Background: During feeding, mosquito saliva modulates the vertebrate host's hemostasis and inflammation response.
Results: Anopheles gambiae Sgs4 and Sgs5 are major immunogenic proteins in mosquito saliva.
Conclusion: Sgs4 and Sgs5 appear to play an essential role in blood feeding.
Significance: Sgs4 and Sgs5 could serve as markers of human exposure to mosquito bites and in the development of disease control strategies.
Keywords: Infectious Diseases, Innate Immunity, Insect, Invertebrates, Malaria, Aedes, Anopheles, Mosquito, Blood Feeding
Abstract
Mosquitoes transmit Plasmodium and certain arboviruses during blood feeding, when they are injected along with saliva. Mosquito saliva interferes with the host's hemostasis and inflammation response and influences the transmission success of some pathogens. One family of mosquito salivary gland proteins, named SGS, is composed of large bacterial-type proteins that in Aedes aegypti were implicated as receptors for Plasmodium on the basal salivary gland surface. Here, we characterize the biology of two SGSs in the malaria mosquito, Anopheles gambiae, and demonstrate their involvement in blood feeding. Western blots and RT-PCR showed that Sgs4 and Sgs5 are produced exclusively in female salivary glands, that expression increases with age and after blood feeding, and that protein levels fluctuate in a circadian manner. Immunohistochemistry showed that SGSs are present in the acinar cells of the distal lateral lobes and in the salivary ducts of the proximal lobes. SDS-PAGE, Western blots, bite blots, and immunization via mosquito bites showed that SGSs are highly immunogenic and form major components of mosquito saliva. Last, Western and bioinformatic analyses suggest that SGSs are secreted via a non-classical pathway that involves cleavage into a 300-kDa soluble fragment and a smaller membrane-bound fragment. Combined, these data strongly suggest that SGSs play an important role in blood feeding. Together with their role in malaria transmission, we propose that SGSs could be used as markers of human exposure to mosquito bites and in the development of disease control strategies.
Introduction
The saliva of hematophagous arthropods is a mixture of proteins and small molecules that assists in the acquisition of a blood meal by interfering with the host's hemostasis and inflammation response (1, 2). The injection of arthropod saliva into the vertebrate body also elicits an adaptive immune response that produces antibodies against some salivary constituents. For these reasons, the salivary components of blood-feeding arthropods have been proposed as markers used for surveying exposure to vector bites (3), as targets for transmission blocking vaccines (4), and as a pharmacopoeia of novel therapeutic agents (1). Before such applications can be developed, a better understanding of the “sialomes” of major disease vectors is necessary so that the best candidates for any of these uses can be identified. This task is compounded by the fact that functionally analogous salivary proteins are often not orthologous between groups of arthropods, so it is important to identify proteins from each relevant organism (1).
Mosquitoes are the most menacing arthropod disease vectors, transmitting a broad range of viral, protozoan, and metazoan pathogens. Perhaps the most practical use for mosquito saliva is as a tool for surveying human exposure to mosquito bites (3). A large percentage of the world's population lives in areas where the rate of mosquito-borne disease transmission is not accurately reflected by standard entomological measures of mosquito activity (5, 6). Recent efforts have aimed to improve the sensitivity of vector exposure methods by identifying mosquito-specific saliva proteins that are immunogenic following delivery via bite. A candidate is gSG6, a protein found only in mosquitoes of the genus Anopheles that has shown promise in several laboratory and field studies (6, 7). The identification of other mosquito-specific (and genus-specific) salivary immunogens should improve this epidemiological method, and published data suggest that there are other yet unidentified saliva proteins that are more immunogenic than gSG6 (8, 9).
The saliva of mosquitoes and other arthropods has also been shown to modulate vertebrate immune responses, resulting in the enhanced transmission of certain pathogens (4, 10). Accordingly, vaccines that help incite the host antibody response against salivary immunomodulatory factors have been shown to lower the transmission rates of Leishmania parasites and have been proposed for use against other arthropod-vectored diseases (4, 11). Although mosquito saliva has conclusively been shown to enhance the transmission of several viruses (10), its role in the transmission of malaria parasites remains a point of contention (12–14). Thus, the identification of novel immunomodulatory components of mosquito saliva may aid in the development of disease control strategies.
In the past decade, transcriptomic and proteomic studies have greatly increased our knowledge of the sialomes of a number of arthropod vectors (1, 2), with transcriptomic studies in Anopheles gambiae, Aedes aegypti, and Culex pipiens specifically increasing our understanding of the mixture of proteins found in mosquito salivary glands (15–17). During a transcriptomic analysis of female A. gambiae salivary glands, Arcà et al. (16) identified a contiguous series of large genes of unknown function (Vector base IDs: ENSANGP00000027299, ENSANGP00000027791, and ENSANGP00000029569; later named SGS2 by Korochkina et al. (18)) that had not been predicted during earlier genome scans and that appear to have been horizontally transferred into the mosquito genome from a bacterial source whose only known living relatives are Wolbachia proteobacteria (19). A member of this gene family was independently identified while screening a panel of monoclonal antibodies produced against A. aegypti salivary gland surface proteins. This protein, named aaSgs1 (GenBankTM number AAV28546), was shown to be required for the successful invasion of A. aegypti salivary glands by Plasmodium gallinaceum sporozoites (18). Thus, the biology of SGSs is intriguing, because they have been implicated in the Plasmodium life cycle and represent a rare case of transdomain horizontal gene transfer.
Using an array of complementary techniques, we conclusively show that two members of the A. gambiae SGS gene family, Sgs4 and Sgs5, are associated with blood feeding behavior, form prevalent components of mosquito saliva, and are major salivary immunogens. Further, we show that SGSs are not restricted to A. gambiae saliva, because they are primary saliva constituents in both the anopheline and culicine lineages. Based on these and other data (20), we propose a putative role for SGSs as major immunomodulatory factors in mosquito saliva and offer an explanation for why SGSs have largely been overlooked in previous sialomic studies.
EXPERIMENTAL PROCEDURES
Animal Rearing and Tissue Collection
A. gambiae (G3) and A. aegypti (LVP) mosquitoes were reared as described (21). Briefly, larvae were hatched in plastic water containers and fed a mixture of koi food and yeast. Pupae were separated by size and allowed to eclose in 4.73-liter plastic containers with marquisette tops, and the adults were maintained on a 10% sugar solution at 27 °C, 75% relative humidity, and a 12-h light/12-h dark photoperiod with 30-min crepuscular periods that preceded and followed each light cycle. Unless otherwise stated, 5-day-old adult female mosquitoes were used for all experiments.
Salivary glands were collected by submerging mosquitoes in phosphate-buffered saline (PBS) and pulling the heads off with watchmaker's forceps. Minuten pins held by pin vices were then used to separate the glands from the head or to search through the anterior thorax in the cases when the glands did not pull cleanly from the thorax.
Mosquito saliva was collected by inducing salivation using pilocarpine. For A. gambiae, this was done in a manner similar to that of Remoue et al. (3), except that topical application of 1% (w/v) pilocarpine with 0.2% Tween 20 in PBS (22) was used instead of malathion, and saliva was collected into mineral oil rather than water. Because A. aegypti are more heavily pubescent, salivation in this mosquito was instead induced by intrathoracic injection of 0.2 μl of a 0.01% (w/v) solution of pilocarpine in PBS. Only saliva from mosquitoes that visibly produced small droplets of saliva and appeared healthy after the end of the procedure was used in these experiments.
A. gambiae bite blots were done by attaching a nitrocellulose membrane to the bottom of a water-filled glass bottle heated to 58 °C and pressing the membrane against the freshly replaced marquisette cover of a mosquito cage. Within seconds, female mosquitoes were attracted to the heat and began probing the membrane with their proboscis, and this was allowed to continue in the dark for 10 min. Because heat is a principal cue for blood feeding (23), we believe that this method stimulates the production of saliva in a manner similar to natural blood feeding, as opposed to the sugar-based collection methods used in prior studies (24).
