on the surfaces of sperm and egg. In many examples presented herein, fertilization proteins evolve rapidly and show the signature of positive selection (adaptive evolution). This review describes the molecular evolution of fertilization proteins in sea urchins, abalone, and oysters, animals with external fertilization that broadcast their gametes into seawater. Theories regarding the selective forces responsible for the rapid evolution driven by positive selection seen in many fertilization proteins are discussed. This strong selection acting on divergence of interacting fertilization proteins might lead to prezygotic reproductive isolation and be a significant factor in the speciation process.
Since only a fraction of all eggs are fertilized and only an infinitesimal fraction of male gametes succeed in fertilizing an egg, gametes are obviously a category of entities subjected to intense selection. It is curious that this is never mentioned in the literature dealing with selection, perhaps because we know so little about fitness differences among gametes.
(ErnstMayr, 1997)
Gamete recognition systems in marine invertebrates evolve rapidly and show positive selection. This phenomenon could be involved in speciation.
Animal fertilization remains one of the least understood fundamental biological processes. Through fertilization, generations are bridged by fusion of two haploid cells. Fusion restores the diploid genome and sets the zygote on the path of embryogenesis. There are five events in sperm-egg interaction; not all can be shown in every species. These are chemotaxis of sperm to egg, attachment of sperm to the egg envelope, induction of the sperm acrosome reaction, penetration of sperm through the egg envelope, and fusion of gamete plasma membranes (Vacquier 1998). Forty years ago, researchers began to identify and isolate proteins mediating fertilization. The thinking then was that fertilization was such an important process that “fertilization proteins” would be conserved as a distinct family of genes common among animals. Today, the converse is the reality; proteins involved in all phases of sexual reproduction generally evolve rapidly and show positive Darwinian selection (adaptive evolution), meaning there is adaptive value in altering their sequences. By all phases of sexual reproduction we mean genes encoding protein transcription factors acting on reproductive genes (Wu and Su 2008), female and male genital secretions (Swanson et al. 2001), pregnancy (Xie et al. 1997; Chuong et al. 2010), and proteins on sperm and egg surfaces (Swanson and Vacquier 2002; Dorus et al. 2010).
The evolution of fertilization proteins has an extensive literature (reviewed in Swanson and Vacquier 2002; Clark et al. 2006; Turner and Hoekstra 2008; Turner et al. 2008; Palumbi 2009). We narrow this review to sea urchin, abalone, and oyster proteins that mediate sperm-egg interaction. We present data supporting theories attempting to explain selective forces responsible for driving rapid evolution and positive selection of fertilization proteins.
MARINE INVERTEBRATES AS MODEL ORGANISMS
The most used invertebrates for fertilization research are sea urchins, starfish, clams, oysters, and worms. In most experiments, in which gametes of two congeneric species are mixed in cross-species fertilizations, either no or only a low percentage of eggs are fertilized. In most cases, the barrier to cross-species fertilization is not absolute; if eggs are flooded with hetero-specific sperm, some cross-species fertilization usually occurs. However, when quantitative experiments are performed in which gametes of two species are cross-mixed, the percentage of hybrid zygotes formed can range from 0%–100% depending on the species combinations. Thus, in general, at equivalent gamete concentrations, homo-specific mixtures of sperm and eggs yield more zygotes than hetero-specific mixtures. This indicates that gamete interaction is mediated by sperm and egg proteins that bind each other before plasma membrane fusion.
GAMETE RECOGNITION AND SPECIATION
Species-specific fertilization in externally fertilizing marine invertebrates led to the idea that divergence of gamete recognition systems could be involved in speciation (Coyne and Orr 2004). Could the divergence of fertilization proteins lead the way in the speciation process, or would such molecular differentiation occur only after the genomes of two populations had diverged to the point of incompatibility? Here, we discuss some possible causes for the rapid evolution of gamete recognition proteins. One or all of these mechanisms could be acting simultaneously on the evolution of a gamete recognition system. Other, yet unknown selective forces could be causing the rapid evolution of fertilization proteins and the differentiation of populations into new species. In addition to habitat separation, there are no doubt many different ways that speciation can occur.
TYPES OF SELECTION THAT MAY DRIVE THE RAPID EVOLUTION OF FERTILIZATION PROTEINS
Reinforcement
One population could become two species by geographic separation (allopatry) and in time, their two genomes become incompatible, yet their two fertilization systems might remain compatible. If these two sister species make contact again in sympatry (same habitat), the gamete recognition systems might allow fertilization, leading to hybrid embryo death. To prevent cross-species fertilizations, natural selection would “reinforce” differences in the gamete recognition systems to prevent hetero-specific fertilization (Dobzhanski 1940). This could result in the evolution of species-specific fertilization in closely related marine invertebrate species (Minor et al. 1991; Palumbi and Metz 1991; Coyne and Orr 2004). Reinforcement could select for nonoverlapping breeding seasons, making inter-species hybrids impossible. For example, the two Mexican sand dollar species, Encope grandis and Encope californicus, which occur intermingled in equal numbers, have gametes at exactly opposite times of the year (V Vacquier, unpubl.).
Specialization Following Gene Duplication
New proteins may arise by gene duplication and selective pressure imposed to specialize the functions of each paralog. Gene duplication is thought to be a reason for positive selection in ungulate pregnancy associated glycoproteins (Xie et al. 1997). It may also be involved in the rapid divergence of the two abalone sperm acrosomal paralogs discussed below (Kresge et al. 2001a).
Sexual Conflict: The Balance between Sperm Competition and Egg Polyspermy Avoidance
Polyspermy is the fusion of more than one sperm with the egg, resulting in embryo death. Monospermy requires eggs to evolve mechanisms to insure fusion with only one sperm. Some species block polyspermy by rapidly reversing the electrical potential of the egg membrane after the first sperm fuses, which blocks fusion with a second sperm by an unknown mechanism (Gould and Stephano 2003). In sea urchins, the electrical block is followed by elevation of the fertilization envelope, a physical barrier to sperm penetration (Wong and Wessel 2008). Sexual conflict (Rice and Holland 1997) comes into play because sperm competition is always selecting for the fastest sperm to fuse with the egg. Eggs must evolve ways to slow sperm fusion to avoid polyspermy, and one of the ways they might do this is to alter their sperm surface receptors. Altered receptors, having a “poorer fit” with their cognate sperm proteins, slow sperm fusion. In theory, eggs survive to become adults by slowing sperm fusion by altering their egg surface sperm receptors. However, as a counter force, sperm competition continuously selects for sperm with the “best fit” to the altered receptor. To summarize, the egg surface protein changes first and its cognate sperm ligand adapts to the ever-changing female target. This concept has been termed “the coevolutionary chase” of female and male GRPs that could lead to the splitting of one population into two new species (Gavrilets 2000; Gavrilets and Waxman 2002; Gavrilets and Hayashi 2005).
Sexual Selection
This occurs solely if it is a benefit to reproduction. It can drive the rapid evolution of reproductive traits (Andersson 1994). Because of differences in sperm surface alleles, eggs might be fertilized more readily with certain alleles of the cognate sperm surface protein. Such a molecular recognition system could be termed “cryptic female choice” (Palumbi 1999).
RAPID EVOLUTION AND POSITIVE SELECTION IN MARINE INVERTEBRATE FERTILIZATION PROTEINS: ECHINODERM SPERM BINDIN
Sea urchin (su) bindin was the first gamete recognition protein isolated (Vacquier and Moy 1977). After the acrosome reaction, su-bindin coats the acrosomal process that attaches the sperm to the egg vitelline layer (Moy and Vacquier 1979; Vacquier et al. 1995). Su-bindin binds species-specifically with EBR1 (egg bindin receptor-1) (Kamei and Glabe 2003), a large, repetitive glycoprotein. Su-bindin is expressed in spermatocytes (Cameron et al. 1990) as a preproprotein of ∼51-kDa (Gao et al. 1986). Mature su-bindin is packaged into the acrosomal vesicle. Su-bindin (mature su-bindins are 193–418 residues) has a central domain of ∼60 residues that is conserved among sea urchin species. Inside this region are 29 residues that have been completely conserved for ∼250 million years (Zigler and Lessios 2003a; Ziegler 2008). Within this sequence is an 18-residue peptide (B18) that is highly fusagenic (mediates the fusion of lipid bilayers) to biomembranes (Rocha et al. 2008). Thus, su-bindin has two functions, first to species-specifically attach the sperm to the egg, and second, to fuse the gamete membranes. The conserved domain is flanked by species-specific sequences, which are usually repeats of 7–9 residues (Minor et al. 1991; Biermann 1998). Correlating the percent inter-species zygotes formed with su-bindin sequences, shows that 10 differences out of ∼240 residues are enough to produce complete reproductive isolation (Zigler et al. 2005).
