Background: An enigma in the bioenergetics of methanogens is how the generation of proton and sodium gradients are used to synthesize ATP.
Results: Purified methanogen ATP synthase was stimulated by sodium ions that also provided pH-dependent protection against DCCD.
Conclusion: Methanobrevibacter ruminantium harbors an A-type enzyme with the ability to switch between sodium ions and protons.
Significance: ATP synthesis by methanogens depends on the environmental conditions that prevail.
Keywords: Archaea, ATP Synthase, Bioenergetics, Microbiology, Sodium Transport
Abstract
An unresolved question in the bioenergetics of methanogenic archaea is how the generation of proton-motive and sodium-motive forces during methane production is used to synthesize ATP by the membrane-bound A1Ao-ATP synthase, with both proton- and sodium-coupled enzymes being reported in methanogens. To address this question, we investigated the biochemical characteristics of the A1Ao-ATP synthase (MbbrA1Ao) of Methanobrevibacter ruminantium M1, a predominant methanogen in the rumen. Growth of M. ruminantium M1 was inhibited by protonophores and sodium ionophores, demonstrating that both ion gradients were essential for growth. To study the role of these ions in ATP synthesis, the ahaHIKECFABD operon encoding the MbbrA1Ao was expressed in Escherichia coli strain DK8 (Δatp) and purified yielding a 9-subunit protein with an SDS-stable c oligomer. Analysis of the c subunit amino acid sequence revealed that it consisted of four transmembrane helices, and each hairpin displayed a complete Na+-binding signature made up of identical amino acid residues. The purified MbbrA1Ao was stimulated by sodium ions, and Na+ provided pH-dependent protection against inhibition by dicyclohexylcarbodiimide but not tributyltin chloride. ATP synthesis in inverted membrane vesicles lacking sodium ions was driven by a membrane potential that was sensitive to cyanide m-chlorophenylhydrazone but not to monensin. ATP synthesis could not be driven by a chemical gradient of sodium ions unless a membrane potential was imposed. ATP synthesis under these conditions was sensitive to monensin but not cyanide m-chlorophenylhydrazone. These data suggest that the M. ruminantium M1 A1Ao-ATP synthase exhibits all the properties of a sodium-coupled enzyme, but it is also able to use protons to drive ATP synthesis under conditions that favor proton coupling, such as low pH and low levels of sodium ions.
Introduction
Microorganisms have been shown to harbor a number of membrane-bound ATP synthases that are used to synthesize ATP via a proton or sodium ion gradient. These enzymes can be divided into three distinct classes as follows: F-type (F1Fo)-ATPases, A-type archaeal (A1Ao)-ATPases, and V-type (V1Vo)-ATPases. The A1Ao-ATP synthase is composed of at least nine different subunits with a stoichiometry of A3B3DE2FH2 for the A1 domain and Cac1–10 for the Ao domain and are structurally more closely related to the V1Vo-ATPases (1, 2). In most described A1Ao-type enzymes, multiple copies of the membrane-bound subunit c form the proteolipid ring of the Ao domain. The proteolipids of A1Ao-ATPases show considerable variability in size (2, 4, 6, or 26 transmembrane helices) and are proposed to have variable coupling stoichiometries (number of ions translocated per ATP synthesized) based on the number of conserved ionizable groups per monomer (2).
Methanogenic archaea are a group of strictly anaerobic microorganisms that produce methane from a limited group of substrates, such as H2 and CO2, formate, methanol, methylamine, and/or acetate by a pathway involving unique coenzymes (3). Methanogens produce both proton and sodium ion gradients during methanogenesis through ion-translocating mechanisms (4, 5). These gradients are then used to synthesize ATP by a chemiosmotic mechanism involving a membrane-bound A1Ao-ATP synthase (4, 6, 7). An unresolved issue in this bioenergetic scheme is how these two primary ion gradients are use to synthesize ATP. Examples of proton-coupled (e.g. Methanosarcina mazei Gö1) (8) and sodium-coupled (e.g. Methanothermobacter marburgensis strain Marburg) (9, 10) A1Ao-ATP synthases have been reported, despite the presence of a sodium ion-binding signature in the c subunits of all methanogen enzymes (11). However, it should be noted that even for bacterial F1Fo-ATP synthases that are sodium ion-coupled, protons are still translocated by these enzymes under conditions that favor proton transport (e.g. low pH and sodium ion concentrations) (12).
To date, very few A1Ao-ATP synthases from non-cytochrome-containing methanogens have been examined with respect to coupling ion specificity, and the enzymes from mesophilic methanogens have proved difficult to purify from the host organism. The genome of the rumen methanogen Methanobrevibacter ruminantium M1 was recently sequenced (13), and this provided us with a molecular platform to address the ion specificity of the A1Ao-ATP synthase from this archaeon.
EXPERIMENTAL PROCEDURES
Growth of M. ruminantium M1
M. ruminantium M1 (DSM 1093) was grown in medium RM02, and the effects of inhibitors were tested as described by Wedlock et al. (14). All inhibitors were dissolved in 0.1 ml of ethanol (or an appropriate control of ethanol), except for amiloride and 5-(N-ethyl-N-isopropyl)-amiloride (EIPA),2 which were dissolved in DMSO. Inhibitor or the appropriate diluent was added to 10-ml cultures in the exponential phase (absorbance at 600 nm (A600) ∼0.1, 16-mm path length).
Preparation of M. ruminantium M1 Inverted Membrane Vesicles
Typically 2 g of cell pellets were washed and resuspended with pre-cooled TMGT buffer (50 mm Tris·Cl (pH 7.5), 5 mm MgSO4, 10% (w/v) glycerol, and 0.1 mm tris[2-carboxyethyl]phosphine (TCEP)). Pancreatic DNase I and phenylmethylsulfonyl fluoride (PMSF) were added to 0.1 mg/ml and 0.1 mm, respectively, and the cells were disrupted by sonication on ice five times for 2 min with a tip sonicator. The lysate was cleared of debris by centrifugation at 8,000 × g for 10 min, and the inverted membrane vesicles were pelleted from the supernatant at 180,000 × g for 1 h at 4 °C and resuspended in TMGT buffer to a concentration of 5 mg/ml.
E. coli Strains, Plasmids, and Growth Conditions
Escherichia coli DH10B (15) was used for all cloning experiments, and E. coli DK8 (16), lacking the ATP synthase genes encoding the unc operon (Δatp), was used to overproduce the A1Ao-ATP synthase of M. ruminantium M1. Other common E. coli expression strains, including C41(DE3), C43(DE3), and BL21(DE3), were also tested (17). Plasmids used were pUC19 (18) for cloning and pTrc99A (Amersham Biosciences) for overexpression of A1Ao-ATP synthase. To overproduce the A1Ao-ATP synthase, transformants of E. coli DK8 Δatp were routinely grown at 37 °C with shaking at 200 rpm in 2× YT medium (19) containing 2 g/liter glucose and 100 μg of ampicillin/ml.
Construction of an Expression Plasmid for A1Ao-ATP Synthase
The genes encoding for the subunits of the M. ruminantium M1 A1-ATPase (ahaECFABD) were cloned into the expression vector pTrc99A (Amersham Biosciences) as a 6.2-kb BamHI-XbaI PCR product, amplified utilizing the forward primer MbrA1FWD (5′-AAATTTGGATCCGGAATCTTAGGTTAGGAGGTCAAT-3′) containing a BamHI site and the reverse primer MbrA1Rev (5′-AAATTTTCTAGATAACAAGCAAAATATGAATTGC-3′) containing an XbaI site. M. ruminantium M1 genomic DNA served as the template. The amplicon was digested with BamHI and XbaI and cloned into pTrc99A digested with the same restriction enzymes, creating the plasmid pTrMbrA1. To facilitate purification, an N-terminal hexa-histidine tag was introduced into subunit A (atpA) by PCR overlap extension (20). The plasmid pTrMbrA1 was used as the template for primers MbrA1HisMid (5′-TTAGACAAGTTCTTAGTCGACTCTG-3′), overlapping a natural SalI site, and the primer MbrA1HisRev (5′-ATGATGATGATGATGATGCATCCCATCTGCGACGATAACAGG-3′), generating a 0.55-kb PCR product. A second PCR was performed with the primers MbrA1HisFWD (5′-ATGCATCATCATCATCATCATAGAGGAACTCAAATGTATGAA-3′) and MbrA1REV, generating a 3.9-kb fragment. The 0.55- and 3.9-kb fragment were then used for PCR overlap extension with the external primers MbrA1HisMid and MbrA1Rev. The 4.45-kb product obtained was digested with SalI and XbaI. Plasmid pTrMbrA1 was digested with BamHI and SalI, and the 1.7-kb BamHI-SalI fragment generated from this digestion along with the 4.45-kb SalI-XbaI PCR overlap product fragment were ligated simultaneously into pTrc99a digested with BamHI and XbaI to create the plasmid pTrMbrA1His. To construct a plasmid containing the entire atp operon, the genes (atpHIK) encoding the M. ruminantium Ao subunits were cloned by PCR using the forward primer MbrAoFWD containing an NcoI site (5′-ATTTAATTACCATGGTGATTTATTATGGCA-3′) and the reverse primer MbrAoREV (5′-AGAGACAATTTTATCTGCCCCAGAGCTCAT-3′) containing a SacI site. The 3.3-kb fragment was digested with NcoI and SacI and ligated into the plasmid pTrMbrA1HIS digested with NcoI and SacI, thereby creating the plasmid pTrMbrA1AoHIS containing the full-length M. ruminantium M1 atp operon with an N-terminal hexa-His-tagged subunit A.