Following early suspicions that SGS levels might be affected by feeding behavior or circadian changes in expression, tissue collections were done between hours 6 and 8 of the light photoperiod, unless otherwise stated, and collection began at exactly the same times within each experiment.
Protocols used in this study for the maintenance and use of vertebrate animals were approved by Vanderbilt University's Institutional Animal Care and Use Committee. Vanderbilt University's Division of Animal Care oversaw and provided veterinary care for this study and is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Gene Expression Analyses
For non-salivary gland tissues, RNA from 10 mosquitoes was isolated using TRIzol reagent (Invitrogen) and repurified using the PurelinkTM Micro-to-Midi total RNA purification System (Invitrogen). For salivary gland samples, RNA from 50 glands was isolated directly using the Purelink system, per the manufacturer's protocol for animal tissue samples. Up to 5 μg of RNA per sample was then treated with RQ1 RNase-free DNase (Promega, Madison, WI), and first strand cDNA was synthesized from poly(A) + RNA using the SuperScript® III first-strand synthesis system (Invitrogen). A standard phenol/chloroform extraction was used to purify the cDNA, and samples were then quantified and normalized. Standard PCR was conducted on a Bio-Rad DNA Engine® thermal cycler using Choice-TaqTM DNA polymerase (Denville Scientific, Metuchen, NJ). Real-time quantitative PCR was done using Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA) on an ABI 7300 real-time PCR system, and relative quantification was carried out using the 2−ΔΔCT method (25). For both standard PCR and quantitative PCR, rpS7 was used as the reference gene. Primers used for standard PCR were Sgs4_d_F (5′-GCTGTTCCTTCGAAACTTGC-3′), Sgs4_d_R (5′-TCGCCGTGTATACCAATGAA-3′); Sgs5_d_F (5′-ACCGCAACCGTAGCAATAAC-3′), Sgs5_d_R (5′-TCGAACAATCTGGGAGGTTC-3′), rpS7_01F (5′-CGTGAGGTCGAGTTCAACAA-3′), and rpS7_01R (5′-GCTGCAAACTTCGGCTATTC-3′). Primers used for quantitative PCR were Sgs4_q01F (5′-TAACAACCCGCAGGAGATTC-3′), Sgs4_q01R (5′-GCTGCTGAATCGTTTCCTTC-3′), Sgs5_q01F (5′-GCATCGGATCGTGGAACTAT-3′), Sgs5_q01R (5′-GTGGTGCTTGGGATGAAACT-3′), rpS7_02F (5′-GACGGATCCCAGCTGATAAA-3′), and rpS7_02R 5′-GTTCTCTGGGAATTCGAACG-3′.
Antibody Production
Anti-SGS antibodies were produced by challenging rabbits with recombinant fragments of Sgs4 and Sgs5. Three polyclonal antibodies were developed. The first, referred to throughout this work as α-SGSRHS, recognizes both Sgs4 and Sgs5 and was produced against a region of Sgs4 that shares 96% amino acid identity with Sgs5. This region, residues 2248–2571 of Sgs4, is located directly N-terminal of the predicted transmembrane domain and includes the RHS (retrotransposon hot spot) domain. The other two antibodies, referred throughout this manuscript as α-Sgs4 and α-Sgs5, specifically recognize Sgs4 or Sgs5 by targeting unique regions located 88–549 and 512–822 residues from the N terminus, respectively. Briefly, the SGS regions detailed above were amplified by PCR using Accuprime Pfx SuperMix (Invitrogen). Resulting amplicons were cloned into the pET-46 Ek/LIC expression vector (Novagen, Madison, WI), and proteins were expressed in BL21(DE3) Escherichia coli cells. N-terminal His-tagged protein fragments were purified using BD TALON metal affinity resin (Clontech), and polyclonal antibodies were produced in rabbits (Affinity Bioreagents, Golden, CO). The IgG fraction of immune and preimmune sera was purified using a KPL Protein A-agarose kit (KPL, Gaithersburg, MD), concentrated using a 30-kDa cut-off Microcon® filter (Millipore Corp., Billerica, MA), and buffer-exchanged to PBS. Antibody concentrations were determined by measuring A280 (26). Primers used to amplify the cloned fragments were as follows: for α-SGSRHS, Sgs4&5Pos_VS (5′-GACGACGACAAGATGTGTGGAAAACGACTCGCTCTGAAT-3′) and Sgs4&5Neg_VS (5′-GAGGAGAAGCCCGGTCTTACCCCGAACCTGCTTGAATGT-3′); for α-Sgs4, Sgs4_5P2_F (5′-GACGACGACAAGATCAAGGTGAAAGGGTTCGTGT-3′) and Sgs4_5P2_R (5′-GAGGAGAAGCCCGGTCGTCGCTTGCTCAAGTTTTCC-3′); for α-Sgs5, Sgs5_Pos_VS (5′-GACGACGACAAGATGCGACAAACGCAAACCTTCACATCG-3′) and Sgs5Neg_VS (5′-GAGGAGAAGCCCGGTTGCTTTCACGGTTACCCATTCTCC-3′) (vector sequences are underlined).
Antibodies against A. gambiae saliva were produced in female Swiss Webster mice via exposure to mosquito bites. Mice were placed on top of cages containing ∼150 starved female mosquitoes and fed upon for 5 min. Halfway through the procedure, mice were lifted off the cage and quickly repositioned, which induced the mosquitoes to reprobe. Blood was collected by cardiac puncture, and serum was purified (26) at the end of each of three bite regimens: 1 week after a single bite exposure, 1 week after the last of three weekly bite exposures, and 1 week after the last of six weekly bite exposures. Non-immune control sera were purified from age- and cage-matched naive mice.
SDS-PAGE, Immunoblots, and Total Protein Staining
In order to accommodate the large size of SGSs, a variety of gels and protein standards were used throughout the study. These varied depending on the level of resolution and the protein mass inclusion best suited for an individual experiment. Whole tissues (salivary glands, thoraces, etc.) were either homogenized in 1% Nonidet P-40 buffer containing Complete® protease inhibitors (Roche Applied Science) prior to mixing with denaturing NuPage® LDS loading buffer (Invitrogen) containing 2.5% β-mercaptoethanol or were homogenized directly in denaturing buffer with β-mercaptoethanol. Collected saliva was pooled, vigorously mixed with LDS sample buffer, and separated from the mineral oil by centrifugation at 8,600 × g for 5 min. Whole tissue or saliva samples were then electrophoresed in either 4% Tris/glycine polyacrylamide gels for 2 h at 125 V, 3–8% Tris acetate polyacrylamide gels for 1 h at 150 V, or 4–12% BisTris polyacrylamide gels for 1 h at 200 V, and separated proteins were transferred to PVDF membranes (Invitrogen) using the manufacturer's protocol. Western blotting was subsequently performed using the KPL Protein DetectorTM LumiGLO Western blotting kit (Gaithersburg, MD). When necessary, Western blots were stripped using RestoreTM Western blot stripping buffer (Thermo Scientific, Rockford, IL) prior to reprobing.
Bite blots of mosquito saliva were treated in a manner similar to our Western blots and the bite blots of previous authors (24). A step similar to a standard transfer was included to ensure that saliva proteins were strongly bound to the membrane.
In order to assess the presence of total proteins in salivary samples, an imidazole-zinc negative detection protocol was used (27). Although imidazole-zinc staining is very sensitive, it does not allow for the quantification of relative band content. When this was necessary, a quantitative Coomassie staining protocol was used (28). Gels were digitally scanned in a manner that avoided signal saturation, and densitograms were produced in ImageJ (National Institutes of Health, Bethesda, MD).