Sea urchins and starfish (sf) diverged ∼500 million years ago. The sf-bindin sequence is known for one starfish species (Patiño et al. 2009). The conserved features between su- and sf-bindins include: placement of cysteine residues in a long prepro region, a furin cleavage site between the prepro and mature sequences, repeating sequence elements, and the B18 fusagenic peptide that differs from sea urchin by one residue. Unique features of sf-bindin include: two collagen-like domains, longer more numerous repeats that are unrelated to other sequences, a larger mature mass of ∼88-kDa and placement of the fusagenic B18 peptide at the carboxyl terminus. The structural similarities of these two echinoderm bindins over such long evolutionary time suggests that the B18 peptide will be found in bindins of the three other extant classes of the echinoderms.
EBR1, THE SEA URCHIN EGG RECEPTOR FOR SU-BINDIN
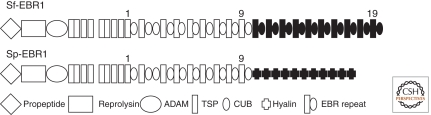
Su-bindin particles adhere species-specifically to unfertilized eggs (Glabe and Vacquier 1977; Glabe and Lennarz 1979; Metz et al. 1994), indicating the presence of species-specific egg receptors. cDNAs for EBR1 from two spe cies, Strongylocentrotus purpuratus (Spurp) and Strongylocentrotus franciscanus (Sfran) were sequenced (Kamei and Glabe 2003). These glycoproteins are homologous in their amino-terminal halves, yet strikingly different in their carboxy-terminal thirds. Sf-EBR1 is 4595 and Sp-EBR1 3710 residues (Fig. 1). From the amino terminus, both have signal sequences, propeptides, reprolysin, and ADAM domains. Next, both have nine tandem TSP domains (thrombospondin type-1), each being 53–59 residues. Progressing toward the carboxyl terminus, both have eight tandem repeats of CUB domains (C1s/Clr, uEGF, bone morphogenic protein) of 104–123 residues. The combination of one TSP domain and one CUB domain is an “EB repeat,” averaging 171 residues. Spurp-EBR1 has 8.5 EBR repeats, whereas Sfran has 19. To this point, the amino-terminal halves of these two proteins are >80% identical. In Sfran, the terminal 10 EB repeats of the carboxyl terminus are different from the upstream EB repeats and are species-specific. In Spurp, after the 8.5 EB repeats, come 11 tandem hyalin repeats each of 81 residues. Hyalin repeats are not present in Sfran-EBR1. Thus, the carboxy-terminal thirds of these two EBR1s show no evolutionary resemblance to each other, except that they contain tandem repeats of roughly the same size and number (Kamei and Glabe 2003). The hyalin repeats are polymorphic in two Spurp individuals, one Echinometra mathaei and one Echinometra oblonga individual (Palumbi 2009). In the two Spurp individuals, 13 and 14 polymorphic hyalin repeats were found in 26 and 27 clones. This polymorphism in Spurp hyalin repeats could provide the raw material for selection to act on to insure fertilization success under a variety of sperm concentrations (Palumbi 2009).
Figure 1.
The protein domains of the sea urchin egg bindin receptor, EBR1, from Strongylocentrotus franciscanus and S. purpuratus. After the ninth EB repeat, the Sf TSP/CUB domains are species-specific in sequence (black). However, after the ninth EB repeat in Sp, there are 11 hyalin repeats of 81 residues each, which have no relationship to TSP/CUP sequences. (Figure redrawn from Kamei and Glabe [2003] and reprinted, with permission, from Cold Spring Harbor Press © 2003.)
Unfortunately, sequences of EBR1 from many individuals of one species, from the same and different populations, and from many different species, have not yet been determined. The EB repeats in Sfran-EBR1 are all homologous. The possession of terminal hyalin repeats in Spurp-EBR1 suggests that exon shuffling occurred to produce a chimeric EBR1, which was advantageous to fertilization success.
RAPID BINDIN EVOLUTION, SEQUENCE POLYMORPHISM, AND POSITIVE SELECTION IN SYMPATRIC SPECIES OF SEA URCHINS
The first su-bindin sequences of congeneric, sympatric sea urchin species were those of Strongylocentrotus purpuratus and S. franciscanus. The sequences were different in the regions flanking su-bindin’s conserved central domain (Minor et al. 1991; Biermann 1998). These two are the most divergent species in this genus (Biermann et al. 2003; Lee 2003). Subsequent sequencing work on Hawaiian sea urchins, Echinometra oblonga and E. mathaei, living intermingled on the same reefs, showed that these two species diverged 0.5–2.0 million years ago. Cross-species mixtures of their gametes showed a strong block to hybrid fertilization (Palumbi and Metz 1991), caused by sperm su-bindin failing to attach the sperm to the egg (Metz et al. 1994).
These two Hawaiian Echinometra species and four unnamed incipient Echinometra species from the western Pacific and Panama (Metz and Palumbi 1996; McCartney and Lessios 2002, 2004; Zigler et al. 2005), and species of the genus Strongylocentrotus (Biermann 1998) all show that positive selection has acted on the rapid evolution of su-bindin in a region ∼40 amino acids upstream of su-bindin’s conserved central core. The greatest su-bindin rate of evolution and highest su-bindin sequence polymorphism correlate positively with the greatest number of sympatric species, suggesting that reinforcement might be the selective force operating on the evolution of su-bindins (Palumbi and Lessios 2005; Lessios 2007).
In the sea urchin, Echinometra oblonga, individuals from Guam and Okinawa have dramatically different sperm sizes and cross-fertilizations between these two locations are less successful than within the same location. These differences have arisen after an estimated divergence time of ∼250,000 years (Landry et al. 2003). Likewise, within the circumpolar species, Strongylocentrotus droebachiensis, sperm dimensions are different between the Atlantic and Pacific oceans and also between various islands (Manier and Palumbi 2008; Marks et al. 2008). Thus, sperm morphometrics, su-bindin sequences and reproductive isolation can evolve rapidly depending on the sea urchin populations in question. It would be interesting to have EBR1 repeat sequences from these sea urchins.
Paracentrotus gaimardi is one of the two sea urchin species of this genus, which occurs in five distinct sympatric color morphs on the coast of Brazil. Genes for mitochondrial ATPase 8 and 6, a histone intron and sperm bindin were sequenced. Regarding the mitochondrial genes, statistically significant differences are seen between only one color morph and the other four. Essentially no differences were seen in the histone gene among the morphs. However, in bindin all color morphs were differentiated from each other with nine positions under positive selection (Calderon et al. 2010). Whether this represents the first stages of reproductive isolation differentiation in incipient speciation, or merely accelerated evolution of a reproductive protein, remains conjectural at this time.
SELECTION IN THE MAINTENANCE OF RARE SU-BINDIN ALLELES
Fifteen su-bindin alleles are known in Strongylocentrotus franciscanus: one represents 58% of alleles, one 15%, one 14%, and the remaining 13% comprise 12 rare alleles (Levitan and Ferrell 2006). When sperm densities were high, embryos that survived were found to have been fertilized with sperm carrying rare su-bindin alleles. Conversely, when sperm densities were low, the common su-bindin alleles were found in the surviving embryos. These results can be explained by sexual conflict, the balance between sperm competition and egg avoidance of polyspermy (Rice and Holland 1997; Frank 2000; Haygood 2004). Theoretically, at high sperm densities, eggs with altered EBR1s survive to become embryos because rare su-bindin alleles fit altered EBR1s poorly, the result being the slowing of sperm fusion and monospermy. Thus, one explanation for these data is that sexual conflict in polyspermy avoidance maintains the rare alleles in this species and hence explains the extensive su-bindin polymorphism (Levitan and Ferrell 2006). The positive selection seen in su-bindin evolution could result from sperm competition for the “best fit” for binding to an altered egg EBR1 receptor (Kamei and Glabe 2003; Palumbi 2009).
It is difficult to distinguish the above explanation from sexual selection (cryptic female choice) in that eggs that are favored to become embryos are those that have “selected” to fuse with sperm carrying rare su-bindin alleles (Palumbi 1999). Mutants in the tandem repeats of EBR1 could have a better fit with mutant su-bindins and be passed on in the population. Because of its tandem construction, a mutant EBR1 repeat could be spread through the repeat array by concerted evolution, the process mediated by unequal crossing over and gene conversion that homogenizes the sequences of tandem repeats (Elder and Turner 1995). However, the high amount of sequence polymorphism in hyalin repeats of EBR1 shows that concerted evolution has not influenced evolution of this protein (Palumbi 2009).