Expression and Purification of MbbrA1Ao
E. coli strain DK8 (Δatp) harboring plasmid pTrMbbrA1AoHis was grown at 37 °C with shaking at 200 rpm in 2× YT medium (19) containing 2 g of glucose/liter and 100 μg of ampicillin/ml. At an A600 (10-mm path length) of 0.4, the culture was induced with 1 mm isopropyl β-d-thiogalactopyranoside (IPTG) and incubation continued for 4 h. Cells were harvested, washed with pre-cooled TMGT buffer, and resuspended in the same buffer. PMSF was added to 0.1 mm, and the cells were disrupted by three passages through a French pressure cell at 20,000 p.s.i. Pancreatic DNase I was added to 0.1 mg/ml, and the mixture was kept on ice for 1 h or until viscosity decreased. The lysate was cleared of debris by centrifugation at 8,000 × g for 10 min, and the inverted membrane vesicles were pelleted from the supernatant at 180,000 × g for 1 h at 4 °C. Inverted membrane vesicles were washed twice in TMGT buffer and resuspended in 50 mm Tris·Cl (pH 7.5), 5 mm MgSO4, 10% (w/v) glycerol, 0.1 mm PMSF, and 0.1 mm TCEP. To extract the MbbrA1Ao, membrane vesicles were diluted to 5 mg of protein/ml in solubilization buffer (50 mm Tris·Cl (pH 7.5), 5 mm MgSO4, 10% (w/v) glycerol, 1% n-dodecyl-β-d-maltoside (DDM), 0.1 mm PMSF, and 0.1 mm TCEP) and incubated with gentle stirring at 4 °C for 2 h. The nonsolubilized material was removed by ultracentrifugation (180,000 × g, 1 h, 4 °C). The supernatant was applied to an IMAC column containing high performance nickel-nitrilotriacetic acid-Sepharose (Amersham Biosciences) that was equilibrated with purification buffer A (50 mm Tris·Cl (pH 7.5), 5 mm MgSO4, 10% (w/v) glycerol, 0.05% (w/v) DDM, 500 mm NaCl, 0.1 mm PMSF, and 0.1 mm TCEP) containing 10 mm imidazole. To remove contaminating proteins, the column was washed first with purification buffer A containing 10 mm imidazole and second with purification buffer A containing 40 mm imidazole. The MbbrA1Ao was eluted with purification buffer containing 100 mm imidazole. Further removal of other protein contaminants and excess salts was performed by precipitation with polyethylene glycol 6000 (PEG6000). The eluate was incubated for 1 h at room temperature with 10% (w/v) PEG6000, and the precipitate was subsequently removed by centrifugation at 54,000 × g for 20 min at 4 °C. The MbbrA1Ao was then precipitated by incubating the supernatant for 1 h with 15% (w/v) PEG6000. The precipitate was pelleted by centrifugation at 54,000 × g for 20 min at 4 °C and resuspended in 0.5 ml of resuspension buffer A (50 mm Tris·Cl (pH 7.5), 5 mm MgSO4, 10% (w/v) glycerol, 0.05% (w/v) DDM, 0.1 mm PMSF, and 0.1 mm TCEP). PEG6000 was pelleted by a 1-min centrifugation at 17,000 × g, the pellet discarded, and the supernatant containing the purified protein retained. To extract the c-oligomer (proteolipid), a chloroform/methanol extraction was performed using purified MbbrA1Ao. This was carried out as described by Dmitriev et al. (21) using 10% (w/v) citric acid as the buffer.
SDS-PAGE and Immunoblotting
MbbrA1Ao preparations were routinely analyzed on 14% (w/v) polyacrylamide gels in the presence of 0.1% SDS using the buffer system of Laemmli (22). Polypeptide bands were visualized using either Simply Blue® Safe Stain (Invitrogen) or by silver staining (23). Where trichloroacetic acid (TCA) precipitation was applied, TCA was added to purified MbbrA1Ao solution to a final concentration of 10% (v/v) and incubated on ice for 5 min. The precipitate was pelleted by centrifugation for 5 min at 16,000 × g to ensure all protein was collected, and the pellets were washed with 200 mm Tris·Cl (pH 8.9). The pellet was resuspended in resuspension buffer A, heated for 5 min at 96 °C, and analyzed by SDS-PAGE. During immunoblotting, membrane vesicles were subjected to 14% SDS-PAGE followed by electroblotting onto a polyvinylidene difluoride membrane, ensuring efficient transfer by including 0.02% (w/v) SDS in the running buffer of Laemmli (22). Detection was achieved using a penta-His antibody conjugate (Qiagen) directed against the hexa-histidine tag of the A subunits of the recombinant A1Ao as described previously (24). The antibody-specific bands were visualized using the SuperSignal® West Pico chemiluminescence system. The identification of the A1Ao-ATP synthase subunits was further confirmed by matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry (MALDI TOF/TOF MS) on a 4800 MALDI TOF/TOF analyzer (AB Sciex, MA). Protein bands, corresponding to the predicted sizes of the A1Ao subunits, were excised from the gel, and proteins in-gel were digested with trypsin following the protocol of Shevchenko et al. (25). All MS spectra were acquired in positive ion mode with 800 laser pulses per sample spot. The strongest 15 precursor ions per sample spot were selected for collision-induced dissociation MS/MS with 1600 laser pulses per precursor. For protein identification, collision-induced dissociation MS/MS peak lists were searched against the Swiss-Prot amino acid sequence data base using the Mascot search engine.
Functional Assays
ATP hydrolysis activity was determined at pH 6.5 in an assay containing 100 mm MES, 40 mm sodium acetate, 40 mm Na2S2O5, 10 mm MgSO4, and 10% (w/v) glycerol. For determination of ATPase activity in the absence of sodium ions, the assay buffer contained potassium acetate and K2S2O5 to replace the respective sodium salts. For pH range examination (pH 5–9), 100 mm MES, 100 mm MOPS, and 100 mm Tris·Cl were included. The reaction was started using either sodium-ATP or Tris-ATP to a final concentration of 2.5 mm. ATPase activity was measured at 37 °C using a colorimetric assay that detects inorganic phosphate liberated from ATP as described previously by Heinonen and Lahti (26). Nonenzymatic degradation of ATP under these conditions was less than 10% of the total phosphate. One unit of ATPase activity is defined as the amount of enzyme liberating 1 μmol of Pi or ADP/min at 37 °C. Approximately 50–100 μg of recombinant A1Ao-ATP synthase was used for measurements.
ATP synthesis experiments were performed using inverted membrane vesicles of E. coli DK8 (Δatp) expressing MbbrA1Ao and prepared at pH 7.8. Assays were performed at 30 °C in 2 ml of assay buffer A containing 50 mm MES (pH 6.50), 40 mm potassium acetate, 40 mm K2S2O5, 10 mm MgSO4, 160 mm KCl, 1.5 mm ADP, 5 mm KH2PO4. Inverted membrane vesicles (0.5 mg) were diluted 40-fold into assay buffer, and ATP synthesis reaction was initiated by the addition of 2 μm valinomycin to induce a potassium diffusion potential of ∼100 mV as calculated using the Nernst equation: 59 × log10([K+]out/[K+]in). To create a chemical gradient of sodium ions (ΔpNa+) of 95 mV (59 × log10([Na+]out/[Na+]in), NaCl-loaded (125 mm) vesicles were diluted 40-fold into the assay buffer. Vesicles not loaded with NaCl were subjected to the same treatment. At various time intervals, 100-μl aliquots were removed and transferred to 400 μl of stop solution (1% (w/v) trichloroacetic acid, 2 mm EDTA). Each sample was diluted 250-fold in water prior to the measurement of ATP using the luciferin-luciferase assay (27). To measure ATP, each sample was diluted into 400 μl of Tris acetate buffer (50 mm Tris acetate (pH 7.8), 2 mm EDTA, 50 mm MgCl2) in a luminometer tube. Fifty μl of luciferin-luciferase reagent (Sigma) was added to the tube, and the chemiluminescence was monitored with an FB12 chemiluminometer (Berthold). The amount of ATP synthesized was calculated from a standard curve performed on the day of each set of ATP measurements. For each individual experimental set, the presence of background ATP was measured using nonenergized vesicles and subtracted from total ATP measured. Protein concentrations were determined using a bicinchoninic acid protein assay kit (Sigma) with bovine serum albumin as the standard.
