Abstract
In Saccharomyces cerevisae, expanded polyglutamine (polyQ) fragments are assembled into discrete cytosolic aggregates in a process regulated by the molecular chaperones Hsp26, Hsp70, Hsp90, and Hsp104. To better understand how the different chaperones might cooperate during polyQ aggregation, we used sequential immunoprecipitations and mass spectrometry to identify proteins associated with either soluble (Q25) or aggregation-prone (Q103) fragments at both early and later times after induction of their expression. We found that Hsp26, Hsp70, Hsp90, and other chaperones interact with Q103, but not Q25, within the first 2 h. Further, Hsp70 and Hsp90 appear to be partially released from Q103 prior to the maturation of the aggregates and before the recruitment of Hsp104. To test the importance of this seemingly ordered process, we used a chemical probe to artificially enhance Hsp70 binding to Q103. This treatment retained both Hsp70 and Hsp90 on the polyQ fragment and, interestingly, limited subsequent exchange for Hsp26 and Hsp104, resulting in incomplete aggregation. Together, these results suggest that partial release of Hsp70 may be an essential step in the continued processing of expanded polyQ fragments in yeast.
Keywords: Chaperone Chaperonin, Chemical Biology, Heat Shock Protein, Mass Spectrometry (MS), Polyglutamine
Introduction
The heat shock proteins (HSPs)3 are abundant molecular chaperones that have been shown to be important for maintaining proteostasis under both normal conditions and during times of cellular stress (1–3). Together, these factors play multiple roles in quality control, especially related to polypeptide synthesis, folding, and turnover (4–7). Moreover, HSPs are emerging as drug targets in cancer, neurodegenerative disorders and other diseases (8, 9). Thus, there is interest in better understanding how they coordinate protein quality control decisions.
The major families of HSPs are named based on their approximate kDa molecular weights, including Hsp60, Hsp70, Hsp90, Hsp104, and the small heat shock proteins (e.g. Hsp26), (1). Most organisms express multiple members of each of the HSP families; for example, Saccharomyces cerevisae expresses both heat inducible and constitutive forms of cytosolic Hsp70s (Ssa2 and Ssa1, respectively) and Hsp90s (Hsp82 and Hsc82, respectively). In addition to having different sizes, the individual members of the HSP families have vastly different structures. For example, Hsp70 is a flexible, two-domain chaperone that largely carries out its functions as a monomer, Hsp90 is a “V-shaped” dimer, Hsp104 is a cyclindrical dodecamer and Hsp26 assembles into an ensemble of spherical homo-oligomers (10). As might be expected based on their diverse structurs, these classes of HSPs also possess some specialized chaperone functions. For example, Hsp70 is commonly thought to be a major triage chaperone (6), while Hsp90 has a more restricted set of substrates that it maintains in a stabilized, active state (6, 11). Hsp104 specifically disassembles large aggregates (5, 12), while Hsp26 protects its substrates from acute denaturation (13). Despite these substantial differences, one common feature of the HSPs is that they bind unfolded or partially folded regions of their protein substrates. Thus, the HSPs have both similarities and differences, which has led to a model in which they might cooperate during protein quality control (4, 6, 14). In this model, each of the chaperones is expected to play some specific roles, while collectively protecting bystander proteins from exposure to misfolded intermediates.
Our understanding of chaperone cooperation has significantly benefitted from a large number of studies in S. cereviasae that have focused on understanding the aggregation of prions and polyglutamine (polyQ) fragments (2, 3, 15). These model substrates share some similarities in that both are aggregation-prone and they form large inclusions in yeast. Prions, such as [RNQ1]+, are required for polyQ toxicity (16, 17) and, likewise, polyQ aggregation influences prion formation (18). These results and many others (2, 3, 15). highlight the complex interplay between these aggregation systems in yeast. Importantly for the current study, genetic experiments have clearly shown that the HSPs regulate assembly of both prions and polyQ fragments (2, 3, 5, 19–21). For example, Hsp70 activity appears to be required to facilitate Hsp104 disaggregase activity during the propagation and inheritance of prions (22–24) and other model substrates (25). In addition, Hsp26 works with Hsp104 to help break large fibrils into inheritable fragments (26), while Hsp70 and Hsp90 also assist in this process (27). Some of these shared activities may involve direct interactions between chaperones; for example, Hsp70 binds directly to Hsp104 (28) and Hsp70 and Hsp90 can be physically linked via Sti1 (29). Together, these studies suggest that HSP-class chaperones, especially members of the Hsp26, Hsp70, Hsp90, and Hsp104 families, collaborate to regulate protein aggregation.
Despite these illuminating studies, there are a number of key questions to be asked about how the chaperones cooperate. Do they all converge on the misfolded substrates simultaneously? Do they “hand-off” substrates in an assembly line-type process? If so, which chaperones come first? Because of the rather specific role of Hsp104 in disaggregation (5, 12), it is logical to predict that it might associate with substrates at relatively later stages. However, what about the other chaperones? Do they need to dissociate before Hsp104 can bind? Understanding these issues might help us better comprehend protein quality control.