Thus at low sperm densities, common bindin alleles are used more frequently to produce embryos to avoid egg loss (Levitan and Ferrell 2006). In addition, sea urchin species that are more resistant to polyspermy have fewer bindin alleles than species that are more susceptible to polyspermy (Levitan et al. 2007). However, other work on sexual conflict in polyspermy avoidance shows that even at low sperm densities, a measurable fraction of eggs become polyspermic (Franke et al. 2002). In summary, sexual conflict, resulting from the balance between the egg’s interest in monospermy and the sperm’s interest to compete with other sperm to be the first sperm to fuse with the egg, is the likely selective force for the rapid evolution, maintenance of polymorphic alleles, and positive selection of su-bindin in S. franciscanus. Although sexual conflict in polyspermy prevention is an explanation for maintenance of multiple su-bindin alleles, the possibilities that reinforcement to prevent cross-species fertilizations, and sexual selection, could also be selective forces acting on su-bindin’s evolutionary trajectory.
However, the strengths of the blocks against polyspermy, adult sea urchin density, egg reception of bindin, and bindin genotype all influence fertilization success. This is a complex situation that is difficult to quantitatively parse into the components of the selective forces acting on each element. In the sea urchin Strongylocentrotus purpuratus, spawned in the sea, both negative and positive selection impinge on the evolution of sperm bindin. This species is sympatric with S. franciscanus mentioned above. S. purpuratus occurs in higher population densities than S. franciscanus and also requires higher sperm densities to achieve fertilization and its eggs are also more resistant to polyspermy. Such complex species differences have been discussed from field experiment fertilization data (Levitan and Stapper 2010) and from the results of theoretical modeling (Tomaiuolo and Levitan 2010).
SU-BINDIN EVOLUTION IN ALLOPATRIC SEA URCHIN SPECIES: SLOW EVOLUTIONARY RATES WITHOUT POSITIVE SELECTION
Four species of sea urchins, genus Arbacia, inhabit different coasts, each being separated from another Arbacia species by thousands of kilometers. Mitochondrial DNA and su-bindin sequences show they are distinct species with divergence times of ∼4–9 million years. Gametes of the Pacific species, Arbacia incisa, show no barrier to fertilization when crossed with gametes from the Atlantic species, Arbacia punctulata (Metz et al. 1998a). Few amino acid substitutions occur in su-bindins of these four species and only three substitutions out of 235 residues are nonconservative. No evidence of positive selection in su-bindin evolution was found among these four species. Su-bindin evolution has been slow and purifying in the absence of sympatric, congeneric species.
A similar study was performed on su-bindin from three singly occurring (allopatric) species of the genus Tripnuestes. Two of these species, that diverged ∼3 million years ago, because of the rise of the Isthmus of Panama, differ by only four amino acids and one insertion/deletion. Among these three su-bindins, there is no sign of rapid evolution or positive selection (Zigler and Lessios 2003b). A third study of eight allopatric species of the genus Lytechinus, also showed little variation in su-bindin sequences and no signal of rapid evolution by positive selection (Zigler and Lessios 2004).
In all three genera of allopatric sea urchin species mentioned above, su-bindin sequence evolution has been slow and not influenced by positive selection, which is the converse to that of the sympatric species. These findings are indirect evidence supporting the theory that reinforcement to prevent hybrid fertilization among sympatric species might explain the rapid evolution of bindin in sympatric species (Palumbi and Lessios 2005; Lessios 2007). Among the allopatric species of these three genera, egg loss because of inter-species hybrid embryo formation is impossible, so there is no selective pressure to reinforce differences in their gamete recognition systems. Another study of Echinometra oblonga supports reinforcement in that sympatric populations of this species have more variation in the su-bindin gene than do allopatric populations (Geyer and Palumbi 2003). Other research on two species of the genus Heliocidaris supports the reinforcement hypothesis in that su-bindin evolution may have been driven by the selective pressure to prevent fertilization between the direct developing sea urchin Heliocidaris erythrogramma and the indirect developing Heliocidaris tuberculata (Zigler et al. 2003).
POSITIVE SELECTION IN SU-BINDIN IN ALLOPATRIC SEA URCHINS
Paracentrotus is the other allopatric species of this genus. The species Paracentrotus lividus, in the Mediterranean and west coast of Africa is highly variable in su-bindin sequences. In 63 full-length sequences from different locations, 23 su-bindin haplotypes were found (Calderon et al. 2009). Positive selection was detected in 12 sites on either side of the single intron 5′ to the ∼60 residue constant domain. Variation was mainly in the number of glycine rich short repeats flanking the constant domain. Because no other congener is present in the Mediterranean, reinforcement cannot account for the positive selection. Sexual conflict in polyspermy avoidance is probably a more likely explanation, in which a variety of su-bindin alleles are adapting to a variety of EBR1 repeat sequences.
Echinometra lucunter is a sea urchin occurring sympatrically with its sister species, Echinometra viridis, in the Caribbean, and allopatrically in other parts of the Atlantic. The sympatric and allopatric populations show mtCO1 sequence differences indicative of ∼200,000 years of divergence. However, both populations show equal positive selection in su-bindin, indicating again that reinforcement to prevent intra-species hybrids is probably impossible. More likely hypotheses to account for positive selection in su-bindin are the intra-specific processes of sexual selection, sexual conflict, and sperm competition (Geyer and Lessios 2009).
POSITIVE SELECTION IN THE SEA URCHIN SPERM RECEPTOR FOR EGG JELLY PROTEINS (suREJ)
When sea urchin sperm contact egg jelly (EJ), the fucose sulfate polymer of EJ induces the required acrosome reaction (AR), which consists of the polymerization of actin to form the finger-like acrosomal process coated with su-bindin (Moy and Vacquier 1979; Vacquier and Hirohashi 2004; Darszon et al. 2007; Vilela-Silva et al. 2008). The AR is induced by the opening of two distinct calcium channels and the elevation of sperm pH by ∼0.2 units (Darszon et al. 2001; Hirohashi and Vacquier 2002). The sperm plasma membrane has at least two EJ receptors, suREJ-1 and -3, that localize to the plasma membrane covering the acrosome vesicle. suREJ-1 has two carbohydrate recognition domains (CRDs) and completes with EJ for binding to sperm (Moy et al. 1996). suREJ-3 has one CRD and one galactose recognizing lectin domain (Mengerink et al. 2002). Sea urchin REJ proteins are homologs of polycystin-1, the protein mutated in autosomal dominant polycystic kidney disease, the most common monogenic disease of humans (Gunaratne et al. 2007).
suREJ-1 and -3 were sequenced from six Strongylocentrotus species. A maximum likelihood analysis showed the CRD modules are subjected to positive selection. The positively selected residues map onto the carbohydrate binding region of each CRD, indicating that they might be important in species-specific induction of the AR by EJ (Mah et al. 2005). Thus, in Strongylocentrotus, positive selection can act both on su-bindin, a sperm ligand binding an egg receptor (EBR1) and suREJ-1, a sperm receptor for an egg ligand. Thus, selection can act simultaneously on two distinct sequential events in the fertilization cascade, first acrosome reaction induction, and second, sperm binding to the egg. The reason for the selective pressure underlying the positive selection in the CRDs of the sperm suREJ proteins is not readily apparent. Is it reinforcement to prevent zygote loss, or is it sexual conflict in the balance between sperm competition and egg avoidance of polyspermy?
EXTREME POSITIVE SELECTION IN ABALONE SPERM LYSIN AND 18-kDa PROTEIN
Abalone sperm have two major acrosomal proteins, 16-kDa lysin and 18-kDa protein (sp18) that are single copy genes in the red abalone species. Lysin dissolves the elevated vitelline envelope (VE) surrounding the egg by a nonenzymatic mechanism that probably involves competitively displacing hydrogen bonds that hold the fibrous VE glycoproteins together. sp18 coats the acrosomal process membrane and is thought to mediate sperm-egg fusion (Swanson and Vacquier 1995a; Kresge et al. 2001b). Deduced amino acid sequences of lysin were obtained from 20 abalone species. The amino-terminal sequence of lysin is always species-unique. Other regions are hypervariable, but not necessarily species-unique (Lee and Vacquier 1992; Lee et al. 1995; Kresge et al. 2001). Between species, comparisons show that lysin is extremely divergent in certain regions, yet highly conserved in others. Nucleotide substitution analysis showed that lysin’s divergent regions have been subjected to strong positive Darwinian selection (protein adaptive evolution). Using mitochondrial (mt) CO1 as a standard for divergence time, it was estimated that the four most closely related eastern Pacific abalone species diverged from each other 1–2 million years ago. In pairwise comparisons of these four species, both acrosomal proteins appeared to be evolving faster than mtDNA, and sp18 evolves 2.5 times faster than lysin (Metz et al. 1998b). In one comparison between the sympatric white and pinto abalone species, mtCO1 was 1% different in nucleotide sequence, whereas lysin amino acid sequences differed by 15% and sp18 sequences by 23% (Metz et al. 1998b). These are some of fastest rates of divergence known for any metazoan protein. Comparison of three species showed that the five exons of both genes evolve much faster than the four introns. Sequencing ∼29,000 base pairs of lysin and sp18 genomic DNA from red abalone individuals separated by ∼1200 kilometers showed almost no nucleotide polymorphism, whereas the intron of a G-protein coupled receptor, showed ∼7% nucleotide variation among individuals. These data suggest that the lysin and 18-kDa sequences in today’s red abalone population are the results of many selective sweeps of favorable mutations in both genes (Metz et al. 1998b). Analysis of maximum likelihood models of 25 lysin sequences showed that the divergence rate (and thus selection pressure) varied in different lineages and that positive selection was most intense among closely related, sympatric species (Yang et al. 2000). Abalone sperm lysin remains one of the most robust examples of positive Darwinian selection known to date (Nunney and Schuenzel 2006).