Immunohistochemistry
Salivary glands were dissected, fixed for 1 min in 100% acetone, and washed three times for 5 min each in PBS. Washed samples were blocked with 2% bovine serum albumin in PBS (B-PBS) for 1 h, rinsed three times for 5 min each in PBS, and incubated in rabbit α-Sgs4, α-Sgs5, or preimmune IgG in B-PBS for 1 h. Samples were then washed as above, incubated in goat α-rabbit Alexa Fluor 568 (Invitrogen) with 0.01 mg/ml Hoescht 33342 in B-PBS for 1 h, washed, and mounted on slides using Aqua-Poly/Mount (Polysciences Inc., Warrington, PA). Salivary glands were imaged using differential interference contrast and fluorescence illumination on a Nikon 90i light microscope (Nikon Corp., Tokyo, Japan) connected to a Photometrics CoolSNAP HQ2 high sensitivity monochrome CCD camera (Roper Scientific, Ottobrunn, Germany), and Z-stack images were captured and analyzed using Nikon Advanced Research NIS-Elements software. To determine the precise location of SGS staining, Z-stacks were processed using the AQ 3D Blind Deconvolution module of NIS-Elements, and for the purpose of publication, Z-stacks were flattened into two dimensions using the Maximum Intensity Projection module of NIS-Elements. Throughout, Protein A-purified primary (immune and preimmune) and secondary antibodies were used at concentrations of 10 and 2.6 μg/ml, respectively.
Bioinformatic Analyses
SGS sequences were scanned for functional sites that occur in non-structured regions using the Eukaryotic Linear Motif server (available on the World Wide Web), applying A. gambiae as the taxonomic range filter. The ExPASy PeptideCutter tool was used to scan for putative caspase-1 family cleavage sites (see the ExPASy Web site), and SignalP 3.0 and SecretomeP 2.0 (both at the Center for Biological Sequence Analysis Web site) were used to search for classical and non-classical signal peptide cleavage sites, respectively. Following prediction of protease cleavage sites, the predicted molecular weights of SGS fragments were calculated using the Compute pI/Mw tool in ExPASy, and ExPASy ProtScale was used to plot hydrophobicity using the Rao and Argos scale (29).
RESULTS
Sgs4 and Sgs5 Are Expressed Exclusively in the Salivary Glands
To determine the tissue-specific transcription of Anopheles SGSs, cDNA synthesized from salivary glands, heads, thoraces, thoraces from which the salivary glands had been removed, or abdomens of female mosquitoes was used as template for PCR using gene-specific primers. PCR revealed that sgs4 (GenBankTM number AAV28544) and sgs5 (GenBankTM number AAV28545) are only transcribed in the salivary glands and in the thorax but that when the salivary glands are removed from the thorax, this signal disappears (Fig. 1A). Quantitative PCR confirmed this finding and further showed that mRNA levels of sgs4 and sgs5 are 120- and 143-fold higher, respectively, in the salivary glands when compared with the whole body (Fig. 1B). When mRNA levels of sgs2 and sgs3 were assayed, expression was inconsistently detected and only in non-salivary gland tissues (not shown). For this reason, Sgs2 and Sgs3 were not studied further.
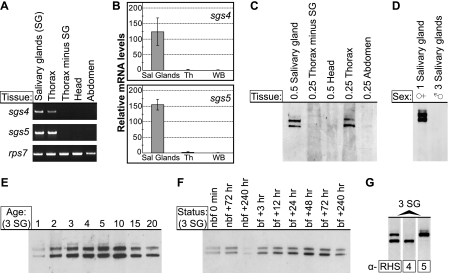
FIGURE 1.
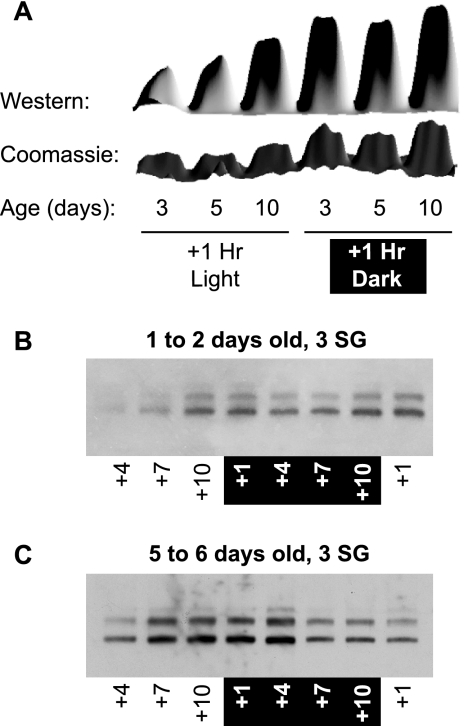
Sgs4 and Sgs5 are produced specifically in the salivary glands of adult female A. gambiae, and their production is associated with blood feeding. A and B, conventional PCR (A) and real-time quantitative PCR (B) comparing sgs transcript levels in tissues from 5-day-old adult mosquitoes. sgs4 and sgs5 are only transcribed in the salivary glands. C and D, Western blots of tissue samples from 5-day-old adult A. gambiae, showing that Sgs4 and Sgs5 are only present in the salivary glands of female mosquitoes. E, Western blot of A. gambiae salivary glands at different times posteclosion, showing that SGS levels are minimal in newly emerged adults and peak around 10 days of age. F, Western blot of A. gambiae salivary glands at different times following blood feeding (bf) along with non-blood fed controls (nbf). Time 0 represents non-blood-fed 5-day-old mosquitoes, and these data show that blood feeding enhances SGS production. G, the same lane of a Western blot stripped and reprobed with each of the antibodies produced against recombinant SGSs, showing their binding specificities and that Sgs5 is slightly more massive than Sgs4. Western blots in C–F were probed using α-SGSRHS (which recognizes both Sgs4 and Sgs5) and were performed after electrophoresing samples in 4% Tris/glycine SDS-polyacrylamide gels. For an explanation of SGS mass size, see Fig. 6.
Given the salivary gland specificity of sgs4 and sgs5, a polyclonal antibody that recognizes both Sgs4 and Sgs5 was developed (α-SGSRHS). Western analyses revealed that, much like the transcription data, Sgs4 and Sgs5 are salivary gland-specific (Fig. 1C), and interestingly, Sgs4 and Sgs5 are present exclusively in the salivary glands of female mosquitoes (Fig. 1D). Within female salivary glands, Sgs4 and Sgs5 levels are minimal in freshly eclosed mosquitoes and peak around 10 days of age (Fig. 1E). When a blood meal is provided, levels of Sgs4 and Sgs5 immediately increase and remain elevated for the lifetime of the mosquito (Fig. 1F). Given that these data suggest that Sgs4 and Sgs5 are involved in some aspect of blood feeding, antibodies that selectively recognize Sgs4 or Sgs5 were constructed. Stripping and reprobing of the same Western blot with each of the three α-SGS antibodies confirmed the band identities and showed that Sgs5 is slightly more massive than Sgs4 (Fig. 1G).
Sgs4 and Sgs5 Are Present in the Distal-Lateral Acinar Cells and the Salivary Ducts
Analysis of quantitatively deconvolved Z-stacks acquired from salivary glands immunolabeled with α-Sgs4 or α-Sgs5 revealed that both of these proteins are present on the basal and apical cellular surfaces of the distal lateral lobes (Fig. 2, A–J). Lower intensity staining was also detected in the salivary ducts of the proximal lateral lobes, a pattern that also suggests a role in blood feeding. Preimmune controls (Fig. 2, G–I) and midguts labeled with α-Sgs4 and α-Sgs5 showed no staining.