Molecular Modeling and Inhibitor Docking
The M. ruminantium c-oligomer was modeled in Coot (28) using the rotor ring structure of ATP synthase subunit K of Enterococcus hirae (Protein Data Bank codes 2CYD and 2DB4) as a template. Using the Clustal sequence alignment of the deduced amino acids, individual residues were mutated taking care to correctly fit rotamers to produce the most accurate homology model. Molecular docking experiments were carried out using the program Genetic Optimization for Ligand Docking (GOLD), version 5.0 (29), and favoring the Piecewise Linear Potential (CHEMPLP) fitness function. The binding site was composed of residues that fell within 10 Å of the conserved active site residue Glu-140. The GOLD runs employed a 100% search efficiency, and the diverse solutions options was turned on using a cluster size of 2 and root mean square deviation of 2 Å. Hydrogen bonding interactions were encouraged with the active site glutamate and the guanidinium moiety of inhibitors amiloride and 5-(N-ethyl-N-isopropyl)amiloride (EIPA) (Scheme 1) by allowing the residue to rotate freely during the docking process. Twenty binding poses were generated and inspected for each inhibitor, and conformations were chosen taking into account their ranking, interactions with active site residues, and by monitoring per atom scores generated by GOLD and the CHEMPLP fitness function.
SCHEME 1.
Structures of the MbbrA1Ao-ATP synthase inhibitors amiloride and EIPA.
RESULTS
Growth of M. ruminantium M1 on H2 and CO2 Is Dependent on the Membrane Potential and a Chemical Gradient of Sodium Ions
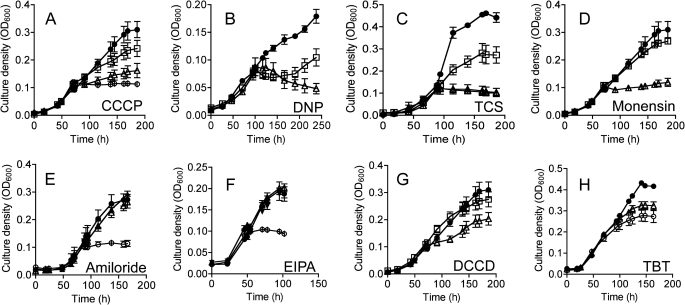
M. ruminantium M1 is found in ruminant animals fed a wide variety of diets (30) and grows with H2 plus CO2 and formate, producing methane (31). To study the role of different electrochemical gradients in the growth of M1, we tested the effect of various uncouplers, ionophores, and ATP synthase inhibitors on the growth of M. ruminantium M1. M. ruminantium M1 was inhibited by the protonophores CCCP, 2,4-dinitrophenol (DNP), and 3,3′,4′,5-tetrachlorosalicylanilide (TCS), but the sensitivity to these agents varied, with DNP and TCS being the most potent (Fig. 1, A–C). Monensin, an electroneutral Na+/H+ antiporter that dissipates the chemical gradient of Na+ ions, was a potent growth inhibitor of M. ruminantium M1 (Fig. 1D). Amiloride, and its more hydrophobic derivative EIPA, are blockers of sodium channels and Na+/H+ antiporter activity, and both compounds inhibited growth of M. ruminantium M1 (Fig. 1, E and F). The ATP synthase inhibitors dicyclohexylcarbodiimide (DCCD) and tributyltin chloride (TBT-Cl) slowed growth of M. ruminantium M1, but only TBT-Cl completely arrested growth (Fig. 1, G and H). Taken together, these data demonstrate that M. ruminantium M1 requires both a proton-motive force and sodium-motive force to grow and that classical ATP synthase inhibitors slow growth of M1. To study the role of these gradients in membrane bioenergetics, we purified and characterized the A1Ao-ATP synthase from M. ruminantium M1.
FIGURE 1.
Effect of protonophores, ionophores, and ATP synthase inhibitors on the growth of M. ruminantium M1 in batch culture. A, CCCP was added at 71 h to give final concentrations of 100 μm (open squares), 200 μm (open triangles), and 500 μm (open circles). B, DNP was added at 97 h to give final concentrations of 2 μm (open triangles) and 20 μm (open squares). C, TCS was added at 91 h to give final concentrations of 2 μm (open squares), 5 μm (open triangles), and 10 μm (solid diamonds). D, monensin was added at 71 h to give final concentrations of 1 μm (closed circles), 5 μm (open squares), and 10 μm (open triangles). E, amiloride was added at 92 h to give final concentrations of 20 μm (closed circles), 200 μm (open triangles) and 2 mm (open circles). F, EIPA was added at 48 h to give final concentrations of 20 μm (closed circles), 50 μm (open triangles), and 100 μm (open squares). G, DCCD was added at 71 h to give final concentrations of 100 μm (open squares) and 200 μm (open triangles), and 100 μl of ethanol (closed circles). H, TBT-Cl was added at 49 h to give final concentrations of 2 μm (closed circles), 20 μm (open triangles), and 50 μm (open circles). For all ethanol/DMSO-soluble inhibitors, 100 μl was also added to another series of cultures as a control. The symbols are means of three replicates, and the thin vertical bars represent one standard error on either side of the mean.
A1Ao-ATP Synthase from M. ruminantium M1
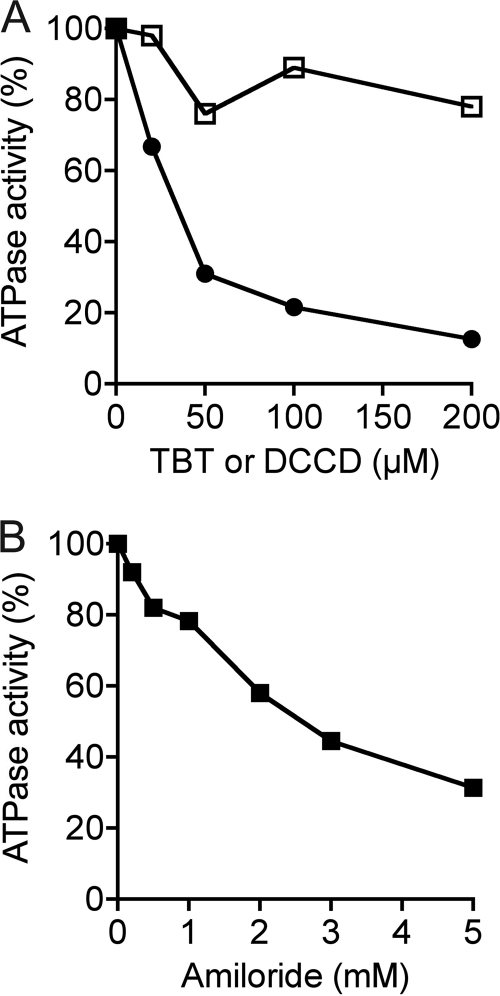
Membrane vesicles of M. ruminantium M1 were prepared from cells grown in anaerobic RM02 medium and assayed for ATPase activity by measuring the liberation of Pi from ATP hydrolysis. The specific activity of ATP hydrolysis in membranes of M. ruminantium M1 was low, 0.080 ± 0.009 units/mg protein, but comparable with the ATPase activity of other A1Ao enzymes (8, 32–35). The ATPase activity of M1 membrane vesicles was stable at 4 °C for up to 5 days, and activity declined after this point (data not shown). The ATPase activity of M1 membranes was sensitive to TBT-Cl and amiloride (Fig. 2, A and B), but DCCD was without significant effect (Fig. 2A).
FIGURE 2.
ATP hydrolysis activity of inverted membrane vesicles of M. ruminantium M1. Effect of TBT-Cl and DCCD (A) and amiloride (B) on ATPase activity. Inverted membrane vesicles were preincubated in assay buffer for 30 min at 37 °C in the presence of either TBT-Cl (closed circles) or DCCD (open squares) or amiloride (closed squares) before the reaction was started by the addition of 2.5 mm (final concentration) of sodium-ATP. Controls received solvent only. ATPase activity was determined using a colorimetric assay that measured the amount of inorganic phosphate liberated at 37 °C and pH 6.5. The values are the means of duplicate determinations, and the experimental error associated with these values was less than 15%. One hundred percent activity was equivalent to 28–34 milliunits/mg protein.