WORK ON TURBAN SNAILS AND MUSSELS
Sperm of other archeogastropod mollusks, such as turban snails (Tegula), also use small cationic acrosomal proteins to dissolve a hole in the egg envelope by a nonenzymatic, species-specific mechanism. Interspecies comparisons show that Tegula sperm lysin is also subjected to strong positive selection (Hellberg and Vacquier 1999; Hellberg et al. 2000). The M7 acrosomal lysin of marine mussels, that dissolves the egg envelope, is a C-type lectin that also shows rapid evolution and positive selection (Riginos and McDonald 2003; Springer and Crespi 2007). Two species of mussels, Mytilus edulis (Me) and Mytilus trossulus (Mt), occur separately or together, and where they co-occur, hybrids exist. Cross species fertilizations of Me eggs and Mt sperm show that where the two species occur together there is greater success of Me eggs being fertilized by Mt sperm as compared to Me eggs from females living in the absence of Mt adults. These data are contrary to what would be expected if reinforcement was acting as a selective force to differentiate the gamete recognition system. These results could be explained by ingression of Mt genes for the egg’s sperm receptor system into the Me gene pool in regions of sympatry (Slaughter et al. 2008).
CRYSTAL STRUCTURES OF ABALONE SPERM LYSIN
Lysin crystal structures show that the species unique amino terminus and the hypervariable carboxyl terminus lie on the same face of the molecule away from the helical bundle that comprises most of the protein. Regions of high amino acid substitution appear to be on the outer edges of the helical bundle, whereas conserved residues are on the bundle’s inner surfaces (Shaw et al. 1993, 1995). Maximum likelihood analysis of lysin shows that 21 positions out of ∼134 have been subjected to positive selection, with the amino terminus being the most highly substituted (Yang et al. 2000). Using recombinant DNA methods to switch the two termini of lysins from two species proved that these regions are indeed involved in species recognition needed for VE dissolution (Lyon and Vacquier 1999).
LYSIN AND THE sp18 AROSE BY GENE DUPLICATION
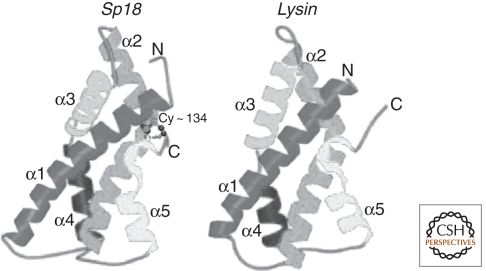
The sp18 sequence is known for five California abalone species. These five sequences are so different from lysin sequences that BLAST searches of GenBank using sp18 as input do not identify the lysin sequences as homologs. However, secondary structure predictions show that both proteins have the same number, length, and position of α-helices (Swanson and Vacquier 1995b). Both genes have four introns, which are in the same positions and interrupt the codons in the same phase (Metz et al. 1998b). Crystal structures of both proteins showed that both have nearly identical α-carbon ribbon structures, which is excellent evidence that they arose by a gene duplication event followed by their progressive differentiation (Fig. 2) (Kresge et al. 2001a). However, charge distribution and hydrophobic regions of the surfaces of both proteins are very different, possibly reflecting extreme specialization of function. In theory, the primordial abalone had one acrosomal protein that performed both functions of dissolving a hole in the egg VE and fusing the gamete plasma membranes. Because the ancestral gene duplication, lysin has specialized to dissolve egg vitelline envelopes (VE), whereas sp18 has specialized to fuse gamete plasma membranes (Swanson and Vacquier 1995a). The rapid evolution and positive selection seen in these two proteins could be driven by: specialization of function following gene duplication and sperm competition, which would be continuously selecting for the fastest sperm to dissolve a hole in the VE and fuse the gamete plasma membranes.
Figure 2.
Abalone sperm lysin and sp18 arose from a gene duplication. The α-carbon ribbon diagrams are proof of their common ancestry. Both genes also have the same intron-exon structure. However, the surface properties of the two proteins are very different. (Figure adapted from Kresge et al. [2001a] and reprinted, with permission, from the American Chemical Society © 2001.)
DUPLICATION AND EVOLUTION OF AN ABALONE LYSIN GENE
One allopatric abalone species, Haliotis tuberculata coccinea, lives on the Azores Islands. Another subspecies, Haliotis tuberculata tuberculata is allopatric on the Brittany Coast of France. The lysin genes of these subspecies are duplicated and both paralogs are expressed in sperm. The exons of the two paralogs are 83% identical and low polymorphism is found within a small population sampling (Clark et al. 2007). Phylogenetic analysis shows that the gene duplication occurred before the two subspecies arose and that both lysin copies are subjected to positive selection. Because these two subspecies are allopatric, there is no chance for cross-species fertilizations, therefore reinforcement cannot account for the positive selection driving the differentiation of the two paralogous lysins (Clark et al. 2007). Selection pressure on the two lysins might be because they bind different regions of VERL, the egg Vitelline Envelope Receptor for Lysin. Currently, nothing is known about VERL of these subspecies. A second hypothesis is that different alleles of VERL may exist in the population and each lysin paralog is specialized to interact with one VERL allele. Sexual conflict, in the balance between sperm competition and polyspermy avoidance, could explain the positive selection seen in the paralogous lysins as the lysins are continuously adapting to two different alleles of VERL. Such a scenario could result in prezygotic reproductive isolation and speciation (Clark et al. 2007).
VERL, THE ABALONE EGG VITELLINE ENVELOPE RECEPTOR FOR SPERM LYSIN
There are few examples of sperm and egg cognate pairs of interacting fertilization proteins. Abalone sperm lysin and its egg VE receptor, VERL, are the most informative cognate pair. Because sperm lysin dissolves isolated egg VEs, the molecule interacting with lysin was obviously in the VE. Radiolabeled lysin and sucrose density gradients were used to identify VERL, the VE Receptor for Lysin, of mass ∼1 million Daltons. Electron microscopy showed VERL to be unbranched rods of undetermined length. VERL bound lysin with high affinity and also species-selectivity (Swanson and Vacquier 1997). Amino acid sequences were obtained and PCR used to amplify the VERL sequence from cDNA and genomic DNA. VERL was composed of tandem repeats of ∼153 amino acids. The repeats were very homogeneous within a species, but differed between species. The repeats evolved by “concerted evolution,” which homogenizes the sequences so that they are nearly identical (Elder and Turner 1995; Swanson and Vacquier 1998). This finding was contrary to theoretical expectations that, like lysin, VERL should show that positive selection had acted on its divergence (Gavrilets 2000; Gavrilets et al. 2001; Van Doorn et al. 2001; Gavrilets and Waxman 2002; Gavrilets and Hayashi 2005; Hayashi et al. 2007). The most recent work shows that VERL and lysin coevolve at similar rates, that is to say that relatively large changes in lysin sequences are mirrored by similar large changes in VERL sequences suggesting that lysin is “chasing” a rapidly evolving VERL (Clark et al. 2009).