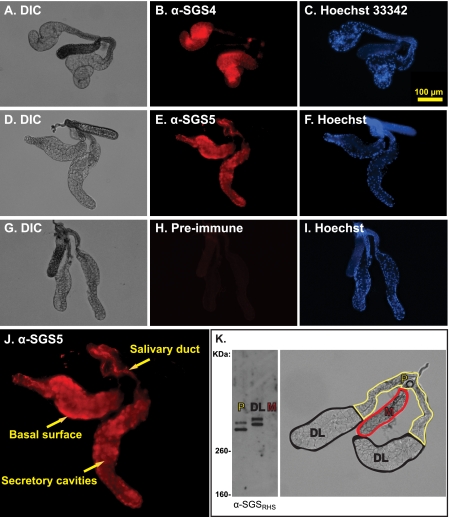
FIGURE 2.
Sgs4 and Sgs5 are present in the distal-lateral acinar cells and the salivary ducts. A–I, immunohistochemistry of female salivary glands using α-Sgs4 (A–C), α-Sgs5 (D–F), or preimmune antibodies (G–I; from a rabbit later immunized with Sgs4). Serial micrographs show imaging through differential interference contrast (DIC) (A, D, and G), Texas Red (B, E, and H; antibody labeling), and Hoechst 33342 (C, F, and I; DNA stain) channels. J, close-up of E showing labeling of the acinar cells of the distal lateral lobes and the salivary ducts of the proximal lobes. K, Western blot of disarticulated salivary glands, confirming that Sgs4 (bottom bands) and Sgs5 (top bands) are present only in the lateral lobes and showing that both are slightly less massive in the proximal lateral lobes than they are in the distal lateral lobes (4% Tris/glycine SDS-PAGE).
Because of the unexpected finding that Anopheles SGSs are present in the salivary ducts, Western blots were conducted in salivary glands that were disarticulated into their three anatomical regions: the distal lateral lobes, the proximal lateral lobes, and the median lobes. Western analyses confirmed the immunolabeling results and suggest that both Sgs4 and -5 are subjected to processing in the proximal lateral region of the salivary glands because in these regions, their mass is reduced by ∼10 kDa (Fig. 2K).
Finally, during the immunolabeling experiments, unambiguous differences between α-SGS samples and preimmune controls were only obtained after fixation with organic solvents (acetone or ethanol) and not following fixation with formaldehyde, regardless of permeabilization with Triton X-100 or heat-induced antigen retrieval. This suggests that SGSs are either located in a region inaccessible to the antibodies following aldehyde fixation or that the antibodies do not efficiently bind the native proteins.
SGSs Form a Major Component of A. gambiae and A. aegypti Saliva
Because expression and localization data suggested that Sgs4 and -5 may be involved in blood feeding, mosquito salivation was artificially induced, and the collected saliva was analyzed for the presence of Sgs4 and Sgs5. Western analyses conclusively detected both Sgs4 and -5 in mosquito saliva (Fig. 3, A and B). In order to ensure that the presence of SGSs in saliva was not the result of pilocarpine treatment, we used heat to entice mosquitoes to probe a nitrocellulose membrane and performed “bite blots” using α-SGS antibodies. Bite blots revealed that Sgs4 and Sgs5 are released with the saliva during probing. Sgs4 and Sgs5 were only detected at the probing sites; no staining was observed outside the probing areas or when 3 times as much preimmune antibody was used instead of α-SGS antibodies (Fig. 3C). Like SGSs found in the proximal lateral salivary lobes (Fig. 2K), SGSs in saliva are ∼10 kDa less massive than SGSs found in the distal lateral lobes.
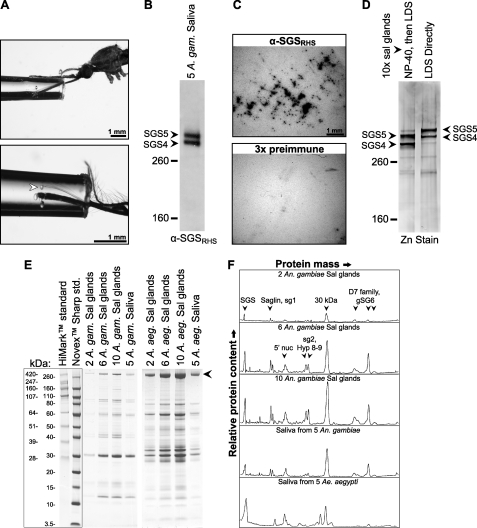
FIGURE 3.
Sgs4 and Sgs5 are the only prevalent proteins in their size range found in mosquito salivary glands and form major constituents of mosquito saliva. A, saliva collection in A. gambiae (top) and A. aegypti (bottom). Droplet formation can be seen in both images, with the A. aegypti image showing saliva being ejected from the hypopharynx (arrowhead). B, Western blot of A. gambiae saliva collected following pilocarpine treatment, showing that Sgs4 and Sgs5 are a component of mosquito saliva. C, bite blots probed with α-SGSRHS (top) or 3 times the amount of preimmune serum (bottom), showing that SGSs are released with the saliva during probing. D, 4% Tris/glycine SDS-polyacrylamide gel stained with zinc-imidazole, showing that Sgs4 and Sgs5 are the only proteins in their size range present in mosquito salivary glands. Note that when glands were immediately immersed in denaturing LDS buffer, their size was ∼10 kDa more massive than when they were incubated in Nonidet P-40 detergent prior to denaturation. Sgs4 and -5 in the saliva match the smaller SGS forms seen in whole salivary glands initially soaked in non-denaturing buffer. E, Coomassie-stained 4–12% SDS-PAGE of salivary glands and saliva from A. gambiae and A. aegypti. The combined SGS bands are marked by an arrowhead. F, densitogram of select lanes from the gel in D, showing relative protein content. In both salivary glands and saliva, SGSs form one of three major peaks in A. gambiae and form the major peak in A. aegypti. Peak identities, other than SGS, are inferred using data from a previous study (45).
To determine the relative levels of SGSs in mosquito saliva, we first stained 4% SDS-polyacrylamide gels with zinc-imidazole and assessed protein bands in the range of 200–500 kDa (Fig. 3D). This experiment yielded two distinct results. First, it showed that SGSs are the only detectable proteins in this size range. Second, it showed that soaking salivary glands in detergent prior to denaturation results in a 10-kDa size reduction. Next, proteins from salivary glands as well as saliva collected by pilocarpine treatment were separated by SDS-PAGE on 4–12% BisTris gradient gels, and relative protein mass content was quantified by Coomassie analysis (Fig. 3, E and F). Within the range of 3.5 kDa to over 500 kDa, SGSs are one of the three most prevalent proteinaceous components of A. gambiae saliva and the most prevalent component of A. aegypti saliva. Taken altogether, these data conclusively show that SGSs are primary components of mosquito saliva in both major mosquito lineages.
SGSs Are a Major Immunogen in Mosquito Saliva
Given the prevalence of Sgs4 and Sgs5 in A. gambiae saliva, we sought to determine whether these proteins elicit an antibody response following blood feeding. Experiments where antisera from mice exposed to mosquito bites were used as primary antibodies in Western blots corroborated the discovery of Sgs4 and Sgs5 as major components of mosquito saliva and also showed that they are highly immunogenic, eliciting a strong IgG response (Fig. 4). Neither antisera from non-immune mice nor antisera from mice exposed to a single round of mosquito bites that occurred ≤1 week before blood collection detected any Anopheles salivary gland protein. However, after three or six weekly exposures to mosquito bites, SGSs were found to be one of two consistently recognized proteins in mosquito saliva, a result that was confirmed by both molecular weight and by stripping membranes and reprobing them with α-SGSRHS, α-Sgs4, or α-Sgs5. Only one other highly immunogenic band was consistently recognized by our anti-saliva antibodies, which was ∼100 kDa under non-reducing conditions and ∼50 kDa under reducing conditions. One other band was observed in one instance under non-reducing conditions and was ∼70 kDa.