In the present study, we asked two specific questions. First, we wondered whether cellular proteins, especially chaperones, bind expanded polyQ fragments in a defined order. Second, we wanted to probe the specific roles of Hsp70 in this process. This second question is important because of the particularly striking connections between Hsp70 and protein aggregation (8, 15, 30). Hsp70 family members have slow ATP turnover rates that are accelerated by binding to co-chaperones, including J proteins (or Hsp40s) that stimulate ATP turnover and nucleotide exchange factors (NEFs) that promote nucleotide release (Fig. 1A) (1, 8). These co-chaperones are important because the ADP-bound form of Hsp70 has a tighter affinity for substrates than the ATP-bound state (31). Thus, J proteins are expected to promote tight binding of Hsp70s to their substrates and these co-chaperones have been found to play multiple roles during protein aggregation (21, 32). Thus, one promising approach to chemically controlling the activities of Hsp70s may be to selectively target the interaction with the J proteins (8, 33). Toward that goal, the Brodsky group originally identified dihydropyrimidines as chemical reagents for specifically promoting the J-stimulated activity of Hsp70s (34, 35). Following on this work, we recently found that one promising example of this chemical class, 115-7c, stimulates ATP turnover by directly favoring interactions between Hsp70s and J proteins (Fig. 1A) (36, 37). Moreover, 115-7c is membrane permeable, selective for Hsp70 and does not induce a stress response (36), so it can be used to selectively tune Hsp70 activity in cells without impacting chaperone expression. Together, these observations suggest that 115-7c can be used as a chemical probe to ask how Hsp70 impacts polyQ fragment assembly.
FIGURE 1.
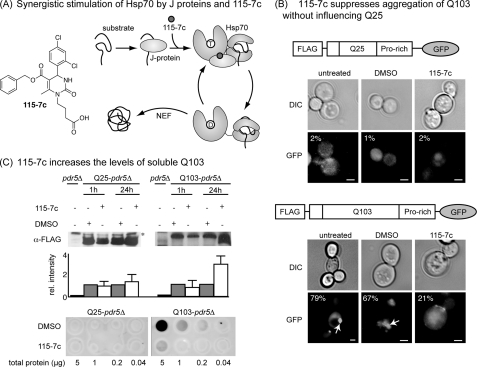
The Hsp70 activator 115-7c suppresses aggregation of an expanded polyglutamine in yeast. A, chemical structure of the dihydropyrimidine, 115-7c, and a schematic of its ability to enhance J protein-mediated ATP turnover in Hsp70. B, 115-7c (100 μm) blocks formation of puncta in cells expressing Q103. A schematic of the polyQ-GFP fusions is shown. The arrow indicates a representative puncta. The percentage of cells with visible puncta is indicated (n = at least 250 cells). Scale bar, 1 μm. C, 115-7c enhances the solubility of Q103. Top, Q25 bands were observed at ∼50 kDa and the Q103 bands were ∼100 to 120 kDa. Cells of the same strain lacking the polyglutamine (pdr5Δ) were used as a control. *, nonspecific band. Results are representative of experiments performed in triplicate and error bars represent standard error of the mean (S.E.). Bottom, filter trap assays were also performed on the lysates, and the amount of aggregated polyQ retained on the membrane was measured. Results are representative of independent duplicates.
In this study, we used immunoprecipitations and mass spectrometry to identify a set of ∼320 proteins that bind to short and long polyQ fragments at early and later times after induction. We also employed 115-7c to stimulate Hsp70 activity, suppress polyQ aggregation and probe how this change influenced the polyQ-associated proteome. Finally, we specifically focused on how the HSPs, including Hsp26, Hsp70, Hsp90 and Hsp104, associate with an aggregation-prone polyQ fragment. Together, these studies suggest that HSP assembly is an ordered process, with partial Hsp70 and Hsp90 release occurring prior to the final steps of aggregation and recruitment of Hsp104. These results provide mechanistic insights into the process of polyQ aggregation in yeast.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Growth Conditions
Yeast strains: W303 (MATa, leu2–3,112 his3–11 trp1–1 ura3–1 can1–100 ade2–1); YKO (pdr5Δ) B4147; YKO WT, pdr5Δ (MATa/MATα orf::kanMX4/orf::kanMX4 ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 met15Δ0/MET15 lys2Δ0/LYS2. Yeast cells were grown overnight at 30 °C in rich medium (YPD) or synthetic minimal medium (SMD) containing 2% glucose, raffinose, or galactose supplemented with the appropriate amino acids and vitamins. For experiments using 115-7c, the compound was dissolved in DMSO at a stock concentration of 10 mm and added to the cell culture 1 h after polyQ induction to a final concentration of 100 μm (36).