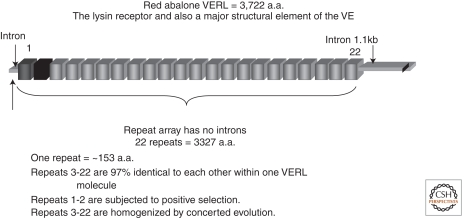
Full-length VERL cDNA (11,166 bp) was cloned from the red abalone (Galindo et al. 2002). About 90% of the amino acid sequence consists of 22 tandem repeats of ∼153 residues, which are predicted to be β-sheets (Fig. 3). Following repeat 22 are 353 residues containing a zona pellucida domain (ZP) and a hydrophobic COOH-terminus. The most interesting feature is that repeats 1 and 2 have not been homogenized with repeats 3–22 (Galindo et al. 2002). Sequencing VERL repeats 1–4 from eight species showed that repeats 1 and 2 are subjected to positive selection, and have not been homogenized with repeats 3–22, since the divergence of these eight species (Galindo et al. 2003). The ZP domain was originally found in the three glycoproteins that make up the mammalian egg envelope, the zona pellucida (Wassarman and Litscher 2008). This domain of about 250 amino acids is found in many extracellular proteins and is involved in the polymerization and assembly of structures such as egg envelopes (Jovine et al. 2005; Monné et al. 2008). Further work on the VE has identified at least 30 other ZP domain containing proteins of smaller size than VERL. One of these VEZP proteins, VEZP14, is a paralog of VERL and contains only one VERL repeat that evolves by positive selection twice as rapidly as do repeats 1 and 2 of VERL. VEZP14 also shows preferential binding to the sp18 acrosomal protein and is thus the putative sp18 receptor (Aagaard et al. 2010). Several of these 30 VEZP proteins show positive selection. The selective forces of polyspermy avoidance, specialization following gene duplication and reinforcement of molecular differences between species might account for such strong selection (Aagaard et al. 2006, 2010). What is exciting is that ZP domain proteins are found in both mammalian and abalone egg envelopes. Could this be a case of orthologous evolution from primordial animals, or is this a case of convergent evolution?
Figure 3.
The structure of the red abalone egg vitelline envelope receptor for lysin (VERL). There are 353 residues after the last repeat, which contains one ZP domain. Repeats 1 and 2 evolve by positive selection, whereas repeats 3–22 are subjected to concerted evolution. (Figure adapted from Galindo et al. [2002] and reprinted, with permission, from Elsevier © 2002.)
COMPARING SEA URCHIN EGG EBR1 AND ABALONE EGG VERL
Sea urchins and abalone diverged at least 500 million years ago. Why should these two distantly related animals have sperm receptors on their egg surfaces that have such similar structures? Sea urchin Sf-EBR1 has 19 tandem CUB/TSP repeats, each dual-domain repeat being ∼171 amino acids. In Sf, the last 10 CUB/TSP repeats are species-specific. In Sp, the terminal 11 repeats are hyalin repeats of 81 residues that are not homologous to the CUB/TSP repeats of Sf-EBR1 (Kamei and Glabe 2003). This lack of carboxy-terminal homology might have arisen by exon shuffling that created a new carboxy-terminal EBR1 gene. The hyalin repeats are also found in Echinometra, which is far diverged from Strongylocentrotus (Palumbi 2009). CUB, TSP, and hyalin domains are all known to be involved in protein–protein binding. The tandem repeats of sea urchin EBR1 are separated by introns, which could promote exon shuffling. The 22 tandem repeats of abalone egg VERL are each ∼153 amino acids. VERL repeats 1 and 2 do not homogenize with repeats 3–22, and there are no introns between any of the VERL repeats, which makes exon shuffling improbable (Galindo et al. 2002, 2003). Thus, both these broadcast spawning invertebrates evolved sperm receptors on their eggs that are constructed of repeat arrays of similar sizes and numbers of amino acids. Unequal crossing over was most probably involved in their expansion. The secondary structures of both suggest IgG-like β-sandwich folds.
OYSTER SPERM BINDIN
Oyster (Crassostrea gigas) sperm have a ring-shaped acrosomal granule containing a bindin (oy-bindin) that attaches sperm to egg. The rings of insoluble oy-bindin can be isolated (Brandriff et al. 1978). When the acrosomal rings of single males are separated by SDS-PAGE, one or two protein-staining bands appear per individual, of relative molecular masses 35-, 48-, 63-, 75-, and 88-kDa. Translating cDNA sequences of oy-bindins show the protein has a signal sequence of 24 residues, followed by a 97 residue unique amino terminus, which is followed by from one to five tandem fucose-binding lectin repeats (FLR) of 134 residues. Internal FLRs are separated by a spacer sequence of either PGAK or PGAKGK. The differences in SDS-PAGE mobilities of oy-bindin proteins represent different numbers of FLRs, the extremes being the 35-kDa isoform with one FLR and the 88-kDa isoform with five FLRs. The presence of one or two molecular mass classes of bindin per individual represents the two alleles (Moy et al. 2008, Springer et al. 2008).
Southern blots show that oy-bindin is a single copy gene. Genomic DNA contains oy-bindin genes possessing from 1–5 FLRs. Such variation in repeat number is usually created by unequal crossing over. The spermatogenic tissue of individual males having oy-bindin genes with 5 FLRs has oy-bindin mRNAs with 1-5 FLRs; however, only one or two isoforms are translated per individual, indicating that an unexplained mechanism governs translation. The five isoforms of mRNA show that alternative splicing of the primary transcript must occur (Moy et al. 2008, Springer et al. 2008). The amino-terminal 122 residues of the oy-bindin protein is highly conserved in sequence. In 39 full-length sequences from 16 males from five worldwide locations, there were only five substituted amino acids out of 4,315 (Moy et al. 2008). In the exact middle of each FLR is one intron of highly variable size, which is the site of recombination between FLR halves. In 39 full-length oy-bindin cDNA sequences, there were 107 FLRs that contained 41 different FLR sequences, resulting from the recombination of 24 unique amino-terminal FLR halves with 18 unique carboxy-terminal halves. This is the most intra-specific variation that has ever been found in a fertilization protein.
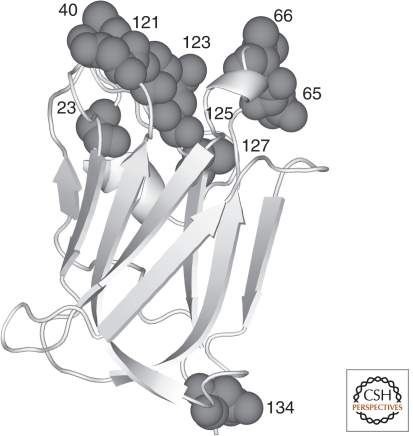
The F-lectin domain is an ancient protein module present in both pro- and eukaryotes. The oy-bindin FLR sequence is about 45% identical to the consensus F-lectin sequence. Maximum likelihood analysis of oy-bindin identifies nine sites subjected to positive section. Threading the oy-bindin F-lectin sequence onto the crystal structure of FLR shows that eight of these sites are clustered in the fucose binding groove (Fig. 4), indicating there is a survival benefit to altering the fucose binding affinity of oy-bindin for its yet unknown egg surface receptor (Moy et al. 2008; Springer et al. 2008; Wu et al. 2010).
Figure 4.
The oyster bindin fucose lectin repeat threads onto the known fucose lectin crystal structure. Dark residues are eight sites under positive selection that surround the fucose-binding groove, indicating the importance of adaptive evolution in binding fucose. Number 138 is the ninth positively selected site, which is not in the fucose binding groove. (Figure adapted from Moy et al. [2008] and reprinted, with permission, from the National Academy of Sciences © 2008.)
As mentioned above, the number of FLRs contained in one oy-bindin gene can vary. One gene with three tandem FLRs had a 3.6 kb retroposon immediately upstream of the first FLR. However, two oy-bindin genes, which both had only one FLR, lacked this retroelement. Thus, insertion of a retroposon might be another way to increase the number of FLRs in an oy-bindin gene. The presence of upstream GA, and complementary downstream CT, microsatellites between FLR repeats also suggests that loopout strand duplex formation, copying and copy integration may be yet another way of altering the number of FLRs in an oy-bindin gene (Moy and Vacquier 2008).
WHY IS OYSTER BINDIN SO VARIABLE?
One male C. gigas can synthesize hundreds and maybe thousands of forms of the oy-bindin protein (Moy et al. 2008; Springer et al. 2008). Experimental evidence suggests this species uses every known mechanism to create variation in the FLR sequence. These mechanisms are: alternative splicing to create mRNAs with different numbers and combinations of FLRs, recombination of halves of FLRs to create variation in FLR sequences, retrotransposition, and loopout strand copying to increase the number of FLRs in the tandem repeat array. One big difference between oysters on one hand, and sea urchins and abalone on the other hand, is that oyster eggs do not have an electrical block against polyspermy (Alliegro and Wright 1983; Stephano and Gould 1988; Togo and Morisawa 1999; Gould and Stephano 2003). Because of the lack of an electrical block, polyspermy prevention might be more dependent on the initial events of sperm-egg interaction before the gametes fuse (Moy et al. 2008). We do not know anything about the egg’s oy-bindin receptor. We can assume that it is a glycoprotein with exposed fucose residues. Those eggs that survive to pass on their oy-bindin genes might be only those with altered oy-bindin receptors, which slow sperm fusion. However, sperm competition is continuously selecting for the fastest sperm to fuse. To guarantee that fertilization will succeed, males make a wide variety of oy-bindins. These experimental facts support the hypothesis that sexual conflict is the driving force in the great variation seen in oyster sperm bindin. Because the oysters used in these experiments were raised in culture facilities as a single species, reinforcement is not a plausible theory to explain such extreme variation in this fertilization protein. Further knowledge will come from identifying and sequencing the egg receptor for this most interesting protein (Wu et al. 2010).