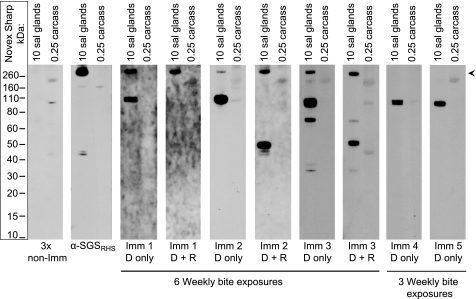
FIGURE 4.
SGSs are one of two major immunogenic components of A. gambiae saliva. Shown are Western blots of salivary glands (sal glands) and carcasses excluding salivary glands (carcass) electrophoresed in 4–12% BisTris gels and probed with the following (from left to right): sera from non-immune mice (non-Imm; 1:80 dilution), α-SGSRHS, sera from mice exposed to weekly mosquito bites for 6 weeks (Imm 1–3; 1:500 dilution), and sera from mice exposed to weekly mosquito bites for 3 weeks (Imm 4–5; 1:500 dilution). Results are shown for extracts run under denaturing (D only) and denaturing plus reducing (D + R) conditions (for sera shown only under denaturing conditions, reducing yielded identical results). Overall, the major immunogenic components of mosquito saliva are SGSs (arrowhead) and another protein that migrates at 110 or 55 kDa, depending on whether it has been reduced.
Salivary Gland SGS Content Is Highest in the Early Evening
Because mosquito feeding behavior follows a circadian cycle, we investigated the prevalence of Sgs4 and Sgs5 at different times of the day. Western blots and Coomassie staining of salivary glands collected every 3 h for 24 h or 1 h into both the light and dark cycles demonstrated that salivary gland SGS content fluctuates with the time of day and that SGS levels are highest in the late afternoon and early evening (Fig. 5, A–C). The SGS circadian gradient was not always as consistent as what is seen in Fig. 5C, but multiple replicates of this experiment support this trend.
FIGURE 5.
SGS levels fluctuate within 24-h periods and are most prevalent during the late afternoon and early evening. A, surface plot of relative combined band intensities of both Sgs4 and Sgs5 from Western (top) and Coomassie (bottom) analyses of salivary glands from mosquitoes of the same cohort, at different ages, collected at +1 h into the light and dark cycles. B and C, Western blots of salivary glands (SG) extracted from 1–2- and 5–6-day-old mosquitoes showing SGS levels over a 24-h period.
Moreover, from these experiments, two additional trends became apparent. First, the initial onset of SGS expression occurs during the mosquito's first night as an adult (Fig. 5B), although protein levels remain relatively low until the second afternoon (Fig. 1E). Second, circadian expression is most accentuated in younger mosquitoes, and the circadian effect diminishes once peak SGS expression is reached at around 10 days of age (Fig. 5A). Given that A. gambiae have a nighttime feeding preference and that newly eclosed mosquitoes do not effectively blood-feed, these circadian data again suggest that Sgs4 and Sgs5 are involved in blood feeding.
Western and Bioinformatic Analyses Suggest That Sgs4 and Sgs5 Are Proteolytically Processed Prior to Secretion
Because SGSs are extremely large and are predicted to have multiple transmembrane regions, it was unexpected to find them in mosquito saliva. Hence, Western and bioinformatic analyses were performed in efforts to explain this phenomenon. The initial question pertained to the mode of secretion. Signal-P detected no classical signal peptides in A. gambiae Sgs4 or Sgs5, or in A. aegypti Sgs1. However, SecretomeP analysis using a Gram-negative model predicted significant likelihoods of non-classical protein secretion for both Sgs4 and Sgs1 (NN-scores >0.5). When a mammalian model was used instead, Sgs4 and -5, as well as Sgs1, all had scores that approached the 0.5 cut-off (an insect model for non-classical secretion does not exist). These findings suggest that SGSs are secreted by non-classical means, a process that is known to involve proteolytic pathways.
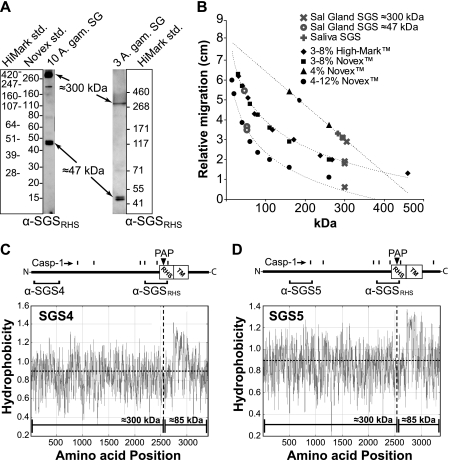
The second question pertained to protein size. The Compute pI/Mw tool in ExPASy predicts that full-length Sgs4 and Sgs5 are >380 kDa. However, Western blots conducted using the 3–8% Tris-acetate polyacrylamide gels that are recommended for high molecular weight proteins, along with the HiMarkTM high weight protein standard, showed that Sgs4 and Sgs5 are ∼300 kDa (Fig. 6, A and B) instead of the ∼220 kDa previously reported for A. aegypti Sgs1 (18). Western blots using 4–12% BisTris and 4% Tris/glycine gels and two high mass standards confirmed the ∼300 kDa mass of Sgs4 and Sgs5 (Fig. 6B). Analyses using gradient gels also detected a smaller doublet of immunoreactive bands that are ∼47 kDa (Fig. 6, A and B), and interestingly, this smaller doublet was detected when using α-SGSRHS but not when using α-Sgs4 or α-Sgs5 (not shown). These data suggest that Sgs4 and Sgs5 are each cleaved into ∼300- and ∼47-kDa fragments.
FIGURE 6.
The masses of Sgs4 and Sgs5, along with bioinformatic analyses, suggest that they are cleaved into a 300-kDa soluble N-terminal fragment and a smaller, membrane-bound fragment. A, Western blots of 10 female salivary glands run on 4–12% BisTris (left) or 3–8% Tris/acetate (right) SDS-polyacrylamide gels and probed with α-SGSRHS. A single band is seen at around 300 kDa that has been shown to correspond to a large fragment of Sgs4 and -5 (one band seen here due to lack of resolution on these gels). A doublet of ∼47 kDa is also observed. B, plot of observed protein migration distances versus two sets of standards on various gel types. Gray symbols represent SGS band migration distances, whereas black symbols and trend lines represent the observed migration of different standards. In all cases, SGS mass is calculated to be 300 ± 10 kDa. C and D, graphical representation of Sgs4 (C) and Sgs5 (D) showing the locations of the putative RHS domains, transmembrane regions (TM), PAPs, caspase-1 cleavage sites (Casp-1), and the regions that the anti-SGS antibodies recognize. Below these gene diagrams, hydrophobicity plots show that cleavage at the PAP sites would result in an N-terminal fragment of ∼300 kDa and a highly hydrophobic membrane-bound fragment of ∼85 kDa (higher values represent higher hydrophobicity).