Immunoprecipitations and Western Blots
The protocol for the collection of yeast fractions suitable for proteomics was described by Wiederhold et al. (38). Briefly, yeast cells of a 30 ml exponentially growing culture were harvested by centrifugation at 3000 revolutions/min for 5 min at 18 °C, suspended in lysis buffer (Tris-HCl 50 mm, 200 mm sorbitol, 1 mm MgCl2, 0.1% Tween 20, 1 mm PMSF, and protease mixture inhibitor) and disrupted by vortexing with glass beads (6 × 30 s, 1 min ice in between). These lysates were cleared by centrifugation at 5,000 × g to remove unlysed cells, nuclei and membrane fractions. A portion of these lysates was subject to filter trap analysis as previously described (16). Next, 1 ml of this supernatant (normalized to A600 units = 0.8) was incubated with 2 μg of anti-Flag polyclonal antibody (Sigma-Aldrich) and IgG-Sepharose beads (Sigma) for 1 h at 4 °C. The beads were washed with lysis buffer eight times and the protein complexes eluted in 30 μl of SDS-loading buffer by incubating at 95–100 °C for 5 min. Afterward, the samples were subjected to electrophoretic separation on 10–20% Tricine-SDS-polyacrylamide gels (Novex®, Invitrogen). The gels were stained with Coomassie Blue (Invitrogen) and further analyzed by the Mass Spectrometry-Based Resources core facility (Department of Pathology, University of Michigan). For the Western blot analyses, the separated proteins were transferred to a polyvinylidene difluoride membrane (Immobilon) and probed with the appropriate antibodies. For those experiments, the amount of precipitated polyQ was first normalized. The primary antibodies were diluted as follows: 1:1000 for Flag (F7425, Sigma-Aldrich), GFP (Roche), Hsp70 (3A3, Santa Cruz Biotechnology, Inc.); 1:3000 for Hsp90 (yF-17, Santa Cruz Biotechnology., Inc.); 1:8000 for Hsp104 and Hsp26. All the secondary antibodies, coupled to horseradish peroxidase, were used at 1:10000. The vendors were: goat anti-rabbit (28177, Ana Spec), rabbit anti-mouse (Jackson ImmunoResearch) and donkey anti-goat (Santa Cruz Biotechnology., Inc.). Signals were developed with SuperSignal® West Pico Substrate (ThermoScientific, Rockford, IL) and exposed to BioMax MR film (Eastman Kodak). Western blots were quantified in Image J (NIH) by first normalizing all the band intensities to the input levels and then comparing the 115-7c treated to the mock control.
Protein Identification by Tandem LC/MS-MS
Standard protocols for in-gel digestion followed by mass spectrometry were carried out as described (39). The lanes were excised into 14 equal sized slices and destained with 30% methanol for 4 h. After reduction (10 mm DTT) and alklylation (65 mm iodoacetamide) of the cysteines at room temperature for 30 min, proteins were digested overnight with trypsin (Promega). Resulting peptides were resolved on a nano-capillary reverse phase column (Picofrit column, New Objective) using a 1% acetic acid/acetonitrile gradient at 300 nl/min and directly introduced into a linear ion-trap mass spectrometer (LTQ XL, ThermoFisher). Data-dependent MS/MS spectra on the 5 most intense ions from each full MS scan were collected (relative CE ∼35%). All mass spectra acquired were analyzed and matched to the S. cerevisiae database (6817 entries, 2008 release) appended with decoy reverse sequences and human polyQ protein using X!Tandem/Trans-Proteomic Pipeline (TPP) software suite (38). All peptides and proteins with a PeptideProphet and ProteinProphet probability score of >0.9 (fdr <2%) were considered positive identifications.
RESULTS
Aggregation of PolyQ Fragments Is Suppressed by the Hsp70 Stimulator 115-7c
We recently reported that the dihydropyrimidine 115-7c stimulates the ATPase activity of Hsp70s by enhancing stimulation by J proteins (36). Using a yeast model, this compound was also found to suppress polyQ aggregation, comparable to what is seen after Hsp70 overexpression (41). These results suggest that 115-7c might be suitable as a probe to better understand critical Hsp70 functions. Thus, in the present studies, we first sought to re-evaluate this compound's activity using a newer set of polyQ expression vectors (42–45). These constructs contain an N-terminal FLAG epitope tag that can be used for immunoprecipitations (43). Also, they contain either 25 or 103 copies of glutamine (herein referred to as Q25 and Q103) controlled by a galactose-inducible promoter. Finally, we choose to use the variants that include a proline-rich motif to avoid growth defects related to removing this region (variant IV; 42).
Using these constructs, we first confirmed the distribution of Q25 and Q103 in untreated cells. For these studies, we used a pdr5Δ background strain because deletion of the PDR5 gene is known to improve cytosolic retention of small molecules (46). Consistent with previous observations (42, 43), we found that the GFP signal from the polyQ fusion was diffusely distributed in the Q25-expressing cells after 24 h, while a single puncta, typically called an aggresome, formed in nearly all (∼80%) of the Q103-expressing cells (Fig. 1B). By counting the number of cells that contained either single or multiple aggregates, we found that treatment with 115-7c (100 μm), but not the solvent control (1% DMSO), significantly suppressed aggregation of Q103 with no obvious effect on Q25 (Fig. 1B). Specifically, only ∼20% of the Q103 cells still had visible aggregates after the treatment.
Next, the cells were lysed with glass beads and supernatants collected using a low speed centrifugation step to remove intact cells and nuclei. The resulting soluble fractions were subject to anti-FLAG Western blots to determine their relative amounts of polyQ. These studies revealed that 115-7c (100 μm) increased the apparent levels of Q103 by ∼3-fold, with little effect on Q25 (Fig. 1C). This result is consistent with the compound enhancing polyQ solubility. Then, we performed filter trap assays on the lysates to examine the amount of aggregated polyQ. These studies confirmed that Q25 lysates lack aggregates, while Q103 lysates contained a significant amount of aggregated material (Fig. 1C). Treatment with 115-7c (100 μm) for 24 h suppressed aggregate formation. Thus, for the purposes of this study, we concluded that 115-7c could be used to perturb the aggregation process.