STRUCTURAL CONSERVATION IN A GAMETE RECOGNITION PROTEIN MODULE IN EUKARYOTES
The mammalian egg envelope, the zona pellucia (ZP), is composed of four glycoproteins (ZP1–4). ZP2 and ZP3 bind sperm and ZP3 can induce the sperm acrosome reaction. ZP proteins are made of two domains, ZP-N and ZP-C. The crystal structure of ZP3’s ZP-N domain defines a novel type of immunoglobulin (Ig) superfamily, having: two invariant 1-4 and 2-3 disulfide bonds, a unique E′ strand and a conserved Tyr residue in strand F′. Comparing sequences of ZP-N domains in mammalian ZPs, abalone VERL repeats and yeast mating agglutinin proteins, shows no similarities among these proteins. However, threading the abalone VERL repeat sequences and the amino terminus of abalone VEZP-14 onto the mammalian ZP-N 3-D structure, results in a statistically robust match. The VEZP-14 model was then used to thread the α-agglutinin mating factor of the yeast Saccharomyces cerevisiae and the invasion glycoprotein of Candida albicans. The yeast IgIII protein identified the ZP-N Ig fold as the top hit. Both yeast proteins show the 1-4, 2-3 disulfide bonding pattern. In conclusion, these sex proteins from mammals, mollusks, and yeast all share the same fold of a novel Ig module. It remains unknown whether this 3-D structural similarity results from an ancestral relationship, or if this is a case of convergent evolution (Swanson et al. 2011). This discovery exemplifies the pitfall of using only primary structure to search for similarities of proteins from diverse species.
CONCLUSION
The quote from Ernst Mayr at the beginning of this review was his guess that selection must be strong on gametes. The rapid rate of sequence divergence, usually driven by positive selection (adaptive protein evolution), witnessed in many fertilization proteins, is testament to his clairvoyance. The factors responsible for such strong selection on fertilization proteins are at present difficult to define with certainty. Sexual conflict, sexual selection, reinforcement, specialization after gene duplication, and yet unknown mechanisms, might work simultaneously in the evolution of gamete recognition systems. Knowledge of sequences of additional cognate pairs of interacting sperm and egg proteins in more animal models should yield the needed information to enable refinement of current hypotheses and the formulation of novel hypotheses to explain this general phenomenon.
ACKNOWLEDGMENTS
This work was funded by NIH Grant HD12986 to V.D.V., NIH Grants HD042563, HD054631, HD057974 and NSF Grants DEB-0743539 and DEB-0918106 to W.J.S.
Footnotes
Editors: Paolo Sassone-Corsi, Margaret T. Fuller, and Robert Braun
Additional Perspectives on Germ Cells available at www.cshperspectives.org
REFERENCES
- Aagaard JE, Yi X, MacCoss MJ, Swanson WJ 2006. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc Natl Acad Sci 103: 17302–17307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard JE, Vacquier VD, Maccoss MJ, Swanson WJ 2010. ZP domain proteins in the abalone egg coat include a paralog of VERL under positive selection that binds lysin and 18-kDa sperm proteins. Mol Biol Evol 27: 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliegro MC, Wright DA 1983. Polyspermy inhibition in the oyster Crassostrea virginica. J Exp Zool 227: 127–137 [DOI] [PubMed] [Google Scholar]
- Andersson M 1994. Sexual selection. Princeton University Press, Princeton, NJ [Google Scholar]
- Biermann CH 1998. The molecular evolution of sperm bindin in sex species of sea urchins (Echinoida: Strongylocentrotidae). Mol Biol Evol 15: 1761–1771 [DOI] [PubMed] [Google Scholar]
- Biermann CH, Kessing BD, Palumbi SR 2003. Phylogeny and development of marine model species: Strongylocentrotid sea urchins. Evol Dev 5: 360–371 [DOI] [PubMed] [Google Scholar]
- Brandriff B, Moy GW, Vacquier VD 1978. Isolation of sperm bindin from the oyster (Crassostrea gigas). Gamete Res 1: 89–99 [Google Scholar]
- Calderón I, Turon X, Lessios HA 2009. Characterization of the sperm molecule bindin in the sea urchin genus Paracentrotus. J Mol Biol 68: 366–376 [DOI] [PubMed] [Google Scholar]
- Calderón I, Ventura CRR, Turon X, Lessios HA 2010. Genetic divergence and assortative mating between colour morphs of the sea urchin Paracentrotus gaimardi. Mol Ecol 19: 484–493 [DOI] [PubMed] [Google Scholar]
- Cameron RA, Minor JE, Nishioka D, Britten RJ, Davidson EH 1990. Locale and level of bindin mRNA in maturing testis of the sea urchin (Strongylocentrotus purpuratus). Dev Biol 142: 44–49 [DOI] [PubMed] [Google Scholar]
- Chuong EB, Tong W, Hoekstra HE 2010. Maternal-fetal conflict: Rapidly evolving proteins in the rodent placenta. Mol Biol Evol 27: 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ 2006. Evolution of reproductive proteins from animals and plants. Reproduction 131: 11–22 [DOI] [PubMed] [Google Scholar]
- Clark NL, Findlay GD, Yi X, MacCoss MJ, Swanson WJ 2007. Duplication and selection on abalone sperm lysin in an allopatric population. Mol Biol Evol 24: 2081–2090 [DOI] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ 2009. Coevolution of interacting fertilization proteins. PLoS Genetics 5: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA 2004. Speciation. Sinauer Associates, Sunderland, MA [Google Scholar]
- Darszon A, Beltrán C, Felix R, Nishigaki T, Treviño CL 2001. Ion transport in sperm signaling. Dev Biol 240: 1–14 [DOI] [PubMed] [Google Scholar]
- Darszon A, Treviño CL, Wood C, Galindo B, Rodríguez-Miranda E, Acevedo JJ, Hernandez-González EO, Beltrán C, Martínez-López P, Nishigaki T 2007. Ion channels in sperm motility and capacitation. Soc Reprod Fertil Suppl 65: 229–244 [PubMed] [Google Scholar]
- Dobzhanski T 1940. Speciation as a stage in evolutionary divergence. Am Nat 74: 302–321 [Google Scholar]
- Dorus S, Wasbrough ER, Busby J, Wilkin EC, Karr TL 2010. Sperm proteomics reveals intensified selection on mouse sperm membrane and acrosomal genes. Mol Biol Evol 27: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder JF, Turner BJ 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol 70: 297–320 [DOI] [PubMed] [Google Scholar]
- Frank SA 2000. Sperm competition and female avoidance of polyspermy mediated by sperm-egg biochemistry. Evol Ecol Res 2: 613–625 [Google Scholar]
- Franke ES, Babcock RC, Styan CA 2002. Sexual conflict and polyspermy under sperm-limited conditions: In situ evidence from field simulations with the free spawning marine echinoid (Evechinus chloroticus). Am Nat 160: 485–496 [DOI] [PubMed] [Google Scholar]
- Galindo BE, Moy GW, Swanson WJ, Vacquier VD 2002. Full-length sequence of VERL, the egg vitelline envelope receptor for abalone sperm lysin. Gene 288: 111–117 [DOI] [PubMed] [Google Scholar]
- Galindo BE, Vacquier VD, Swanson WJ 2003. Positive selection in the egg receptor for abalone sperm lysin. Proc Natl Acad Sci 100: 4639–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Klein LE, Britten RJ, Davidson EH 1986. Sequence of mRNA coding for bindin, a species specific sperm protein required for fertilization. Proc Natl Acad Sci 83: 8634–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S 2000. Rapid evolution of reproductive barriers driven by sexual conflict. Nature 403: 886–889 [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Hayashi TI 2005. Speciation and sexual conflict. Evol Ecol 19: 167–198 [Google Scholar]
- Gavrilets S, Waxman D 2002. Sympatric speciation by sexual conflict. Proc Natl Acad Sci 99: 10533–10538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S, Arnqvist G, Friberg U 2001. The evolution of female mate choice by sexual conflict. Proc Biol Sci 268: 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer LB, Lessios HA 2009. Lack of character displacement in the male recognition molecule, bindin, in Atlantic sea urchins of the genus Echinometra. Mol Biol Evol 26: 2135–2146 [DOI] [PubMed] [Google Scholar]