The third question pertained to the cleavage process. Several sequence analysis programs predict two prophenoloxidase-activating protease (PAP) type cleavage sites (consensus sequence: (I/L/V)XXR(V/F)(G/S)X (30)) that are located N-terminal of the transmembrane domain of both Sgs4 and Sgs5 (between amino acids 2500 and 2650; Fig. 6, C and D) and nowhere else in either protein. In both Sgs4 and Sgs5, one of these sites shares high similarity with the only predicted PAP site in Aedes Sgs1 (consensus sequence from Sgs1, -4, and -5: L(L/T)QRVS(ER)), and is located at the same distance from the N terminus in all three SGSs (from 2533 to 2549). The Compute pI/Mw tool predicts that cleavage of full-length Sgs4 or Sgs5 at this site should result in an N-terminal soluble fragment of ∼300 kDa and a membrane-bound fragment of ∼85 kDa. The ∼300-kDa fragment is the larger mass observed during PAGE analyses (Fig. 6, A and B), whereas the smaller bands recognized by α-SGSRHS migrate at ∼47 kDa rather than the predicted ∼85 kDa (Fig. 6A). However, this smaller fragment is also predicted to contain at least six transmembrane helices and to be highly hydrophobic (Fig. 6, C and D). High hydrophobicity is known to cause excessive SDS binding and faster migration through SDS-polyacrylamide gels, causing a protein to appear much less massive than it actually is (31). As expected under this scenario, the α-SGSRHS antibody that recognizes sequences both N- and C-terminal of the PAP site binds both the 300-kDa and the 47-kDa fragments, but the α-Sgs4 and α-Sgs5 antibodies that recognize sequences located N-terminal of the PAP site recognize only the ∼300-kDa fragment. An alternative possibility for proteolytic processing involves caspase-1-based cleavage because this has been shown to be a regulatory mechanism of non-classical secretion (32). PeptideCutter predicts six and seven caspase-1 type cleavage sites in Sgs4 and Sgs5, respectively, of which four are located between amino acids 2100 and 2670 from the N terminus of both Sgs4 and Sgs5. Again, the ∼47- and ∼300-kDa fragments recognized by α-SGSRHS could be explained by several of the predicted caspase-1 sites (Fig. 6, C and D), offering another plausible explanation for SGS cleavage and subsequent release into the saliva.
Finally, SGSs appear to undergo an additional processing step in the salivary duct of the proximal-lateral lobes. When whole salivary glands are placed in denaturing LDS buffer immediately following dissection, a ∼300-kDa SGS form is by far the most prevalent. However, when salivary glands are soaked in Nonidet P-40 prior to denaturation in LDS buffer, the ∼300-kDa fragment is converted into a slightly less massive form (Fig. 3D). This conversion, which results in an ∼10-kDa reduction in mass, is completed within 30 min and matches precisely the size of SGSs collected during artificial salivation experiments. Interestingly, this effect is only seen when the proximal lateral lobes are included in the non-denaturing lysate solution, showing that this processing requires a component of the proximal lateral lobes (Fig. 2K). The reason for this drop in mass is unknown, but because certain amylases and proteases are only produced in the proximal-lateral lobes (33), together with the prediction that SGSs are glycosylated (18), we hypothesize that the interaction of SGSs with saliva components results in additional proteolytic cleavage or deglycosylation. Taken altogether, Western and bioinformatic analyses suggest that Sgs4 and Sgs5 are secreted via a non-classical pathway following proteolytic cleavage into a ∼300-kDa soluble fragment and an ∼85-kDa membrane-bound fragment. The ∼300-kDa fragments are then processed into slightly less massive forms by an unknown product of the proximal lateral lobes prior to being expelled with the saliva.
DISCUSSION
Most research investigating the salivary glands of arthropod disease vectors has focused on uncovering the protein repertoire of salivary components in efforts to identify candidates for use in the development of epidemiological techniques (3, 7), transmission blocking vaccines (4), or novel therapeutic agents (1). Here we characterized the spatial, temporal, and functional expression of two Anopheles saliva proteins that are involved in blood feeding. The expression and biochemical characteristics of these proteins suggest that they play multiple physiological roles and that they may be useful for the epidemiological surveillance of mosquito activity and in the development of disease control strategies.
The only published study to date that focuses on the biology of SGSs showed that a member of the A. aegypti SGS gene family, Sgs1, is produced exclusively in the salivary glands, where it is embedded in the basal lamina and serves as an inadvertent receptor for P. gallinaceum sporozoites (18). In the present study, we found that A. gambiae Sgs4 and Sgs5 are also present on the basal side of the salivary glands, and because the major portions of these proteins are cleaved from their transmembrane domain, it is likely that they are also embedded in the basal lamina. Thus, Sgs4 and Sgs5 may also function as Plasmodium receptors in this major vector of human malaria. However, an unexpected finding was that Sgs4 and Sgs5 are also secreted apically into the salivary duct, where they form a major component of mosquito saliva. Because A. aegypti proteins in the SGS size range are major saliva components, it is likely that Sgs1 is also secreted into the saliva.
A. gambiae Sgs4 and Sgs5 are produced in salivary gland regions known to produce factors involved in blood feeding (33), and production of Sgs4 and Sgs5 ramps up during the times of the day when these mosquitoes show a higher propensity to bite mammals (34). Together with their substantial release with saliva during probing, these data show that they are involved in blood feeding. The precise role of Anopheles SGSs in this process has not been fully determined, but we show that they are among the most immunogenic proteins injected during natural blood feeding. This was not entirely unexpected, because SGSs are large proteins sharing little homology with any vertebrate protein, and thus, relatively high antibody titers would be expected from a small antigen dose. High immunogenicity opens the possibility that human anti-SGS antibodies could serve as sensitive markers for assessing exposure of humans to mosquito bites (3, 7) and that this could be an important tool in areas of low to moderate mosquito density. Furthermore, because the large N-terminal region of the SGSs is highly variable, small recombinant SGS peptides could be used as species-specific and genus-specific antigens in this epidemiological strategy. Such taxon specificity has often been cited as one justification for the use of gSG6 in surveying bite exposure (3, 7), but gSG6 is only present in one mosquito genus.
When assaying the immunogenicity of mosquito saliva components, two distinct sets of proteins were identified: SGSs and a protein that is ∼100 kDa under non-reducing conditions and 50 kDa when reduced. The electrophoretic pattern of the latter protein suggests that it is a disulfide-linked dimer, and interestingly, this pattern of migration is identical to that seen for Saglin, an Anopheles salivary protein that was shown to be involved in Plasmodium invasion of the salivary glands, along with some evidence that it is a component of mosquito saliva (35, 36). The detection of putative Saglin is strong after only 3 weeks of exposure to bites, suggesting that Saglin may also be a sensitive target for an exposure surveillance strategy. Furthermore, given that Sgs1 and Saglin are the only two molecularly identified candidate receptors for Plasmodium on the salivary gland surface, it is interesting that both are also present in the saliva. These findings suggest that their interaction with the parasite may be long lived, and that besides serving as basal receptors, they may also facilitate sporozoite traversal of acinar cells, survival in the salivary duct, and/or transmission to the vertebrate host.
It has been shown that immunomodulatory factors in arthropod saliva can enhance the transmission of protozoan parasites, and thus, a vaccine that neutralizes such factors might offer protection from pathogens (4). For example, vaccination against a single immunomodulatory and vasodilatory protein from sandfly saliva provides resistance to Leishmania infection (37). There is now a strong body of evidence supporting an immunomodulatory effect by mosquito saliva, although the identity of these immunomodulatory proteins has not been determined (10). One such factor is a large immunogenic protein from Anopheles stephensi that possesses neutrophil chemotactic activity (9). Another large immunomodulatory protein was discovered from A. aegypti saliva, is about 387 kDa, and significantly lowers cytokine release and the proliferation of murine T- and B-cells (20). Based on our proteomic analyses (Figs. 3B and 6) as well as transcriptomic analyses by others (16, 38), SGSs are the only anopheline or culicine saliva proteins whose mass approximates the value predicted for this ∼387-kDa protein. In support of our hypothesis that Sgs4 and Sgs5 are immunomodulatory, it has recently been shown that bacterial RHS/YD-repeat proteins (distant relatives of SGS) play a role during the deactivation of human macrophages by the bacterial pathogen Burkholderia pseudomallei (39). If SGSs are indeed immunomodulators, they should be considered when developing transmission-blocking vaccines.