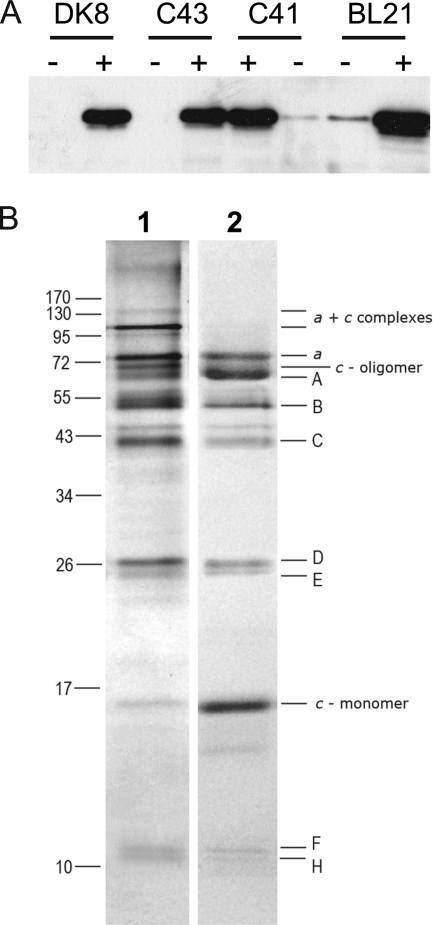
The genome of M. ruminantium M1 has recently been sequenced and harbors genes for an A-type ATP synthase (13). The nine genes are arranged in an operon in the order ahaHIKECFABD (viz. mru695-703) and encode the M. ruminantium M1 A1Ao-ATP synthase subunits H, I (a), K (c), E, C, F, A, B, and D, respectively. The start codon of each gene was designated by alignment of aha gene sequences with those of other microorganisms and the positions of potential ribosome-binding sites. The genome contains only one copy of the aha operon, and no F-type ATP synthases are present in the genome sequence (13). The ahaHIKECFABD operon, coding for MbbrA1Ao with a hexa-histidine tag at the N terminus of the A subunit, was cloned into the expression plasmid pTrc99A, and expression was tested in a number of different E. coli hosts: atp operon deletion mutant E. coli DK8 (Δatp) and common E. coli expression strains C41, C43, and BL21. All cells were induced with 1 mm IPTG, and immediately after induction the growth rate of all E. coli strains slowed, although growth was not completely inhibited (data not shown). The cells were grown for 4 h at 30 °C after induction and harvested. An immunoblot of the inverted membrane vesicles, which targeted the hexa-histidine tag on the N terminus of the A subunit, revealed that the MbbrA1Ao was indeed expressed (Fig. 3A).
FIGURE 3.
Expression and purification of MbbrA1Ao-ATP synthase from E. coli. A, expression of MbbrA1Ao (hexa-histidine tag of the A subunit) in various E. coli hosts. 1st lane, DK8 cells uninduced (−); 2nd lane, DK8 cells induced (+); 3rd lane, C43 cells uninduced; 4th lane, C43 cells induced; 5th lane, C41 cells induced; 6th lane, C41 cells uninduced; 7th lane, BL21 cells uninduced; 8th lane, BL21 cells induced. All cells were induced with 1 mm IPTG and harvested after 4–5 h of incubation at 30 °C. B, SDS-PAGE analysis of the purified MbbrA1Ao-ATP synthase. Samples were resolved on 14% polyacrylamide gel and stained with silver. Lane 1, 5 μg of purified MbbrA1Ao; lane 2, 5 μg of purified MbbrA1Ao treated with TCA. The molecular masses (in kilodaltons) corresponding to a broad range molecular mass marker (Invitrogen) are indicated on the left. The predicted molecular masses of the individual subunits are given on the right.
Various detergents, decyl maltoside, DDM, Triton X-100, CHAPS, sodium cholate, octylglucoside, and FOS-choline, were tested for their ability to extract and solubilize the A1Ao-ATP synthase from E. coli membranes. FOS-choline (0.5% (v/v) final concentration) was the most effective detergent for extracting the A1Ao-ATP synthase from E. coli membranes (>90%), but the enzyme lost activity rapidly in this detergent (data not shown). DDM (1% (w/v) final concentration) was only 40% effective at extracting A1Ao, but the enzyme retained good activity in this detergent. The DDM-solubilized A1Ao was purified using immobilized metal (nickel-Sepharose) ion affinity chromatography exploiting the His tag on the A subunit and eluted with 100 mm imidazole. The A1Ao fraction was further purified and concentrated by PEG precipitation (15% (w/v) PEG6000), and the final preparation was resuspended in 10 mm Tris·Cl (pH 8.0), 2 mm MgSO4, and 0.05% (w/v) DDM. The activity of the purified A1Ao-ATP synthase was 0.92 units/mg protein. SDS-PAGE analysis and MALDI-TOF MS analysis of the purified recombinant enzyme identified all nine subunits (Fig. 3B) as follows: c subunit (atpK) (predicted 15 kDa), D subunit (predicted 26.04 kDa), B subunit (predicted 48.63 kDa), A subunit (predicted 62.3 kDa), F subunit (predicted 11.32 kDa), E subunit (predicted 24.78 kDa), C subunit (predicted 42.45 kDa), H subunit (predicted 11.09 kDa), and a subunit (atpI) (predicted 73.97 kDa). SDS-PAGE revealed a protein running at an apparent molecular mass of 68 kDa (Fig. 3B, lane 1). TCA treatment, a procedure known to dissociate c-oligomers (36, 37), resulted in the disappearance of this 68-kDa protein and the appearance of a protein at 15 kDa, attributed to the size of the monomeric c subunit (Fig. 3B, lane 2).
MbbrA1Ao Is Stimulated by Sodium Ions, and DCCD Inhibition Can Be Prevented via the Addition of Sodium Ions in a pH-dependent Manner
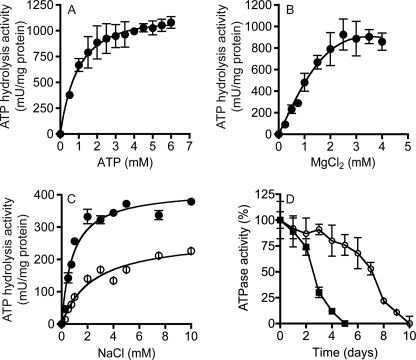
The properties of the purified MbbrA1Ao, unless otherwise stated, were examined measuring the liberation of inorganic phosphate (Pi) from ATP hydrolysis. The purified enzyme had an apparent Km for ATP of 0.55 mm when the Mg2+:ATP ratio was maintained at 2:1 (Fig. 4A). ATP hydrolysis activity was dependent upon the presence of MgCl2, with an apparent Km of 1 mm (Fig. 4B). ATPases capable of translocating Na+ ions show a specific stimulation of their ATP hydrolysis activity in the presence of low concentrations of NaCl (37–40). ATPase activity of the purified A1Ao-ATP synthase was stimulated 3–4-fold (pH 7.5) or 5–6-fold (pH 9.0) with increasing concentrations of Na+ (Fig. 4C). Kinetic analyses of the data in Fig. 4, using the Lineweaver-Burk equation, indicated that the apparent Km for Na+ at pH 7.0 and 9.0 was 0.5 mm (data not shown). In membrane vesicles, ATPase activity at 4 °C appeared stable, retaining nearly 100% of the starting activity for a period extending beyond 4 days (Fig. 4D). This was in contrast to the stability of the enzyme in its purified form in detergent solution, which progressively lost activity over the same time period (Fig. 4D).
FIGURE 4.
Biochemical properties of the purified MbbrA1Ao-ATP synthase. Effect of ATP (A) and MgCl2 (B) on ATPase activity. ATP hydrolysis assays were performed at pH 6.5 with 7.5 μg of purified MbbrA1Ao in the presence of 125 mm NaCl using a colorimetric assay that measured the amount of inorganic phosphate liberated at 37 °C. C, effect of NaCl on ATPase activity at pH 6.5 (open circles) and pH 8.5 (closed circles). ATP hydrolysis was performed with 7.5 μg of purified A1Ao-ATP synthase using the potassium salt of ATP. D, stability of the purified MbbrA1Ao at 4 °C in 0.05% (w/v) DDM (closed squares) and MbbrA1Ao in E. coli membrane vesicles (open circles).
A characteristic property of F-type ATPases is their specific inhibition by DCCD (41). This inhibition is due to the covalent binding of DCCD to a highly conserved carboxyl residue (glutamic acid or aspartic acid) in the c subunit shown to be located in the middle of the cytoplasmic membrane (42, 43). Protection from DCCD inhibition in a pH-dependent manner by Na+ has been observed with the F-type ATP synthases from Propionigenium modestum (44), Acetobacterium woodii (45), Ilyobacter tartaricus (46), Clostridium paradoxum (40), and the A-type enzyme from M. marburgensis strain Marburg (9, 10).