Compound 115-7c Does Not Cure [RNQ1]+
To gain additional insight into the mechanism of treatment-induced suppression of Q103 aggregation, we tested whether 115-7c could cure the Q103 strain of its [RNQ1]+ phenotype (42). This is an important question because this prion is known to be required for polyQ toxicity (17, 47). Thus, we wanted to discern whether 115-7c might limit polyQ assembly via “indirect” effects on this prion. As a first test of this idea, we found that 115-7c (100 μm) did not alter the localization of Rnq1p-YFP, suggesting that prion status is unchanged by this treatment (supplemental Fig. S1a). Further, overexpression of Rnq1p in [RNQ1]+ cells was still toxic (14), even with the addition of 115-7c (100 μm) (supplemental Fig. S1b), again indicating that this compound does not cure [RNQ]+. Finally, 115-7c did not suppress toxicity resulting from the expression of Q103 lacking the proline-rich region (FlagQ103ΔPro; supplemental Fig. S1b). Together, these findings all suggest that 115-7c does not impact Q103 aggregation via effects on [RNQ1]+ status. Perhaps consistent with this general idea, the Chernoff Laboratory has observed that some chaperones impact polyQ toxicity through effects on prions, while others do not (48).
Identification of PolyQ-interacting Proteins by LC-MS/MS
One goal of this study is to understand which proteins change their association with polyQ fragments in response to different polyglutamine lengths, aggregation states and compound treatments. Toward that end, we performed immunoprecipitations from lysates of cells expressing either Q25 or Q103. In these studies, we examined both early (2 h) and late (24 h) times after induction of the polyQ proteins in an attempt to understand the impact of progressive aggregation (Fig. 2A). To enable comparisons between the treatment conditions, we normalized the cleared lysates to total protein levels. After the precipitations, we further normalized the samples to the polyQ content to avoid bias in the subsequent analysis of bound proteins. Finally, we performed identical immunoprecipitations from pdr5Δ cells lacking the polyQ constructs to control for the promiscuous binding of HSPs.
FIGURE 2.
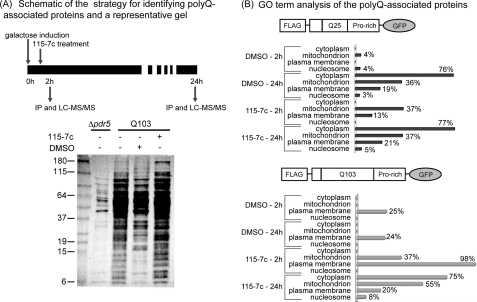
Identification of polyQ-associated proteins by immunoprecipitation and mass spectrometry. A, schematic of the experimental protocol. Anti-FLAG immunoprecipitations were performed either 2 h or 24 h after induction of polyQ expression in cells harboring either Q25 or Q103 constructs. Cells were treated with either DMSO or 115-7c (100 μm) at 1 h after induction. A representative silver-stained gel of the immunoprecipitated material is shown, along with a control immunoprecipitation from cells lacking polyQ expression. B, using ProteinProphet, a total of ∼320 unique proteins were identified (probability score >0.9 and at least 2 peptides). Between ∼140 and 250 proteins were observed in each individual experimental condition. Using this protein set, GO term analysis suggested that polyQ-associated proteins are largely associated with the plasma membrane, mitochondria, and cytosol.
Before performing the mass spectrometry studies on the immunoprecipitated samples, we separated the proteins by gel electrophoresis and examined them by silver staining to roughly compare the total number of associated factors in the control and polyQ-expressing cells. These results suggested that a promising number of interacting proteins were observed in the polyQ pulldowns and that the washing steps were stringent enough to remove most of the proteins from the control samples (Fig. 2A). Next, segments of identical gels were extracted, trypsinized and the resulting peptides subject to identification by tandem LC-MS/MS (see “Experimental Procedures” for details). For the interpretation of the resulting data, we employed a relatively straightforward positive peptide identification method. Specifically, we analyzed the peptides by PeptideProphet and the associated proteins were confirmed if they included >2 unique peptides and scored ≥ 0.9. Although other mass spectrometry methods, such as spectral counting or SILAC, are powerful ways to examine more subtle quantitative and potentially important changes in the patterns of bound proteins, we decided to focus this specific study on first cataloging the most dramatic differences.