- Geyer LB, Palumbi SR 2003. Reproductive character displacement and the genetics of gamete recognition in tropical sea urchins. Evolution 57: 1049–1060 [DOI] [PubMed] [Google Scholar]
- Glabe CG, Lennarz WJ 1979. Species specific sperm adhesion in sea urchins: A quantitative investigation of bindin-mediated egg agglutination. J Cell Biol 83: 595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glabe CG, Vacquier VD 1977. Species specific agglutination of eggs by bindin isolated from sea urchin sperm. Nature 267: 836–838 [DOI] [PubMed] [Google Scholar]
- Gould MC, Stephano JL 2003. Polyspermy prevention in marine invertebrates. Microsc Res Technol 61: 379–388 [DOI] [PubMed] [Google Scholar]
- Gunaratne HJ, Moy GW, Kinukawa M, Miyata S, Mah SA, Vacquier VD 2007. The 10 sea urchin receptor for egg jelly proteins (suREJ) are members of the polycystic kidney disease-1 (PKD1) family. BMC Genomics 8: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi TI, Vose M, Gavrilets S 2007. Genetic differentiation by sexual conflict. Evolution 61: 516–529 [DOI] [PubMed] [Google Scholar]
- Haygood R 2004. Sexual conflict and protein polymorphism. Evolution 58: 1414–1423 [DOI] [PubMed] [Google Scholar]
- Hellberg ME, Vacquier VD 1999. Rapid evolution of fertilization selectivity and lysin cDNA sequences in teguline gastropods. Mol Biol Evol 16: 839–848 [DOI] [PubMed] [Google Scholar]
- Hellberg ME, Moy GW, Vacquier VD 2000. Positive selection and propeptide repeats promote rapid interspecies divergence of a gastropod sperm protein. Mol Biol Evol 17: 458–466 [DOI] [PubMed] [Google Scholar]
- Hirohashi N, Vacquier VD 2002. High molecular mass egg fucose sulfate polymer is required for opening both calcium channels involved in triggering the sea urchin sperm acrosome reaction. J Biol Chem 277: 1182–1189 [DOI] [PubMed] [Google Scholar]
- Jovine L, Darie CC, Litscher ES, Wassarman PM 2005. Zona pellucida domain proteins. Ann Rev Biochem 74: 83–114 [DOI] [PubMed] [Google Scholar]
- Kamei N, Glabe CG 2003. The species specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev 17: 2502–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresge N, Vacquier VD, Stout CD 2001a. The crystal structure of a fusagenic sperm protein reveals extreme surface properties. Biochemistry 40: 5407–5413 [DOI] [PubMed] [Google Scholar]
- Kresge N, Vacquier VD, Stout CD 2001b. Abalone lysin: The dissolving and evolving sperm protein. BioEssays 23: 95–103 [DOI] [PubMed] [Google Scholar]
- Landry C, Geyer LB, Arakak Y, Uehara T, Palumbi SR 2003. Recent speciation in the indo-west Pacific: Rapid evolution of gamete recognition and sperm morphology in cryptic species of sea urchin. Proc R Soc Lond B 270: 1839–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-H 2003. Molecular phylogenies and divergence times of sea urchin species of Strongylocentrotidae, Echinoida. Mol Biol Evol 20: 1211–1221 [DOI] [PubMed] [Google Scholar]
- Lee Y-H, Vacquier VD 1992. The divergence of species specific abalone sperm lysin is promoted by positive Darwinian selection. Biol Bull 182: 97–104 [DOI] [PubMed] [Google Scholar]
- Lee Y-H, Ota T, Vacquier VD 1995. Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol Biol Evol 12: 231–238 [DOI] [PubMed] [Google Scholar]
- Lessios HA 2007. Reproductive isolation between species of sea urchins. Bull Mar Sci 81: 191–208 [Google Scholar]
- Levitan DR, Ferrell DL 2006. Selection on gamete recognition proteins depends on sex, density and genotype frequency. Science 312: 267–269 [DOI] [PubMed] [Google Scholar]
- Levitan DR, Stapper AP 2010. Simultaneous positive and negative frequency dependent selection on sperm bindin, a gamete recognition protein in the sea urchin Strongylocentrotus purpuratus. Evolution 64: 785–797 [DOI] [PubMed] [Google Scholar]
- Levitan DR, terHorst CP, Fogarty ND 2007. The risk of polyspermy in three congeneric sea urchins and its implications for gamete incompatibility and reproductive isolation. Evolution 61: 2009–2016 [DOI] [PubMed] [Google Scholar]
- Lyon JD, Vacquier VD 1999. Interspecies chimeric sperm lysins identify regions mediating species-specific recognition of the abalone egg vitelline envelope. Dev Biol 214: 151–159 [DOI] [PubMed] [Google Scholar]
- Mah SA, Swanson WJ, Vacquier VD 2005. Positive selection in the carbohydrate recognition domains of sea urchin sperm receptor for egg jelly (suREJ) proteins. Mol Biol Evol 22: 533–541 [DOI] [PubMed] [Google Scholar]
- Manier MK, Palumbi SR 2008. Intraspecific divergence in sperm morphology of the green sea urchin, Strongylocentrotus droebachiensis: Implications for selection in broadcast spawners. BMC Evol Biol 8: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks JA, Biermann CH, Eanes WF, Kryvi H 2008. Sperm polymorphism within the sea urchin Strongylocentrotus droebachiensis: Divergence between Pacific and Atlantic Oceans. Biol Bull 215: 115–125 [DOI] [PubMed] [Google Scholar]
- Mayr E 1997. Perspective: The objects of selection. Proc Natl Acad Sci 94: 2091–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney MA, Lessios HA 2002. Quantitative analysis of gamete incompatibility between closely related species of neotropical sea urchins. Biol Bull 202: 166–181 [DOI] [PubMed] [Google Scholar]
- McCartney MA, Lessios HA 2004. Adaptive evolution of sperm bindin tracks egg incompatibility in neotropical sea urchins of the genus Echinometra. Mol Biol Evol 21: 732–745 [DOI] [PubMed] [Google Scholar]
- Mengerink KJ, Moy GW, Vacquier VD 2002. suREJ3, a polycystin-1 protein, is cleaved at the GPS domain and localizes to the acrosomal region of sea urchin sperm. J Biol Chem 277: 943–948 [DOI] [PubMed] [Google Scholar]
- Metz EC, Palumbi SR 1996. Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol Biol Evol 13: 397–406 [DOI] [PubMed] [Google Scholar]
- Metz EC, Kane RE, Yanagimachi H, Palumbi SR 1994. Fertilization between closely related sea urchins is blocked by incompatibilities during sperm-egg attachment and early stages of fusion. Biol Bull 187: 23–34 [DOI] [PubMed] [Google Scholar]
- Metz EC, Gomez-Gutierrez G, Vacquier VD 1998a. Mitochondrial DNA and bindin gene sequence evolution among allopatric species of the sea urchin genus Arbacia. Mol Biol Evol 15: 185–195 [DOI] [PubMed] [Google Scholar]
- Metz EC, Robles-Sikisaki R, Vacquier VD 1998b. Nonsynonymous substitution in abalone sperm fertilization genes exceeds substitution in introns and mitochondrial DNA. Proc Natl Acad Sci 95: 10676–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor JE, Fromson DR, Britten RJ, Davidson EH 1991. Comparison of the bindin proteins of Strongylocentrotus franciscanus, S. purpuratus and Lytechinus variegates: Sequences involved in the species specificity of fertilization. Mol Biol Evol 8: 781–795 [DOI] [PubMed] [Google Scholar]
- Monné M, Han L, Schwend T, Burendahl S, Jovine L 2008. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature 456: 653–657 [DOI] [PubMed] [Google Scholar]
- Moy GW, Vacquier VD 1979. Immunoperoxidase localization of bindin during the adhesion of sperm to sea urchin eggs. Curr Top Dev Biol 13: 31–44 [DOI] [PubMed] [Google Scholar]
- Moy GW, Vacquier VD 2008. Bindin genes of the Pacific oyster Crassostrea gigas. Gene 423: 215–220 [DOI] [PubMed] [Google Scholar]
- Moy GW, Mendoza LM, Swanson WJ, Glabe CG, Vacquier VD 1996. The sea urchin sperm receptor for egg jelly is a modular protein with extensive homology to the human polycystic kidney disease protein, PKD1. J Cell Biol 133: 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy GW, Springer SA, Adams SL, Swanson WJ, Vacquier VD 2008. Extraordinary intraspecific diversity in oyster sperm bindin. Proc Natl Acad Sci 105: 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunney L, Schuenzel EL 2006. Detecting natural selection at the molecular level: A reexamination of some classic examples of adaptive evolution. J Mol Evol 62: 176–195 [DOI] [PubMed] [Google Scholar]