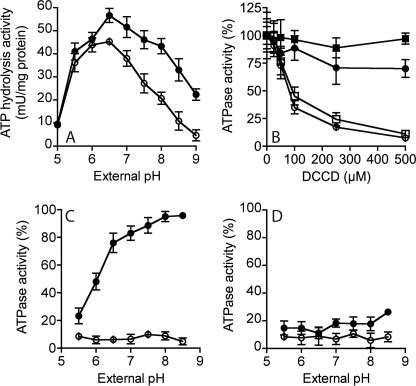
The pH optimum for ATP hydrolysis activity of the purified A1Ao-ATP synthase was broad, with high levels of activity over the pH range 5.5 to 9.0 (Fig. 5A). In the presence of 125 mm NaCl, the A1Ao enzyme had higher levels of ATPase activity in the alkaline pH range. When the A1Ao enzyme was incubated in the presence of increasing concentrations of DCCD for 20 min at pH 6.5 or 8.5 and then assayed for ATPase activity, significant inhibition of ATPase activity by DCCD was observed at 100 μm (Fig. 5B). If 125 mm NaCl was included in the assay with DCCD, the enzyme was protected from DCCD inhibition, and this protection was greatest at pH 8.5 (Fig. 5B). The effect of pH on either DCCD (Fig. 5C) or TBT-Cl (Fig. 5D) inhibition of ATPase activity was tested in the presence and absence of 125 mm NaCl. DCCD inhibited the A1Ao over a broad pH range, but if NaCl was present, the inhibition by DCCD was pH-dependent, and no inhibition was observed at high pH (Fig. 5C). In contrast, TBT-Cl inhibited ATPase activity over a broad pH range, and this inhibition was unaffected by NaCl (Fig. 5D).
FIGURE 5.
Effect of pH, NaCl, and DCCD on the activity of the MbbrA1Ao-ATP synthase in inverted membrane vesicles from E. coli DK8 (Δatp). A, effect of external pH on ATPase activity in the presence (closed circles) and absence (open circles) of 125 mm NaCl. B, effect of DCCD concentration on ATPase activity at pH 6.5 (open circles) and 8.5 (open squares) in the absence (open symbols) or presence (closed symbols) of 125 mm NaCl. C, effect of external pH on DCCD (250 μm) inhibition of ATP hydrolysis activity in the presence (closed circles) and absence (open circles) of 125 mm NaCl. D, effect of external pH on TBT-Cl (150 μm) inhibition of ATP hydrolysis activity in the presence (closed circles) and absence (open circles) of 125 mm NaCl. E. coli DK8 (Δatp) membranes containing MbbrA1Ao (25 μg) were incubated at 37 °C in buffer at either 6.5 or 8.5 with either DCCD or TBT-Cl (plus and minus 125 mm NaCl) for 30 min and then assayed for ATPase activity. Each point is the result of two to three biological replicates and the standard error associated with this measurement is shown.
ATP Synthesis in Inverted Membrane Vesicles Can Be Driven by an Artificially Imposed ΔpNa+ in the Presence of Δψ
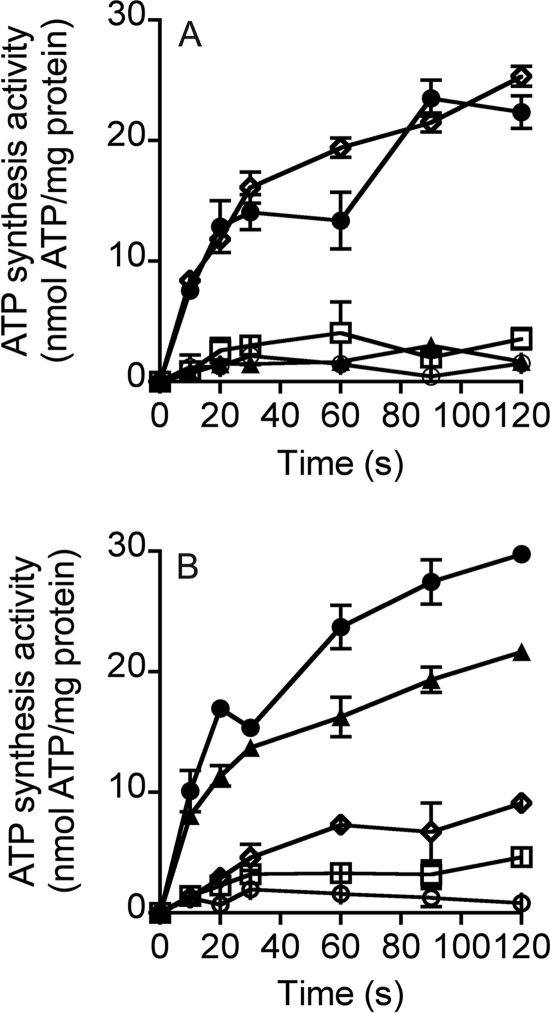
To study vectorial ion transport with the MbbrA1Ao, ATP synthesis was examined in inverted membrane vesicles of E. coli (DK8 Δatp) expressing the enzyme. ATP synthesis in inverted membrane vesicles was studied using a valinomycin-induced potassium diffusion potential (Fig. 6). When inverted membrane vesicles (pH 7.8) were diluted 40-fold into assay buffer A, no ATP synthesis was detected, even though a ΔpH gradient of 1.3 units was present (Fig. 6A). However, if 2 μm valinomycin was added to generate a potassium diffusion potential of 100 mV, ATP synthesis proceeded at a rate of 30 nmol of ATP/min/mg protein (Fig. 6A). ATP synthesis under these conditions was completely eliminated by preincubation of the vesicles with TBT-Cl, DCCD, or CCCP (Fig. 6A). Monensin had no effect on ATP synthesis under these conditions, indicating that ATP synthesis was not dependent on a chemical gradient of sodium ions (Fig. 6A). The effect of sodium ions on ATP synthesis was also studied using membrane vesicles loaded with NaCl and diluted into assay buffer without NaCl to generate a ΔpNa+ of 95 mV. No ATP was synthesized under these conditions (Fig. 6B). However, ATP synthesis was observed under identical conditions if a valinomycin-induced potassium diffusion potential of 100 mV was also applied (Fig. 6B). ATP synthesis under these conditions was inhibited by monensin, TBT-Cl, or DCCD, but not significantly by CCCP (Fig. 6B)
FIGURE 6.
ATP synthesis properties of the MbbrA1Ao-ATP synthase in inverted membrane vesicles from E. coli DK8 (Δatp). Time course ATP synthesis assays at pH 6.5 at 30 °C with 0.5 mg of inverted membrane vesicles. A, ATP synthesis was initiated by the addition of a valinomycin (2 μm)-induced potassium diffusion potential of >100 mV (solid circles), no valinomycin addition (open circles), CCCP (100 μm, solid triangles), TBT-Cl (150 μm, open squares), and monensin (5 μm, open diamonds). B, effect of ΔpNa+ on ATP synthesis by inverted membrane vesicles. To generate a ΔpNa+, sodium-loaded vesicles (125 mm NaCl) were diluted 40-fold into buffer A (50 mm MES (pH 6.50), 40 mm potassium acetate, 40 mm K2S2O5, 10 mm MgSO4, 160 mm KCl, 1.5 mm ADP, 5 mm KH2PO4) containing one of the following: 3.1 mm NaCl (inside NaCl concentration, 125 mm) to generate a ΔpNa+ of 95 mV (open circles); 3.1 mm NaCl and 2 μm valinomycin to generate a generate a ΔpNa+ of 95 mV and a K+/valinomycin diffusion potential of >100 mV (solid circles); 5 μm monensin, 3.1 mm NaCl, and 2 μm valinomycin to generate a generate a ΔpNa+ of 95 mV and a K+/valinomycin diffusion potential of >100 mV (open diamonds); 100 μm CCCP, 3.1 mm NaCl, and 2 μm valinomycin to generate a generate a ΔpNa+ of 95 mV and a K+/valinomycin diffusion potential of >100 mV (solid triangles); 150 μm TBT-Cl, 3.1 mm NaCl, and 2 μm valinomycin to generate a generate a ΔpNa+ of 95 mV and a K+/valinomycin diffusion potential of >100 mV (open squares). Each point is the result of three technical replicates, and the standard error associated with this measurement is shown.
Duplicated c Subunit of MbbrA1Ao Harbors Two Putative Sodium-binding Motifs
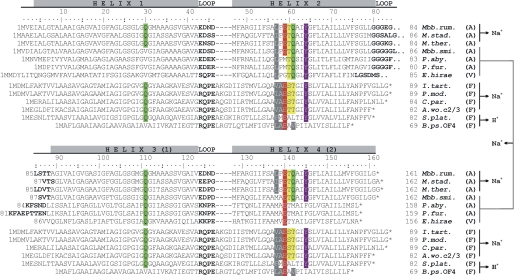
Analysis of the ahaK (c subunit encoding) gene by amino acid sequence alignment, including comparison to the rotor ring subunits from F-, V-, and A-type ATPases/synthases, revealed that the MbbrA1Ao c subunit is duplicated with respect to the two α-helix F-type ATP synthase c subunits and contains four transmembrane helices, forming two α-helical hairpins (Fig. 7). wo conserved carboxylate residues (Glu-59 and Glu-140) are present on the second and fourth helices. Each hairpin displays a complete sodium ion-binding signature (47–49), composed of identical amino acid residues. The first motif on helix 1 (H1) and 2 (H2) includes the amino acids Gln-30 (H1), Leu-57 (H2), Glu-59 (H2), Thr-60 (H2), Gln-61 (H2), and Tyr-64 (H2); the second motif on helix 3 and 4 includes Gln-111 (H3), Leu-138 (H4), Glu-140 (H4), Thr-141 (H4), Gln-142 (H4), and Tyr-145 (H4). The distribution of one Na+-binding site per hairpin resembles the Na+-binding signature, which is usually found in F-type ATP synthases (47, 48). However, in both binding sites, two glutamines are present, which appear be involved in Na+ coordination in the same way it has been described for the K-ring of the E. hirae V-type ATPase (49). Based on this analysis, we hypothesize that M. ruminantium contains duplicated c subunits, consisting of two fused α-helical hairpins, and each of the hairpins forms a complete Na+-binding site, which is equivalent to that found in the K-ring of E. hirae V-type ATPase.