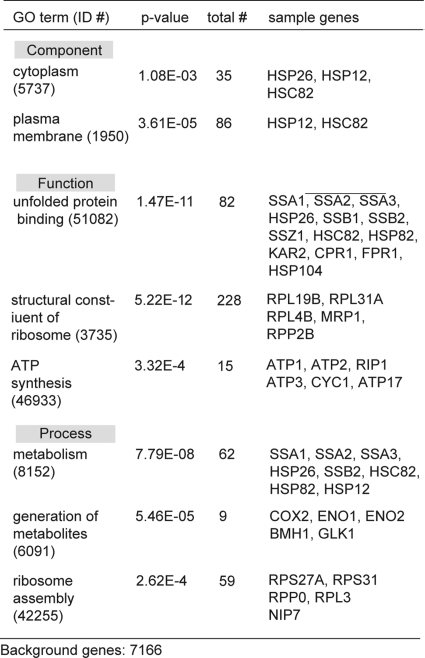
This approach identified a total of ∼320 individual proteins, with between 145 and 254 proteins associated with each condition (see supplemental information). Gene ontology (GO) analysis of the associated proteins in the Saccharomyces Genome Database suggested a relative enrichment in cytoplasmic- and plasma membrane-resident proteins across all the conditions (Fig. 2B) (49). We also noted a number of mitochondrial proteins, consistent with an emerging role for that organelle in polyQ-associated processes (50). Although care must be taken to avoid over-interpreting these results, we also noticed that treatment with 115-7c for 24 h shifted the Q103-associated patterns to more closely resemble those of Q25, perhaps consistent with the ability of this compound to suppress aggregation (see Fig. 1B). Next, we used a GO term search to ask whether any specific components, functions or processes might be statistically enriched in the polyQ-bound proteins. We were encouraged to find that the unfolded protein binding category (GO:51082), which includes a large number of HSPs, was prominently enriched (p value = 1.5 × 10−11) (Fig. 3). We also detected a number of additional chaperones, including Hsp26, which were annotated as being involved in metabolism (GO:8152). The remaining proteins were largely associated with ribosome assembly (GO:42255; GO:3735), metabolism (GO:8152; GO:6091), and ATP synthesis (GO:46933).
FIGURE 3.
GO term analysis of the polyglutamine-associated proteins. GO term analysis of the total protein list revealed a significant prevalence of molecular chaperones and metabolic factors.
Heat Shock Proteins Interact with PolyQ, and Their Binding Patterns Are Sensitive to Polyglutamine Length and Aggregation Time
To clarify the potential roles of HSPs in polyQ aggregation, we generated a list of the chaperones that were identified under each experimental condition (Fig. 4A). This list included those “background” chaperones that were found in the control immunoprecipitations and, because the Hsp70 family members Ssb1 and Ssb2 were abundant in those controls, we discounted them from subsequent analyses. We next compared our results to those of a previous proteomics study in which proteins bound to a Q103 fragment with or without the adjacent proline-rich domain (Q103 and Q103ΔP) were examined (43). Although this earlier study used different experimental conditions, we were encouraged to see a number of similarities (19 of 32 proteins from the previous report were positively identified). Similarly, comparisons to the results from studies on polyQ- (51) and prion-interacting proteins (52) also suggested common features, especially an abundance of metabolic enzymes and HSPs.
FIGURE 4.
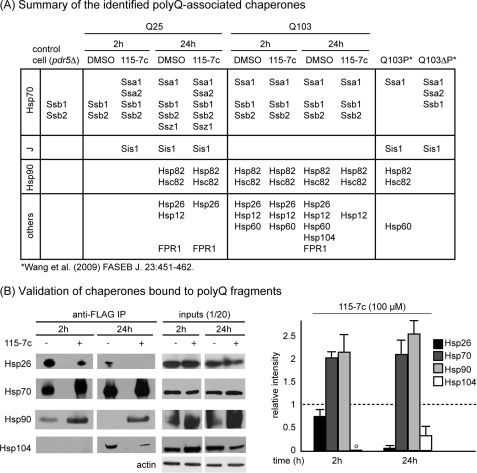
Chaperones associate with polyglutamine fragments in a time, polyQ length and compound-dependent manner. A, summary of the HSPs and chaperones identified in the mass spectrometry experiments. Control cells (pdr5Δ) were the same strain background, but did not express polyQ fragments. For reference, positive partners from a previous study are shown. B, key interactions were verified by Western blots on freshly immunoprecipitated material. The bands were quantified using image analysis, normalized to the input levels and then the results from the drug treated samples were reported relative to the untreated. The results are the average of three independent replicates (e.g. fresh immunoprecipitations). o, quantification could not be performed. See “Experimental Procedures” for additional details.
From the patterns of bound chaperones, a number of interesting observations were readily apparent. First, the molecular chaperones, Hsp26, Hsp40 (Sis1), Hsp70 (Ssz1, Ssa1, and Ssa2) and Hsp90 (Hsp82 and Hsc82), as well as the cis-trans prolyl isomerase, FPR1, appeared to become associated with soluble Q25 over time. More specifically, there was little chaperone associated at 1 h, but multiple HSPs were readily identified at 24 h. These results suggest that the amount of misfolded Q25 at early times may be low. In contrast, soluble Q103 was associated with many chaperones, including Hsp12, Hsp26, Hsp70, Hsp90, and Hsp60, at both the early and later time points, suggesting that this aggregation-prone substrate may be recognized as misfolded relatively soon after its expression. Importantly, we only observed Hsp104 associated with Q103 at 24 h, which is concurrent with the appearance of puncta. Collectively, these proteomic studies lend further support to the model in which the well-known Hsp26, Hsp70, Hsp90, and Hsp104 chaperones are involved in the normal processing of polyQ fragments. In addition, they implicate some relatively under-explored factors, such as Hsp12, Hsp60, and FPR1, as possible additional modifiers.