Another interesting aspect of SGSs is that they share high sequence homology with proteins found in Wolbachia proteobacteria (alignment of Sgs5 and Wolbachia WD0513 yields 28% shared amino acid identity, 47% positives and an E-value of 0.0) and contain regional homology with the viral/bacterial RHS/YD-repeat protein family (16, 18). It is hypothesized that horizontal gene transfer from the intracellular bacterium Wolbachia led to the origin of mosquito SGSs, and the relatively high expression levels of A. aegypti SGSs suggest that some of these genes have evolved to perform essential roles in mosquitoes, unlike what is often observed following transdomain horizontal transfer (19). Our work validates this finding in anopheline mosquitoes by showing that Sgs4 and Sgs5 are specific to salivary tissue and are highly expressed. Although the functions of non-mosquito YD-repeat proteins are not well understood, they may be important in the interactions between microbes and their insect hosts (40, 41). In addition, YD-repeat proteins have been implicated as participants in lectin-like heparin binding, as mediators of cellular interactions, and as cellular toxins with anti-macrophage activity (39, 40, 42, 43). These are roles that share similarities with those proposed for mosquito SGSs, both here and by others (18). As for why a Wolbachia gene would have been adopted as a major salivary gene in mosquitoes, it is tempting to speculate that the ancient horizontal transfer of an immunomodulatory factor could have been a pivotal step during the evolution of hematophagy in mosquitoes. Exaptation and recruitment of endogenous genes into novel functional roles appears to occur often during the evolution of arthropod sialomes (1, 2), but SGSs are now the first examples of a saliva protein originating through horizontal gene transfer in a hematophagous arthropod.
Although we conclusively show that SGSs form a major component of the salivary glands and saliva of anopheline and culicine mosquitoes, these proteins have been largely overlooked in earlier proteomic studies (Table 1). The primary reason for this rests in the size of SGSs because the electrophoretic methods most commonly used in SDS-PAGE studies (e.g. ≥10% polyacrylamide concentration) preclude proteins as large as SGSs from entering the resolving portion of the gel. In all but two studies where a band was seen to approximate the size and relative intensity of SGS, the presence of the protein was not addressed. It is likely either that the large size of SGS made it seem like an artifact or that it was observed but disregarded because it was unrelated to the specific aim of the research. In one study, SGS was invoked as an explanation for a salivary band of 175 kDa that was consistently recognized by IgG from children exposed to high numbers of mosquito bites (8). This same study recognized one other salivary antigen that was estimated to be 72 kDa under non-reducing conditions. It is likely that these bands represent SGS (300 kDa) and Saglin (100 kDa) but that size estimates were incorrect due to the incompatibility of the gels and ladders used. Another study supports our finding that SGS is a component of mosquito saliva, although in that study the authors propose that the presence of SGS in the saliva is a result of contamination (44). These results illustrate that although SGSs are prevalent immunogenic components of mosquito saliva, the methodologies commonly used when studying salivary proteins preclude their detection.
TABLE 1.
Analysis of scientific papers in which mosquito saliva or whole salivary gland proteins were analyzed by SDS-PAGE or by Western blot using antibodies from humans or rodents exposed to mosquito bites
Only studies that could be clearly categorized were included in this analysis.
| SGS not identified: appropriate parametersa | SGS not identified: restrictive parametersb | Band approximating SGS size range but not addressed in the article | SGS explicitly stated in the article | |
|---|---|---|---|---|
| SDS-PAGE | None | Refs. 8 and 46–51 | Refs. 45 and 52–55 | Ref. 44c |
| Western blot (human sera) | None | Refs. 48 and 55–57 | Ref. 58 | Ref. 8d |
| Western blot (rodent sera) | None | Refs. 51 and 59 | Refs. 54 and 60 | None |
a Parameters used should have allowed for the visualization or identification of SGSs.
b Parameters used would preclude the visualization or identification of SGSs. Restrictive parameters include the use of ≥10% SDS-polyacrylamide gels (under normal running conditions, SGSs are too large to enter the resolving portion of the gel), and cropping of gel scans such that the gel portions that would show high molecular weight proteins are not shown.
c In this article, Sgs4 and Sgs5 are erroneously referred to as SG4 and SG5, but the correct GenBankTM accession numbers are listed.
d In this article, putative SGSs are predicted to be 175 kDa.
In summary, this study considerably expands our understanding of a family of mosquito proteins that have now been implicated in blood feeding, Plasmodium transmission, and mosquito-Wolbachia interactions. Given that SGSs form a major component of mosquito saliva, future studies should investigate the role these proteins play in modulating vertebrate immune responses and assess the potential use of SGSs as epidemiological markers for the surveillance of mosquito activity in disease-endemic areas.
Acknowledgment
We thank Tania Estévez-Lao for mosquito rearing and maintenance.
This work was supported by a Vanderbilt University Discovery Grant (to J. F. H.).
- SGS
- salivary gland surface protein
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PAP
- prophenoloxidase-activating protease.
REFERENCES
- 1. Mans B. J., Francischetti I. M. (2011) in Toxins and Hemostasis (Kini R. M., Clemetson K. J., Markland F. S., McLane M. A., Morita T. eds) pp. 21–44, Springer Science + Business Media, New York [Google Scholar]
- 2. Ribeiro J. M. C., Arcà B. (2009) Adv. Insect Physiol. 37, 59–118 [Google Scholar]
- 3. Remoue F., Cisse B., Ba F., Sokhna C., Herve J. P., Boulanger D., Simondon F. (2006) Trans. R. Soc. Trop. Med. Hyg. 100, 363–370 [DOI] [PubMed] [Google Scholar]
- 4. Titus R. G., Bishop J. V., Mejia J. S. (2006) Parasite Immunol. 28, 131–141 [DOI] [PubMed] [Google Scholar]
- 5. World Health Organization (1996) Weekly Epidemiological Record 71, 17–22 8924385 [Google Scholar]
- 6. Poinsignon A., Cornelie S., Ba F., Boulanger D., Sow C., Rossignol M., Sokhna C., Cisse B., Simondon F., Remoue F. (2009) Malar. J. 8, 198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Drame P. M., Poinsignon A., Besnard P., Cornelie S., Le Mire J., Toto J. C., Foumane V., Dos-Santos M. A., Sembène M., Fortes F., Simondon F., Carnevale P., Remoue F. (2010) PLoS One 5, e15596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cornelie S., Remoue F., Doucoure S., Ndiaye T., Sauvage F. X., Boulanger D., Simondon F. (2007) Malar. J. 6, 75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Owhashi M., Harada M., Suguri S., Ohmae H., Ishii A. (2001) Parasitol. Res. 87, 376–382 [DOI] [PubMed] [Google Scholar]