FIGURE 7.
Alignment of the rotor ring subunits (c/K) from A-, V-, and F-type ATPases/synthases. The chains consist either of 1 (F-type) or 2 (A- and V-type) α-helical hairpins (α-helix loop-α-helix motif). The residues involved in Na+ coordination (47–49) or H+ coordination (62, 63) are given in colors. The c subunit of M. ruminantium M1 harbors two hairpins, each displaying one complete Na+-binding signature. The signature is equivalent to the E. hirae Na+ binding signature (1 Na+-binding site per c/K subunit consisting of 4 α-helices), which involves in this case three α-helices. Residue numbering (according to the M. ruminantium gene sequence) and α-helices are indicated. Loop regions are in boldface. Residue numbers of individual sequences are given at the beginning and end of the sequences, and the asterisk indicates the sequence end. The ion-binding type (Na+ or H+), the type of ATPase/synthase (A, V, and F), and the involvement of which hairpins in ion binding for the individual sequences is indicated on the right side after the species name. Sources of amino acid sequences were as follows: M. ruminantium M1 (Mbb. rum.; accession number YP_003423440), Methanosphaera stadtmanae (M. stad.; YP_448163), M. thermautotrophicus (M. ther.; NP_276094), Methanobrevibacter smithii (Mbb. smi.; YP_001273012), Pyrococcus abyssi (P. aby.; NP_127441), Pyrococcus furiosus (P. fur.; NP_577907), E. hirae (E. hirae; BAA04273), I. tartaricus (I. tart.; AAM94908.1), P. modestum (P. mod.; CAA41369.1), Clostridium paradoxum (C. par.; ABB13420.1), A. woodii (A. wo. c2/3; AAC45088.2/AAF01475.1), Spirulina platensis (S. plat.; EF520738), and Bacillus pseudofirmus OF4 (B. ps. OF4; YP_003426325.1).
Modeling Amiloride and EIPA within the c-oligomer of M. ruminantium
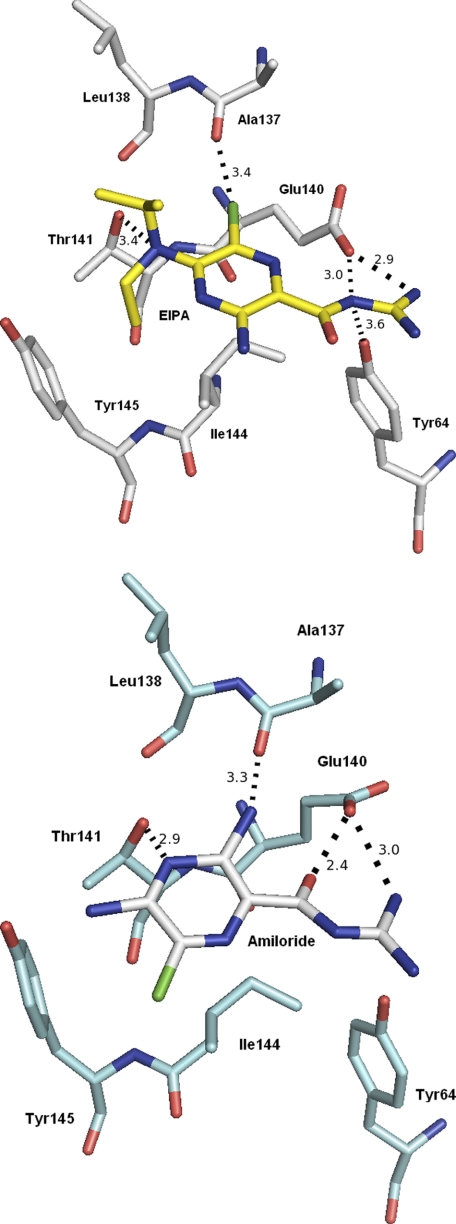
Amiloride inhibited ATPase activity at high concentrations and both amiloride and EIPA inhibited the growth of M. ruminantium M1 (Fig. 1, E and F). Amiloride has been reported to interact with Na+-dependent F-type ATP synthases (50, 51) but not A-type enzymes. To investigate this proposed interaction, amiloride and EIPA were docked within the active site of the c-oligomer of MbbrA1Ao to delineate any clear differences in binding orientation and activity between the inhibitors. Because both Na+ coordination sites within the c subunit of M. ruminantium M1 display significant sequence and structural similarities (Fig. 7) and the docking poses for EIPA at both sites are nearly identical (results not shown), the simulations presented and described here are based exclusively on Na+ coordination site 2 for consistency. The rotameric freedom given Glu-140 was due to the active site alterations thought to occur with the residue during ion translocation and interaction with the stator arginine in the opposing subunit of Na+-dependent ATP synthases (50, 51), which enabled the establishment of critical hydrogen bond interaction between the residue and guanidino group attached to both inhibitors. Our rationale is that the guanidinium group of amiloride and EIPA mimics the interaction of the conserved stator arginine and is in competition for binding to the c-ring sites (50). In both the EIPA and amiloride models, the guanidinium group forms a close hydrogen bond with the acidic carboxylate of Glu-140, as does the carbonyl of amiloride (2.4 Å) and, in the case of EIPA, an additional van der Waals interaction with the hydroxyl moiety of Tyr-64 (3.6 Å; Fig. 8, A and B). The pyrazine ring of EIPA stacks on top of the side chain of Ile-144, whereas the 6-chloro group bound to the pyrazine ring makes a van der Waals contact with the carbonyl of Ala-137 (3.4 Å). A similar interaction is observed with amiloride and the 3-amino group (3.3 Å), although both molecules form van der Waals contacts with the hydroxyl of Thr-141 via the pyrazine nitrogen of amiloride (2.9 Å) or the isopropyl nitrogen of EIPA (3.4 Å). A nonpolar/hydrophobic interaction with the phenyl ring of Tyr-145, the side chain of Ile-144, and the methyl group of Thr-141 is made with the ethyl group of EIPA and to some degree with the pyrazine ring. Amiloride, lacks this hydrophobic moiety, and as a consequence, we observed a larger number of orientations of the molecule within the active site of A1Ao when compared with EIPA orientations post-docking, suggesting that these hydrophobic contacts may provide a more stable enzyme-inhibitor complex and in turn facilitated the additional contacts with residues such as that observed with Tyr-64.
FIGURE 8.
Favored docked orientations of EIPA (A) and amiloride (B) bound within the active site of the c-ring of MbbrA1Ao-ATP synthase. These structures are taken from docking events promoting hydrogen bonding between the carboxylate of Glu-140 and the guanidinium group of the inhibitors. Hydrogen bond and van der Waals contacts are shown as dotted lines. Residues within 4 Å of the inhibitor are shown. All pictures were prepared using PyMOL (69).
Overall, we would assert that the nonpolar/hydrophobic moieties of EIPA and their interaction with active site residues underpin its orientation and efficacy as an inhibitor of A1Ao over amiloride, and both compounds form effective inhibitors due to the promotion of essential hydrogen bond interactions, particularly that with the acidic active site glutamate and guanidinium moieties (52).
DISCUSSION
M. ruminantium inhabits the rumen of ruminant animals, and recent work suggests it is one of the predominant methanogens in this environment (30). M. ruminantium grows by using H2 to reduce CO2 to CH4 and couples this metabolism to the generation of electrochemical gradients using primary pumps for protons (heterodisulfide reductase complex) and sodium ions (membrane-bound N5-methyltetrahydromethanopterin:coenzyme M methyltransferase). The sensitivity of M. ruminantium M1 to both uncouplers and sodium ionophores demonstrates that both gradients are formed during growth and that both are essential for growth. M. ruminantium M1 also harbors genes for a Na+/H+ antiporter, thus providing a mechanism to interconvert sodium and proton gradients, depending on the relative demand for either gradient during growth. One might speculate that Na+/H+ antiporters would be important for methanogens that grow in environments of high salt (e.g. the rumen) and employ a Na+-coupled ATP synthase. Intracellular sodium ions are toxic to microbial cells and therefore a mechanism to keep the intracellular sodium concentration low would be an important transport system. The pH of the rumen is ∼6.5 and is highly buffered, and therefore it is unlikely that strain M1 utilizes a Na+/H+ antiporter as a mechanism for pH homeostasis.