Next, we explored the effects of 115-7c on the chaperones associated with Q25 and Q103. In the Q25-expressing cells, we noted few differences between the samples treated with DMSO or 115-7c. One potentially interesting finding was that the yeast Hsp70, Ssa1, and one of its major J proteins, Sis1, became associated with Q25 at the early time point (2 h) in response to 115-7c. This result may be consistent with the reported ability of this compound to promote the assembly of the high affinity, Hsp70-J protein pair (36). In the Q103-expressing cells, we found that Hsp70 and Hsp90 remained bound at 24 h, while binding to both Hsp26 and Hsp104 was lost. In addition, Hsp60 and FPR1 were no longer identified at 24 h. Together, these results suggested that 115-7c dramatically altered the pattern of chaperones associated with Q103.
Based on these results, we were interested in further exploring the interplay between Q103, Hsp26, Hsp70, Hsp90, and Hsp104. Specifically, we wanted to determine the levels of these HSPs by Western blots to quantify how they bind Q103 in the presence and absence of 115-7c. Using freshly immunoprecipitated material, we confirmed that Hsp104 was associated with Q103 at 24 h, but not at the earlier time point (Fig. 3B). Treatment with 115-7c (100 μm) substantially reduced this interaction (by ∼50 to 60%), consistent with the proteomic studies. 115-7c also reduced the association with Hsp26 by at least 5-fold, concurrent with a greater than 2-fold increase in the amount of bound Hsp70 and Hsp90. Thus, the levels of Hsp26 and Hsp104 appeared to be inversely related to those of Hsp70 and Hsp90. These changes did not appear to be related to any alterations in the total cellular levels of the chaperones, because, consistent with our previous studies (36), 115-7c did not induce HSP expression (Fig. 4B). Together, these results paralleled the LC-MS/MS studies and suggested an essential role for Hsp70 and Hsp90 mobility during polyQ aggregation.
DISCUSSION
Polyglutamine-interacting Proteins
Genetic studies have previously linked polyQ aggregation to disruption of a number of cellular pathways (16, 40, 54). In this study, we wanted to perform an unbiased proteomics analysis to better understand how binding of proteins to polyQ might change as a function of aggregation time and polyQ length. Accordingly, we performed mass spectrometry studies to find proteins associated with Q25 and Q103 fragments.
In all, ∼320 unique proteins were identified as being associated (either directly or indirectly) with Q25 or Q103. Approximately 70 (20%) of these proteins were annotated as being associated with the ribosome, ribosome biogenesis or RNA processing. An additional 12 (4%) were readily identified as chaperones: Ssa1, Ssa2, Ssa3, Ssz1, Sis1, FPR1, Hsp82, Hsc82, Hsp104, Kar2, Phb1, and Hsp60. Additionally, a number of other factors categorized as binding to unfolded proteins were observed, including CPR1. As shown in Fig. 3, the remainder of the proteins included mostly metabolic enzymes (e.g. URA2, SAM1, LYS1, ENO1), cytoskeletal and trafficking components (e.g. ACT1, SLA1, ARF1) and mitochondrial proteins (e.g. VPS1, ATP1, MIR1, RIM1). The preponderance of mitochondrial proteins may be interesting based on the ability of expanded polyQ fragments to disrupt this organelle (50).
How do the Q25-associated proteins differ from those bound to Q103? Surprisingly, we found a number of similarities between the two lists. For example, 49 (∼15%) proteins were shared in at least five of eight treatment conditions and 34 (11%) were shared by at least 6 of 8 conditions, suggesting that a core group of factors might bind both long and short polyQ fragments. These core factors included a number of relatively abundant cellular proteins, such ENO1, PGK1, ADH1, TDH1/2, and BMH1, so caution is certainly warranted in interpreting these findings. One potentially interesting difference between the Q25- and Q103-associated proteins was the presence of HHF1, a core histone H4 component involved in maintenance of genomic integrity, only bound to the longer polyQ.
Expanded Model for the Ordered Recruitment of Molecular Chaperones during Q103 Aggregation
The HSPs are thought to play critical roles in proteostasis and protein quality control (53). Thus, we decided to specifically focus on these molecular chaperones to better understand how they might cooperate during polyQ aggregation. In these studies, 115-7c was an essential tool because it allowed us to, for the first time, promote Hsp70 binding and tune polyQ aggregation without artificially changing chaperone levels. Interestingly, we found that 115-7c prolonged the association of both Hsp70 and Hsp90 with Q103. It is not yet clear why Hsp70 and Hsp90 are co-incident in their accumulation because we did not directly detect the co-chaperone, Sti1, which normally links these chaperones (27, 29). However, it is possible that the stringency of the washing steps removed this factor or that it is present at low levels. Regardless, the result of this prolonged interaction appeared to be an aborted attempt to form puncta (see Fig. 1B). Concurrently, the levels of Hsp26 and Hsp104 bound to Q103 were dramatically reduced. The reduced Hsp104 binding might be expected, as this disaggregase is thought to recognize later-stage aggregates; however the possible impact of reduced binding to Hsp26 is less certain. One possibility is that Hsp26 may compete with Hsp70 and/or Hsp90 for binding sites, such that prolonged interactions directly block Hsp26 recruitment. Alternatively, failure of Hsp70 and Hsp90 to properly facilitate aggregate maturation might reduce apparent levels of bound Hsp26 because this chaperone is incorporated into mature polyQ aggregates (26). Finally, these studies suggest that additional studies might be warranted on FPR1 or Hsp12, chaperones whose potential roles in polyQ aggregation are not as well understood.