- 10. Schneider B. S., Higgs S. (2008) Trans. R. Soc. Trop. Med. Hyg. 102, 400–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morris R. V., Shoemaker C. B., David J. R., Lanzaro G. C., Titus R. G. (2001) J. Immunol. 167, 5226–5230 [DOI] [PubMed] [Google Scholar]
- 12. Kebaier C., Voza T., Vanderberg J. (2010) Infect. Immun. 78, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vaughan J. A., Scheller L. F., Wirtz R. A., Azad A. F. (1999) Infect. Immun. 67, 4285–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donovan M. J., Messmore A. S., Scrafford D. A., Sacks D. L., Kamhawi S., McDowell M. A. (2007) Infect. Immun. 75, 2523–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arcà B., Lombardo F., Francischetti I. M., Pham V. M., Mestres-Simon M., Andersen J. F., Ribeiro J. M. (2007) Insect Biochem. Mol. Biol. 37, 107–127 [DOI] [PubMed] [Google Scholar]
- 16. Arcà B., Lombardo F., Valenzuela J. G., Francischetti I. M., Marinotti O., Coluzzi M., Ribeiro J. M. (2005) J. Exp. Biol. 208, 3971–3986 [DOI] [PubMed] [Google Scholar]
- 17. Ribeiro J. M., Charlab R., Pham V. M., Garfield M., Valenzuela J. G. (2004) Insect Biochem. Mol. Biol. 34, 543–563 [DOI] [PubMed] [Google Scholar]
- 18. Korochkina S., Barreau C., Pradel G., Jeffery E., Li J., Natarajan R., Shabanowitz J., Hunt D., Frevert U., Vernick K. D. (2006) Cell Microbiol. 8, 163–175 [DOI] [PubMed] [Google Scholar]
- 19. Klasson L., Kambris Z., Cook P. E., Walker T., Sinkins S. P. (2009) BMC Genomics 10, 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wasserman H. A., Singh S., Champagne D. E. (2004) Parasite Immunol. 26, 295–306 [DOI] [PubMed] [Google Scholar]
- 21. Hillyer J. F., Estévez-Lao T. Y. (2010) Dev. Comp. Immunol. 34, 141–149 [DOI] [PubMed] [Google Scholar]
- 22. Boorman J. (1987) Med. Vet. Entomol. 1, 211–214 [DOI] [PubMed] [Google Scholar]
- 23. Bowen M. F. (1996) in Ciba Foundation Symposium 200-Olfaction in Mosquito-Host Interactions (Bock G. R., Cardew G. eds) pp. 197–211, John Wiley & Sons, Ltd., Chichester, U.K. [Google Scholar]
- 24. Billingsley P. F., Hodivala K. J., Winger L. A., Sinden R. E. (1991) Trans. R. Soc. Trop. Med. Hyg. 85, 450–453 [DOI] [PubMed] [Google Scholar]
- 25. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 26. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, 1st Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27. Castellanos-Serra L., Hardy E. (2006) Nat. Protoc. 1, 1544–1551 [DOI] [PubMed] [Google Scholar]
- 28. Sasse J., Gallagher S. R. (2009) Curr. Protoc. Mol. Biol., Chapter 10, Unit 10.6 [DOI] [PubMed] [Google Scholar]
- 29. Mohana Rao J. K., Argos P. (1986) Biochim. Biophys. Acta 869, 197–214 [DOI] [PubMed] [Google Scholar]
- 30. Söderhäll K., Cerenius L. (1998) Curr. Opin. Immunol. 10, 23–28 [DOI] [PubMed] [Google Scholar]
- 31. Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keller M., Rüegg A., Werner S., Beer H. D. (2008) Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 33. Juhn J., Naeem-Ullah U., Maciel Guedes B. A., Majid A., Coleman J., Paolucci Pimenta P. F., Akram W., James A. A., Marinotti O. (2011) Parasit. Vectors 4, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saunders D. S. (2002) Insect Clocks, 3rd Ed., pp. 9–35, Elsevier Science, Amsterdam [Google Scholar]
- 35. Ghosh A. K., Devenport M., Jethwaney D., Kalume D. E., Pandey A., Anderson V. E., Sultan A. A., Kumar N., Jacobs-Lorena M. (2009) PLoS Pathog. 5, e1000265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okulate M. A., Kalume D. E., Reddy R., Kristiansen T., Bhattacharyya M., Chaerkady R., Pandey A., Kumar N. (2007) Insect Mol. Biol. 16, 711–722 [DOI] [PubMed] [Google Scholar]
- 37. Reddy V. B., Li Y., Lerner E. A. (2008) J. Mol. Neurosci. 36, 241–244 [DOI] [PubMed] [Google Scholar]
- 38. Calvo E., Sanchez-Vargas I., Kotsyfakis M., Favreau A. J., Barbian K. D., Pham V. M., Olson K. E., Ribeiro J. M. (2010) J. Med. Entomol. 47, 376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dowling A. J., Wilkinson P. A., Holden M. T., Quail M. A., Bentley S. D., Reger J., Waterfield N. R., Titball R. W., Ffrench-Constant R. H. (2010) PLoS One 5, e15693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Degnan P. H., Moran N. A. (2008) Appl. Environ. Microbiol. 74, 6782–6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Iturbe-Ormaetxe I., Burke G. R., Riegler M., O'Neill S. L. (2005) J. Bacteriol. 187, 5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Minet A. D., Rubin B. P., Tucker R. P., Baumgartner S., Chiquet-Ehrismann R. (1999) J. Cell Sci. 112, 2019–2032 [DOI] [PubMed] [Google Scholar]
- 43. Youderian P., Hartzell P. L. (2007) Genetics 177, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Orlandi-Pradines E., Almeras L., Denis de Senneville L., Barbe S., Remoué F., Villard C., Cornelie S., Penhoat K., Pascual A., Bourgouin C., Fontenille D., Bonnet J., Corre-Catelin N., Reiter P., Pagés F., Laffite D., Boulanger D., Simondon F., Pradines B., Fusaï T., Rogier C. (2007) Microbes Infect. 9, 1454–1462 [DOI] [PubMed] [Google Scholar]
- 45. Francischetti I. M., Valenzuela J. G., Pham V. M., Garfield M. K., Ribeiro J. M. (2002) J. Exp. Biol. 205, 2429–2451 [DOI] [PubMed] [Google Scholar]
- 46. Brennan J. D., Kent M., Dhar R., Fujioka H., Kumar N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13859–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jariyapan N., Baimai V., Poovorawan Y., Roytrakul S., Saeung A., Thongsahuan S., Suwannamit S., Otsuka Y., Choochote W. (2010) Parasitol. Res. 107, 509–516 [DOI] [PubMed] [Google Scholar]
- 48. Malafronte Rdos S., Calvo E., James A. A., Marinotti O. (2003) Insect Biochem. Mol. Biol. 33, 63–71 [DOI] [PubMed] [Google Scholar]
- 49. Moreira C. K., Marrelli M. T., Lima S. P., Marinotti O. (2001) J. Med. Entomol. 38, 763–767 [DOI] [PubMed] [Google Scholar]
- 50. Siriyasatien P., Tangthongchaiwiriya K., Jariyapan N., Kaewsaitiam S., Poovorawan Y., Thavara U. (2005) Southeast Asian J. Trop. Med. Public Health 36, 64–67 [PubMed] [Google Scholar]
- 51. Jeon S. H., Park J. W., Lee B. H. (2001) Int. Arch. Allergy Immunol. 126, 206–212 [DOI] [PubMed] [Google Scholar]
- 52. Boisson B., Jacques J. C., Choumet V., Martin E., Xu J., Vernick K., Bourgouin C. (2006) FEBS Lett. 580, 1988–1992 [DOI] [PubMed] [Google Scholar]
- 53. James A. A., Blackmer K., Marinotti O., Ghosn C. R., Racioppi J. V. (1991) Mol. Biochem. Parasitol. 44, 245–253 [DOI] [PubMed] [Google Scholar]
- 54. Racioppi J. V., Spielman A. (1987) Insect Biochem. 17, 503–511 [Google Scholar]
- 55. Waitayakul A., Somsri S., Sattabongkot J., Looareesuwan S., Cui L., Udomsangpetch R. (2006) Acta Trop. 98, 66–73 [DOI] [PubMed] [Google Scholar]
- 56. Peng Z., Li H., Simons F. E. (1998) J. Allergy Clin. Immunol. 101, 498–505 [DOI] [PubMed] [Google Scholar]
- 57. Penneys N. S., Nayar J. K., Bernstein H., Knight J. W., Leonardi C. (1989) Arch. Dermatol. 125, 219–222 [PubMed] [Google Scholar]
- 58. Brummer-Korvenkontio H., Lappalainen P., Reunala T., Palosuo T. (1994) J. Allergy Clin. Immunol. 93, 551–555 [DOI] [PubMed] [Google Scholar]
- 59. Chen Y. L., Simons F. E., Peng Z. (1998) Int. Arch. Allergy Immunol. 116, 269–277 [DOI] [PubMed] [Google Scholar]
- 60. Barreau C., Conrad J., Fischer E., Lujan H. D., Vernick K. D. (1999) Insect Biochem. Mol. Biol. 29, 515–526 [DOI] [PubMed] [Google Scholar]