Methanogens are the only microorganisms reported to produce two primary ion gradients (i.e. a sodium-motive force and a proton-motive force) during growth, and how these two gradients are used to synthesize ATP remains to be explained experimentally (6). Initial hypotheses centered around the idea of two types of ATP synthases in methanogens (e.g. M. mazei and Methanothermobacter thermoautotrophicus), a sodium-coupled F-type (53) and a proton-coupled A-type (54, 55). With the advent of genome sequencing, it was revealed that the majority of methanogen genomes, including M. mazei and M. thermoautotrophicus, harbor only genes for A-type ATP synthases. One notable exception is Methanosarcina acetivorans, which has genes for both F-type and A-type ATP synthases. Saum et al. (56) have recently shown that the F-type ATP synthase is not essential for the growth of M. acetivorans. The M. ruminantium M1 genome contains an ahaHIKECFABD operon, and no genes for an F-type enzyme were identified. The entire operon was successfully expressed in E. coli and the MbbrA1Ao enzyme purified with all subunits present. We propose that this enzyme is able to use both protons and sodium ions to synthesize ATP, with sodium ions being the preferred coupling ion under the physiological conditions of the rumen (high sodium and near neutral pH). The properties of MbbrA1Ao define it as a sodium ion-translocating A-type ATP synthase. ATP hydrolysis activity of the purified complex was stimulated by sodium ions, and sodium ions provided protection against DCCD inhibition (but not TBT-Cl) in a pH-dependent manner. Similar properties have been reported for the A1Ao-ATP synthase from Methanothermobacter marburgensis strain Marburg. Work by Schönheit and co-workers (9, 10) established that sodium ions stimulate ATP synthesis and methanogenesis (at high sodium), and ATP synthesis was insensitive to protonophores. Moreover, DCCD sensitivity was observed only at low sodium concentrations (57). When we assayed the ATP hydrolysis activity of the MbbrA1Ao-ATP synthase in inverted membrane vesicles prepared from M. ruminantium cells, the enzyme was insensitive to DCCD but sensitive to TBT-Cl. We attribute this result to the high sodium content of the native membranes effectively protecting the enzyme from DCCD inhibition as we observed in E. coli membrane vesicles when high salt was added.
The growth of M. ruminantium M1 was not particularly sensitive to classical ATP synthase inhibitors (e.g. DCCD and TBT-Cl). However, amiloride and EIPA were effective inhibitors of growth. The inhibitory effect of amiloride on methanogen growth is proposed to be by inhibition of Na+/H+ antiporter activity (58–60). In support of this contention, amiloride-resistant mutants are defective in Na+/H+ antiporter activity (59). Amiloride and EIPA have also been shown to be inhibitors of Na+-dependent ATP synthases (F-type) (38, 51). The inhibition of F-type enzymes is proposed to be mediated by a mimicking effect of EIPA and amiloride with the side chain of the conserved arginine (stator charge) of subunit a (50, 51). Amiloride and EIPA therefore compete with the subunit a arginine (aR210 in E. coli) for binding to the c subunit in a manner most likely described in our modeling experiments within the active site of the c-ring, leading to enzyme inhibition. In our modeling analysis, we provide an insight into better improving the potential of these newly established inhibitors of the MbbrA1Ao revealing critical interactions and the reasons that they are established.
A major determinant of the ion specificity is the c subunit that assembles to form the proteolipid c ring of Ao. The Ao c subunit has striking variability among methanogenic archaea, with two, four (duplicated), and six (triplicated) transmembrane α-helices reported from sequence comparisons (2, 61). The amino acid analysis of the M. ruminantium M1 c subunit reveals that it consists of two fused α-helical hairpins, as is usually found in V-type/A-type enzymes. The Na+-binding pocket of the E. hirae V-type ATPase involves two α-helical hairpins and contains the residues (E. hirae numbering) Leu-61, Thr-64, Gln-65, and Tyr-68 on helix 2, Gln-110 on helix 3, and Glu-139 on helix 4 (49). However, in contrast to the E. hirae K subunit, the M. ruminantium M1 c subunit shows two complete Na+-binding motifs, each with the same five amino acids as the E. hirae Na+-binding signature (M. ruminantium M1 numbering Gln-30, Leu-57, Glu-59, Thr-60, Gln-61, and Tyr-64 on helices 1/2 and Gln-111, Leu-138, Glu-140, Thr-141, Gln-142, and Tyr-145 on helices 3/4). These two amino acid signatures suggest that the M. ruminantium M1 c-ring binds one Na+ per hairpin, and for each Na+-binding site, five amino acid residues are directly involved in ion coordination. One ion-binding site per hairpin is found in the rotor rings of F-type ATP synthases (47, 62, 63). However, in the case of the F-type Na+-binding site, a water molecule acts as a fifth coordination site (48) but not a glutamine (Gln-60 and Gln-142 for site 1 and 2, respectively). The rotor ring of M. ruminantium M1 ATP synthase therefore shares properties of both the V-type and F-type ATPases/synthases.
A noteworthy feature of the c subunit in methanogen A1Ao-ATP synthases is the presence of a sodium ion-binding signature (11), even in proton-coupled enzymes (8). The natural environment of a methanogen will be a major factor in determining the ion specificity of the A-type enzyme for ATP synthesis. For example, methanogens that inhabit marine environments (high salt) or those growing at extreme temperatures, where the membrane becomes very permeable to protons but not sodium ions (64), have been shown to utilize sodium-coupled enzymes (10, 32, 65, 66). In contrast, ATP synthesis by the A1Ao-ATP synthase of M. mazei (a freshwater species) can be driven by an artificially imposed ΔpH gradient in E. coli membranes but not by ΔpNa+ or ΔμNa+ (8). Studies on the energization of ATP synthases from a wide variety of microorganisms demonstrate that these enzymes can be driven by the membrane potential and/or the pH gradient using either protons or sodium ions. In many cases, the membrane potential appears to be obligatory, and for proton-coupled enzymes, a high concentration of protons (pH < 6.50) at the periplasmic side is required to drive ATP synthesis (67, 68). Data presented here showed that ATP synthesis catalyzed by MbbrA1Ao was dependent on the membrane potential, irrespective of the coupling ion, i.e. protons or sodium ions. M. ruminantium M1 grows over an external pH range of 6.5 to 7.7 at high concentrations of sodium ions that are present in the rumen (>100 mm) (31). Based on these chemical properties, Na+-coupled energetics would be favored in M. ruminantium M1 over proton-coupled processes, assuming bulk-to-bulk coupling, unless the external pH of the rumen was to drop significantly. This will happen from time to time during sub-acute ruminal acidosis, when the pH can drop as low as 5.5, but the rumen can continue to function.
Acknowledgments
We acknowledge the help of Eric Altermann, Bill Kelly, and Sinead Leahy with accessing the M1 genome sequence.
This work was supported by a contract from the Pastoral Greenhouse Gas Research Consortium and in part by a University of Otago research grant and by the AgResearch-University of Otago Collaborative Research Fund.
- EIPA
- 5-(N-ethyl-N-isopropyl)-amiloride
- DCCD
- dicyclohexylcarbodiimide
- DDM
- dodecyl maltoside
- TCEP
- tris[2-carboxyethyl]phosphine
- CCCP
- cyanide m-chlorophenylhydrazone
- DNP
- 2,4-dinitrophenol
- TBT-Cl
- tributyltin chloride
- TCS
- 3,3′,4′,5-tetrachlorosalicylanilide
- GOLD
- Genetic Optimization for Ligand Docking.