Based on these findings, the emerging model by which HSPs influence protein aggregation (2, 3, 5, 19–21, 24, 26) can be further clarified (Fig. 5). Specifically, we observed that Hsp26, Hsp70, and Hsp90 bind Q103, but not Q25, at early time points. This result might suggest that misfolded Q103 may display a molecular “danger signal” relatively early (∼2 h). As aggregation proceeds, Hsp90 and some of the Hsp70 and Hsp26 seemed to partially dissociate in response to an unknown signal, followed by recruitment of Hsp104. Finally, release of Hsp70 and Hsp90 appeared to be important steps in the continued maturation of polyQ aggregates, based on the experiments performed using 115-7c. One possibility is that Hsp70 must iteratively bind and release Q103 as a timing mechanism to promote “hand-off” to Hsp104. If Hsp70 remains tightly bound, it might delay this step and potentially compete with Hsp104 binding sites. Alternatively, Hsp70 release may be coupled with structural transitions that promote Hsp104 binding. Regardless, the net effect of this pharmacologically disrupted timing was aborted aggregation, highlighting that it is an important transition. Together, these findings have interesting implications on how Hsp70 and other HSPs engage in the quality control of misfolded polyglutamine fragments.
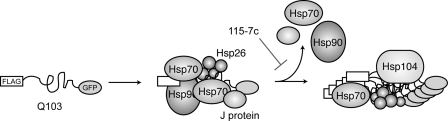
FIGURE 5.
Model for the ordered assembly of chaperones on polyQ during aggregation. Based on the mass spectrometry studies, Hsp26, Hsp70, and Hsp90 appear to recognize misfolded Q103 relatively early in the process (∼ 2 h). Progression to a mature aggregate seems to require at least partial or temporary release of Hsp70 and Hsp90, a process that is sensitive to the Hsp70 stimulator, 115-7c.
Supplementary Material
Acknowledgments
We thank the Andrew Lieberman, Daniel Klionsky, and Lois Weisman Laboratories (University of Michigan) for reagents and helpful advice. The anti-Hsp26 and anti-Hsp104 antibodies were kindly provided by Johannes Buchner (Technische Universität München, Garching) and Stefan Walter (University of Michigan).
This work was supported by NIH Grant NS059690 (to J. E. G.) and grants from AFAR and the Wood Foundation (to M. L. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HSP
- heat shock protein
- polyQ
- polyglutamine
- NEF
- nucleotide exchange factor
- GO
- gene ontology.
REFERENCES
- 1. Hartl F. U., Hayer-Hartl M. (2009) Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 2. Rikhvanov E. G., Romanova N. V., Chernoff Y. O. (2007) Prion 1, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chernoff Y. O. (2007) FEBS Lett. 581, 3695–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frydman J. (2001) Annu. Rev. Biochem. 70, 603–647 [DOI] [PubMed] [Google Scholar]
- 5. Grimminger-Marquardt V., Lashuel H. A. (2010) Biopolymers 93, 252–276 [DOI] [PubMed] [Google Scholar]
- 6. Höhfeld J., Cyr D. M., Patterson C. (2001) EMBO Rep. 2, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Voisine C., Pedersen J. S., Morimoto R. I. (2010) Neurobiol Dis. 40, 12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Evans C. G., Chang L., Gestwicki J. E. (2010) J. Med. Chem. 53, 4585–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Douglas P. M., Dillin A. (2010) J. Cell Biol. 190, 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bukau B., Weissman J., Horwich A. (2006) Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 11. Pratt W. B., Morishima Y., Murphy M., Harrell M. (2006) Handb. Exp. Pharmacol. 111–138 [DOI] [PubMed] [Google Scholar]
- 12. Bösl B., Grimminger V., Walter S. (2006) J. Struct. Biol. 156, 139–148 [DOI] [PubMed] [Google Scholar]
- 13. Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. (2005) Nat. Struct. Mol. Biol. 12, 842–846 [DOI] [PubMed] [Google Scholar]
- 14. Douglas P. M., Treusch S., Ren H. Y., Halfmann R., Duennwald M. L., Lindquist S., Cyr D. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7206–7211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sherman M. Y., Muchowski P. J. (2003) Neuromol. Med. 4, 133–146 [DOI] [PubMed] [Google Scholar]
- 16. Duennwald M. L., Jagadish S., Giorgini F., Muchowski P. J., Lindquist S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11051–11056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meriin A. B., Zhang X., He X., Newnam G. P., Chernoff Y. O., Sherman M. Y. (2002) J. Cell Biol. 157, 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Urakov V. N., Vishnevskaya A. B., Alexandrov I. M., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (2010) Prion 4, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masison D. C., Kirkland P. A., Sharma D. (2009) Prion 3, 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haslberger T., Bukau B., Mogk A. (2010) Biochem. Cell Biol. 88, 63–75 [DOI] [PubMed] [Google Scholar]