- Palumbi SR 1999. All males are not created equal: Fertility differences depend on gamete recognition polymorphisms in sea urchins. Proc Natl Acad Sci 96: 12632–12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi SR 2009. Speciation and the evolution of gamete recognition genes: Pattern and process. Heredity 102: 66–76 [DOI] [PubMed] [Google Scholar]
- Palumbi SR, Lessios HA 2005. Evolutionary animation: How do molecular phylogenies compare to Mayr’s reconstruction of speciation patterns in the sea? Proc Natl Acad Sci 102: 6566–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi SR, Metz EC 1991. Strong reproductive isolation between closely related tropical sea urchins (genus Echinometra). Mol Biol Evol 8: 227–239 [DOI] [PubMed] [Google Scholar]
- Patiño S, Aagaard JE, MacCoss MJ, Swanson WJ, Hart MW 2009. Bindin from a sea star. Evol Dev 11: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR, Holland B 1997. The enemies within: Intergenomic conflict. Interlocus contest evolution, ICE and the intraspecific red queen. Behav Ecol Sociobiol 41: 1–10 [Google Scholar]
- Riginos C, McDonald JH 2003. Positive selection on an acrosomal sperm protein, M17 lysin, in three species of the mussel genus Mytilus. Mol Biol Evol 20: 200–207 [DOI] [PubMed] [Google Scholar]
- Riginos C, Wang D, Abrams AJ 2006. Geographic variation and positive selection on M7 lysin, an acrosomal sperm protein in mussels (Mytilus spp). Mol Biol Evol 23: 1952–1965 [DOI] [PubMed] [Google Scholar]
- Rocha S, Lucio M, Pereira MC, Reis S, Brezesinski G 2008. The conformation of fusagenic B18 peptide in surfactant solutions. J Pept Sci 14: 436–441 [DOI] [PubMed] [Google Scholar]
- Shaw A, McRee DE, Vacquier VD, Stout CD 1993. The crystal structure of lysin, a fertilization protein. Science 262: 1864–1867 [DOI] [PubMed] [Google Scholar]
- Shaw A, Fortes PAG, Stout CD, Vacquier VD 1995. Crystal structure and subunit dynamics of the abalone sperm lysin dimer: Egg envelopes dissociate dimers, the monomer is the active species. J Cell Biol 130: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter C, McCartney MA, Yund PO 2008. Comparison of gamete compatibility between two blue mussel species in sympatry and in allopatry. Biol Bull 214: 57–66 [DOI] [PubMed] [Google Scholar]
- Springer SA, Crespi BJ 2007. Adaptive gamete recognition divergence in a hybridizing Mytilus population. Evolution 61: 772–783 [DOI] [PubMed] [Google Scholar]
- Springer SA, Moy GW, Friend DS, Swanson WJ, Vacquier VD 2008. Oyster sperm bindin is a combinatorial fucose lectin with remarkable intra-species diversity. Int J Dev Biol 52: 759–768 [DOI] [PubMed] [Google Scholar]
- Stephano JL, Gould M 1988. Avoiding polyspermy in oyster (Crassostrea gigas). Aquaculture 73: 295–307 [Google Scholar]
- Swanson WJ, Vacquier VD 1995a. Liposome fusion induced by a M(r) 18,000 protein localized to the acrosomal region of acrosome-reacted abalone spermatozoa. Biochemistry 34: 14202–14208 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD 1995b. Extraordinary divergence and positive Darwinian selection in a fusagenic protein coating the acrosomal process of abalone spermatozoa. Proc Natl Acad Sci 92: 4957–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD 1997. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc Natl Acad Sci 94: 6724–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD 1998. Concerted evolution in an egg receptor for a rapidly evolving abalone sperm protein. Science 281: 710–712 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD 2002. The rapid evolution of reproductive proteins. Nat Rev Genet 3: 137–144 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Yang Z, Wolfner MF, Aquadro CF 2001. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc Natl Acad Sci 98: 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Aagaard JE, Vacquier VD, Monné M, Sadat Al Hosseini H, Jovine L 2011. The molecular basis of sex: Linking yeast to humans. Mol Biol Evol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togo T, Morisawa M 1999. Mechanisms for blocking polyspermy in oocytes of the oyster Crassostrea gigas. J Exp Zool 283: 307–314 [Google Scholar]
- Tomaiuolo M, Levitan DR 2010. Modeling how reproductive ecology can drive protein diversification and result in linkage disequilbrium between sperm and egg proteins. Am Nat 176: 14–25 [DOI] [PubMed] [Google Scholar]
- Turner LM, Hoekstra HE 2008. Causes and consequences of evolution of reproductive proteins. Int J Dev Biol 52: 769–780 [DOI] [PubMed] [Google Scholar]
- Turner LM, Chuong EB, Hoekstra HE 2008. Comparative analysis of testis protein evolution in rodents. Genetics 179: 2075–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier VD 1998. Evolution of gamete recognition proteins. Science 281: 1995–1998 [DOI] [PubMed] [Google Scholar]
- Vacquier VD, Hirohashi N 2004. Sea urchin spermatozoa. Meth Cell Biol 74: 524–540 [DOI] [PubMed] [Google Scholar]
- Vacquier VD, Moy GW 1977. Isolation of bindin: The protein responsible for adhesion of sperm to sea urchin eggs. Proc Natl Acad Sci 74: 2456–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacquier VD, Swanson WJ, Hellberg ME 1995. What have we learned about sea urchin sperm bindin? Dev Growth Differ 37: 1–10 [DOI] [PubMed] [Google Scholar]
- Van Doorn GS, Luttikhuizen PC, Weissing FJ 2001. Sexual selection at the protein level drives the extraordinary divergence of sex related genes during sympatric speciation. Proc R Soc Lond B 268: 2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela-Silva A-CES, Hirohashi N, Mourao PAS 2008. The structure of sulfated polysaccharides ensures a carbohydrate-based mechanism for species recognition during sea urchin fertilization. Int J Dev Biol 52: 551–559 [DOI] [PubMed] [Google Scholar]
- Wassarman PM, Litscher ES 2008. Mammalian fertilization: The egg’s multifunctional zona pellucida. Int J Dev Biol 52: 665–676 [DOI] [PubMed] [Google Scholar]
- Wong JL, Wessel GM 2008. Renovation of the egg extracellular matrix at fertilization. Int J Dev Biol 52: 545–550 [DOI] [PubMed] [Google Scholar]
- Wu HH, Su B 2008. Adaptive evolution of SCML1 in primates, a gene involved in male reproduction. BMC Evol Biol 8: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Li L, Zhang G 2010. Crassostrea angulata bindin gene and the divergence of fucose-binding lectin repeats among three species of Crassostrea. Mar Biotechnol [DOI] [PubMed] [Google Scholar]
- Xie S, Green J, Bixby JB, Szafranska B, DeMartini JC, Hecht S, Roberts RM 1997. The diversity and evolutionary relationships of the pregnancy-associated glycoproteins, an aspartic proteinase subfamily consisting of many trophoblast-expressed genes. Proc Natl Acad Sci 94: 12809–12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Swanson WJ, Vacquier VD 2000. Maximum-likelihood analysis of molecular adaptation in abalone sperm lysin reveals variable selective pressures among lineages and sites. Mol Biol Evol 17: 1446–1455 [DOI] [PubMed] [Google Scholar]
- Zigler KS 2008. The evolution of sea urchin sperm bindin. Int J Dev Biol 52: 791–796 [DOI] [PubMed] [Google Scholar]
- Zigler KS, Lessios HA 2003a. 250 million years of bindin evolution. Biol Bull 205: 8–15 [DOI] [PubMed] [Google Scholar]
- Zigler KS, Lessios HA 2003b. Evolution of bindin in the pantropical sea urchin Tripneustes: Comparisons to bindin of other genera. Mol Biol Evol 20: 220–231 [DOI] [PubMed] [Google Scholar]
- Zigler KS, Lessios HA 2004. Speciation on the coasts of the new world: Phylogeography and the evolution of bindin in the sea urchin genus Lytechinus. Evolution 58: 1225–1241 [DOI] [PubMed] [Google Scholar]
- Zigler KS, Raff EC, Popodi E, Raff RA, Lessios HA 2003. Adaptive evolution of bindin in the genus Heliocidaris is correlated with the shift to direct development. Evolution 57: 2293–2304 [DOI] [PubMed] [Google Scholar]
- Zigler KS, McCartney MA, Levitan DR, Lessios HA 2005. Sea urchin bindin divergence predicts gamete compatibility. Evolution 59: 2399–2404 [PubMed] [Google Scholar]