REFERENCES
- 1. Müller V., Grüber G. (2003) Cell. Mol. Life Sci. 60, 474–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Müller V., Lingl A., Lewalter K., Fritz M. (2005) J. Bioenerg. Biomembr. 37, 455–460 [DOI] [PubMed] [Google Scholar]
- 3. Thauer R. K., Kaster A. K., Seedorf H., Buckel W., Hedderich R. (2008) Nat. Rev. Microbiol. 6, 579–591 [DOI] [PubMed] [Google Scholar]
- 4. Deppenmeier U., Lienard T., Gottschalk G. (1999) FEBS Lett. 457, 291–297 [DOI] [PubMed] [Google Scholar]
- 5. Deppenmeier U. (2002) Prog. Nucleic Acid Res. Mol. Biol. 71, 223–283 [DOI] [PubMed] [Google Scholar]
- 6. Deppenmeier U., Müller V. (2008) Results Probl. Cell Differ. 45, 123–152 [DOI] [PubMed] [Google Scholar]
- 7. Schäfer G., Engelhard M., Müller V. (1999) Microbiol. Mol. Biol. Rev. 63, 570–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pisa K. Y., Weidner C., Maischak H., Kavermann H., Müller V. (2007) FEMS Microbiol. Lett. 277, 56–63 [DOI] [PubMed] [Google Scholar]
- 9. Kaesler B., Schönheit P. (1988) Eur. J. Biochem. 174, 189–197 [DOI] [PubMed] [Google Scholar]
- 10. Schönheit P., Beimborn D. B. (1985) Eur. J. Biochem. 148, 545–550 [DOI] [PubMed] [Google Scholar]
- 11. Müller V. (2004) J. Bioenerg. Biomembr. 36, 115–125 [DOI] [PubMed] [Google Scholar]
- 12. von Ballmoos C., Cook G. M., Dimroth P. (2008) Annu. Rev. Biophys. 37, 43–64 [DOI] [PubMed] [Google Scholar]
- 13. Leahy S. C., Kelly W. J., Altermann E., Ronimus R. S., Yeoman C. J., Pacheco D. M., Li D., Kong Z., McTavish S., Sang C., Lambie S. C., Janssen P. H., Dey D., Attwood G. T. (2010) PLoS One 5, e8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wedlock D. N., Pedersen G., Denis M., Dey D., Janssen P. H., Buddle B. M. (2010) N. Z. Vet. J. 58, 29–36 [DOI] [PubMed] [Google Scholar]
- 15. Hanahan D., Jessee J., Bloom F. R. (1991) Methods Enzymol. 204, 63–113 [DOI] [PubMed] [Google Scholar]
- 16. Klionsky D. J., Brusilow W. S., Simoni R. D. (1984) J. Bacteriol. 160, 1055–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miroux B., Walker J. E. (1996) J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 18. Yanisch-Perron C., Vieira J., Messing J. (1985) Gene 33, 103–119 [DOI] [PubMed] [Google Scholar]
- 19. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 21. Dmitriev O. Y., Altendorf K., Fillingame R. H. (2004) FEBS Lett. 556, 35–38 [DOI] [PubMed] [Google Scholar]
- 22. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 23. Nesterenko M. V., Tilley M., Upton S. J. (1994) J. Biochem. Biophys. Methods 28, 239–242 [DOI] [PubMed] [Google Scholar]
- 24. McMillan D. G., Keis S., Dimroth P., Cook G. M. (2007) J. Biol. Chem. 282, 17395–17404 [DOI] [PubMed] [Google Scholar]
- 25. Shevchenko A., Jensen O. N., Podtelejnikov A. V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heinonen J. K., Lahti R. J. (1981) Anal. Biochem. 113, 313–317 [DOI] [PubMed] [Google Scholar]
- 27. Lundin A., Thore A. (1975) Appl. Microbiol. 30, 713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W., Taylor R. D. (2003) Proteins 52, 609–623 [DOI] [PubMed] [Google Scholar]
- 30. Janssen P. H., Kirs M. (2008) Appl. Environ. Microbiol. 74, 3619–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith P. H., Hungate R. E. (1958) J. Bacteriol. 75, 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pisa K. Y., Huber H., Thomm M., Müller V. (2007) FEBS J. 274, 3928–3938 [DOI] [PubMed] [Google Scholar]
- 33. Lingl A., Huber H., Stetter K. O., Mayer F., Kellermann J., Müller V. (2003) Extremophiles 7, 249–257 [DOI] [PubMed] [Google Scholar]
- 34. Wilms R., Freiberg C., Wegerle E., Meier I., Mayer F., Müller V. (1996) J. Biol. Chem. 271, 18843–18852 [DOI] [PubMed] [Google Scholar]
- 35. Lemker T., Ruppert C., Stöger H., Wimmers S., Müller V. (2001) Eur. J. Biochem. 268, 3744–3750 [DOI] [PubMed] [Google Scholar]
- 36. Matthey U., Kaim G., Dimroth P. (1997) Eur. J. Biochem. 247, 820–825 [DOI] [PubMed] [Google Scholar]
- 37. Neumann S., Matthey U., Kaim G., Dimroth P. (1998) J. Bacteriol. 180, 3312–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laubinger W., Dimroth P. (1988) Biochemistry 27, 7531–7537 [DOI] [PubMed] [Google Scholar]
- 39. Reidlinger J., Mayer F., Müller V. (1994) FEBS Lett. 356, 17–20 [DOI] [PubMed] [Google Scholar]
- 40. Ferguson S. A., Keis S., Cook G. M. (2006) J. Bacteriol. 188, 5045–5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Linnett P. E., Beechey R. B. (1979) Methods Enzymol. 55, 472–518 [DOI] [PubMed] [Google Scholar]
- 42. Dmitriev O. Y., Jones P. C., Fillingame R. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7785–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. von Ballmoos C., Meier T., Dimroth P. (2002) Eur. J. Biochem. 269, 5581–5589 [DOI] [PubMed] [Google Scholar]
- 44. Kluge C., Dimroth P. (1993) J. Biol. Chem. 268, 14557–14560 [PubMed] [Google Scholar]
- 45. Spruth M., Reidlinger J., Müller V. (1995) Biochim. Biophys. Acta Bioenerg. 1229, 96–102 [Google Scholar]
- 46. Meier T., Matthey U., von Ballmoos C., Vonck J., Krug von Nidda T., Kühlbrandt W., Dimroth P. (2003) J. Mol. Biol. 325, 389–397 [DOI] [PubMed] [Google Scholar]
- 47. Meier T., Polzer P., Diederichs K., Welte W., Dimroth P. (2005) Science 308, 659–662 [DOI] [PubMed] [Google Scholar]
- 48. Meier T., Krah A., Bond P. J., Pogoryelov D., Diederichs K., Faraldo-Gómez J. D. (2009) J. Mol. Biol. 391, 498–507 [DOI] [PubMed] [Google Scholar]
- 49. Murata T., Yamato I., Kakinuma Y., Leslie A. G., Walker J. E. (2005) Science 308, 654–659 [DOI] [PubMed] [Google Scholar]
- 50. Kluge C., Dimroth P. (1993) Biochemistry 32, 10378–10386 [DOI] [PubMed] [Google Scholar]
- 51. Vorburger T., Ebneter J. Z., Wiedenmann A., Morger D., Weber G., Diederichs K., Dimroth P., von Ballmoos C. (2008) FEBS J. 275, 2137–2150 [DOI] [PubMed] [Google Scholar]
- 52. de Jonge M. R., Koymans L. H., Guillemont J. E., Koul A., Andries K. (2007) Proteins 67, 971–980 [DOI] [PubMed] [Google Scholar]
- 53. Becher B., Müller V. (1994) J. Bacteriol. 176, 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smigán P., Majerník A., Greksák M. (1994) FEBS Lett. 349, 424–428 [DOI] [PubMed] [Google Scholar]
- 55. Smigán P., Majerník A., Polák P., Hapala I., Greksák M. (1995) FEBS Lett. 371, 119–122 [DOI] [PubMed] [Google Scholar]
- 56. Saum R., Schlegel K., Meyer B., Müller V. (2009) FEMS Microbiol. Lett. 300, 230–236 [DOI] [PubMed] [Google Scholar]
- 57. Sparling R. J., Blaut M., Gottschalk G. (1993) Can. J. Microbiol. 39, 742–748 [Google Scholar]
- 58. Lancaster J. R., Jr. (1989) J. Bioenerg. Biomembr. 21, 717–740 [DOI] [PubMed] [Google Scholar]
- 59. Surín S., Cubonová L., Majerník A. I., McDermott P., Chong J. P., Smigán P. (2007) FEMS Microbiol. Lett. 269, 301–308 [DOI] [PubMed] [Google Scholar]
- 60. Surín S., Cubonová L., Majerník A. I., Smigán P. (2006) Folia Microbiol. 51, 313–316 [DOI] [PubMed] [Google Scholar]
- 61. Müller V., Lemker T., Lingl A., Weidner C., Coskun U., Grüber G. (2005) J. Mol. Microbiol. Biotechnol. 10, 167–180 [DOI] [PubMed] [Google Scholar]
- 62. Pogoryelov D., Yildiz O., Faraldo-Gómez J. D., Meier T. (2009) Nat. Struct. Mol. Biol. 16, 1068–1073 [DOI] [PubMed] [Google Scholar]
- 63. Preiss L., Yildiz O., Hicks D. B., Krulwich T. A., Meier T. (2010) PLoS Biol. 8, e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. van de Vossenberg J. L., Ubbink-Kok T., Elferink M. G., Driessen A. J., Konings W. N. (1995) Mol. Microbiol. 18, 925–932 [DOI] [PubMed] [Google Scholar]
- 65. Dybas M., Konisky J. (1992) J. Bacteriol. 174, 5575–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Crider B. P., Carper S. W., Lancaster J. R. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 6793–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. von Ballmoos C., Wiedenmann A., Dimroth P. (2009) Annu. Rev. Biochem. 78, 649–672 [DOI] [PubMed] [Google Scholar]
- 68. Wiedenmann A., Dimroth P., von Ballmoos C. (2009) Mol. Microbiol. 72, 479–490 [DOI] [PubMed] [Google Scholar]
- 69. DeLano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC, New York [Google Scholar]