- 21. Summers D. W., Douglas P. M., Cyr D. M. (2009) Prion 3, 59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moosavi B., Wongwigkarn J., Tuite M. F. (2010) Yeast 27, 167–179 [DOI] [PubMed] [Google Scholar]
- 23. Schirmer E. C., Lindquist S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13932–13937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shorter J., Lindquist S. (2008) EMBO J. 27, 2712–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zietkiewicz S., Lewandowska A., Stocki P., Liberek K. (2006) J. Biol. Chem. 281, 7022–7029 [DOI] [PubMed] [Google Scholar]
- 26. Cashikar A. G., Duennwald M., Lindquist S. L. (2005) J. Biol. Chem. 280, 23869–23875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reidy M., Masison D. C. (2010) Mol. Cell Biol. 30, 3542–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sielaff B., Tsai F. T. (2010) J. Mol. Biol. 402, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Song Y., Masison D. C. (2005) J. Biol. Chem. 280, 34178–34185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muchowski P. J., Wacker J. L. (2005) Nat. Rev. Neurosci 6, 11–22 [DOI] [PubMed] [Google Scholar]
- 31. Buchberger A., Theyssen H., Schröder H., McCarty J. S., Virgallita G., Milkereit P., Reinstein J., Bukau B. (1995) J. Biol. Chem. 270, 16903–16910 [DOI] [PubMed] [Google Scholar]
- 32. Kampinga H. H., Craig E. A. (2010) Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patury S., Miyata Y., Gestwicki J. E. (2009) Curr. Top Med. Chem. 9, 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fewell S. W., Smith C. M., Lyon M. A., Dumitrescu T. P., Wipf P., Day B. W., Brodsky J. L. (2004) J. Biol. Chem. 279, 51131–51140 [DOI] [PubMed] [Google Scholar]
- 35. Wisén S., Androsavich J., Evans C. G., Chang L., Gestwicki J. E. (2008) Bioorg Med. Chem. Lett. 18, 60–65 [DOI] [PubMed] [Google Scholar]
- 36. Wisén S., Bertelsen E. B., Thompson A. D., Patury S., Ung P., Chang L., Evans C. G., Walter G. M., Wipf P., Carlson H. A., Brodsky J. L., Zuiderweg E. R., Gestwicki J. E. (2010) ACS Chem. Biol. 5, 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang L., Bertelsen E. B., Wisén S., Larsen E. M., Zuiderweg E. R., Gestwicki J. E. (2008) Anal. Biochem. 372, 167–176 [DOI] [PubMed] [Google Scholar]
- 38. Wiederhold E., Veenhoff L. M., Poolman B., Slotboom D. J. (2010) Mol. Cell Proteomics 9, 431–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brady G. F., Galbán S., Liu X., Basrur V., Gitlin J. D., Elenitoba-Johnson K. S., Wilson T. E., Duckett C. S. (2010) Mol. Cell Biol. 30, 1923–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Willingham S., Outeiro T. F., DeVit M. J., Lindquist S. L., Muchowski P. J. (2003) Science 302, 1769–1772 [DOI] [PubMed] [Google Scholar]
- 41. Krobitsch S., Lindquist S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1589–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Duennwald M. L., Jagadish S., Muchowski P. J., Lindquist S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11045–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Y., Meriin A. B., Zaarur N., Romanova N. V., Chernoff Y. O., Costello C. E., Sherman M. Y. (2009) Faseb J. 23, 451–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dehay B., Bertolotti A. (2006) J. Biol. Chem. 281, 35608–35615 [DOI] [PubMed] [Google Scholar]
- 45. Meriin A. B., Zhang X., Alexandrov I. M., Salnikova A. B., Ter-Avanesian M. D., Chernoff Y. O., Sherman M. Y. (2007) FASEB J. 21, 1915–1925 [DOI] [PubMed] [Google Scholar]
- 46. Balzi E., Wang M., Leterme S., Van Dyck L., Goffeau A. (1994) J. Biol. Chem. 269, 2206–2214 [PubMed] [Google Scholar]
- 47. Osherovich L. Z., Weissman J. S. (2001) Cell 106, 183–194 [DOI] [PubMed] [Google Scholar]
- 48. Gokhale K. C., Newnam G. P., Sherman M. Y., Chernoff Y. O. (2005) J. Biol. Chem. 280, 22809–22818 [DOI] [PubMed] [Google Scholar]
- 49. Wiwatwattana N., Landau C. M., Cope G. J., Harp G. A., Kumar A. (2007) Nucleic Acids Res. 35, D810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang H., Lim P. J., Karbowski M., Monteiro M. J. (2009) Hum. Mol. Genet. 18, 737–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y., Meriin A. B., Costello C. E., Sherman M. Y. (2007) Prion 1, 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bagriantsev S. N., Gracheva E. O., Richmond J. E., Liebman S. W. (2008) Mol. Biol. Cell 19, 2433–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. (2008) Science 319, 916–919 [DOI] [PubMed] [Google Scholar]
- 54. Satyal S. H., Schmidt E., Kitagawa K., Sondheimer N., Lindquist S., Kramer J. M., Morimoto R. I. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5750–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.