Background: γ-Secretase modulators (GSMs) bind APP near lysine 624.
Results: Mutation of lysine 624 shifts cleavage toward smaller Aβ with no effect on ϵ cleavage.
Conclusion: The amino acid at 624 in substrates affects the final γ-secretase cut.
Significance: γ-Secretase cleavage likely begins at ϵ and proceeds up the transmembrane until Aβ is released, and GSMs may modulate this process through lysine 624.
Keywords: Alzheimer's Disease, Amyloid Precursor Protein, Aspartic Protease, Cell Biology, Intramembrane Proteolysis, Membrane Proteins, Neurodegenerative Diseases, Presenilin, Protease, Secretases
Abstract
γ-Secretase is a multiprotein intramembrane cleaving aspartyl protease (I-CLiP) that catalyzes the final cleavage of the amyloid β precursor protein (APP) to release the amyloid β peptide (Aβ). Aβ is the primary component of senile plaques in Alzheimer's disease (AD), and its mechanism of production has been studied intensely. γ-Secretase executes multiple cleavages within the transmembrane domain of APP, with cleavages producing Aβ and the APP intracellular domain (AICD), referred to as γ and ϵ, respectively. The heterogeneous nature of the γ cleavage that produces various Aβ peptides is highly relevant to AD, as increased production of Aβ 1–42 is genetically and biochemically linked to the development of AD. We have identified an amino acid in the juxtamembrane region of APP, lysine 624, on the basis of APP695 numbering (position 28 relative to Aβ) that plays a critical role in determining the final length of Aβ peptides released by γ-secretase. Mutation of this lysine to alanine (K28A) shifts the primary site of γ-secretase cleavage from 1–40 to 1–33 without significant changes to ϵ cleavage. These results further support a model where ϵ cleavage occurs first, followed by sequential proteolysis of the remaining transmembrane fragment, but extend these observations by demonstrating that charged residues at the luminal boundary of the APP transmembrane domain limit processivity of γ-secretase.
Introduction
Production and accumulation of the amyloid β (Aβ)2 peptide has been intimately linked to the development of Alzheimer's disease (1). Aβ is produced by the sequential proteolysis of APP, a type-I single-pass transmembrane protein (2) (Fig. 1A). First, β-secretase cleavage of the ectodomain releases secreted sAPPβ, leaving a carboxyl-terminal fragment in the membrane, known as the β-carboxyl-terminal fragment (βCTF or C99). C99 undergoes intramembrane proteolysis by γ-secretase at two primary sites to generate Aβ (termed γ-cleavage) and the APP intracellular domain (AICD, termed ϵ cleavage). The relationship between these two cleavage events by γ-secretase, their regulation, and the overall enzymatic mechanism is still not fully understood and is an active area of investigation.
FIGURE 1.
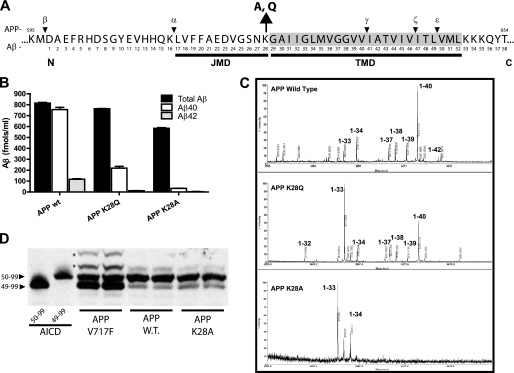
Mutation of Lysine 28 (residue 624 in APP) shifts the production of Aβ to shorter fragments. A, an overview of the portion of APP that is proteolyzed by the α, β, and γ-secretase enzymes. Numbering is shown for full-length APP695 (top) and Aβ (bottom) with the amino acid sequence starting from the luminal amino (N) terminus toward the carboxyl (C) terminus. The predicted APP JMD (17–28) and transmembrane domain (29–52) are underlined. The lysine residue 28 (K28) was mutated to alanine (A) or glutamine (Q). Cleavage of APP695 by β-secretase between methioine 596 and aspartate 597 produces the APP carboxyl-terminal fragment of 99 amino acids (C99). C99 is cleaved by γ-secretase at two main sites, γ (40/41) and ϵ (49/50). γ cleavage produces the secreted Aβ peptide, whereas ϵ produces the AICD (C59). ζ cleavage (46/47) is thought to occur after the ϵ cut, producing an intermediate fragment (Aβ1–46) that is further processed to Aβ1–40 (7, 8, 48). Alternatively, APP can be processed through the non-amyloidogenic pathway by α-secretase (16/17) to produce C83, which is cleaved by γ-cleavage to release the p3 fragment (Aβ 17–40) and AICD (44). B, Aβ levels from H4 cells stably expressing APP WT, K28Q, or K28A were measured from conditioned media (16 h) by ELISA. For APP WT the sum of Aβ 40 and 42 slightly exceeds the estimated total Aβ number. This is due to underestimation of total Aβ inherent in the Aβ standards and sensitivity of the ELISA detection antibody. Values (Aβ fmols/ml, mean ± S.D.). C, the final γ cleavage of APP C99 that releases Aβ is heterogeneous and produces Aβ fragments of different C-terminal lengths. The spectrum of Aβ species (1 to X) secreted by cells expressing APP wild type (WT) was compared with APP mutated at Lys-28 to Glu or Ala by immunoprecipitation of conditioned media and analyzed using mass spectrometry (IP/MS). Aβ1–40 is the major species detected from APP WT cells, with minor amounts of Aβ1–42 and smaller Aβ species (1–39, 38, 37, 34, and 33). In APP K28Q samples, Aβ1–33 followed by Aβ1–40 predominates. For APP K28A, Aβ1–33 and 1–34 become the major Aβ species detected. Mass spectra show the mass-to-charge ratio (m/z) on the x axis. The y axis is signal intensity (arbitrary units). Predicted molecular weights for Aβ were calculated as described under “Experimental Procedures” and take into account the introduced mutations. These Aβ spectra are representative of multiple IP/MS experiments (n = 3–5). D, membrane fractions from cells overexpressing APP WT, APP K28A, and APP V717F were assayed for γ-secretase activity. Assay samples were run on denaturing urea/SDS-PAGE, transferred to membranes, and AICD fragments were visualized with a carboxyl-terminal APP antibody (CT20). Synthetic AICD 50–99 and 49–99 were included as controls. APP WT and APP K28A produced similar ratios of AICD 50–99 to 49–99, whereas APP V717F shifted the ratios of AICD released. Higher molecular weight bands (asterisks) that are immunoreactive for APP were occasionally observed in the APP V717F samples. Identification of these species was not possible because they were not detected by IP/MS and may represent an additional, but low abundant, AICD fragment.
γ-Secretase has a broad substrate specificity and cleaves a number of type-I membrane proteins, typically following ectodomain shedding (3). A major physiologically relevant substrate is Notch1, which has well defined roles in cellular differentiation, survival, and growth. Notch is processed by γ-secretase in a similar manner to APP, producing Notch-β (Nβ, γ cleavage at the S4 site) and the Notch intracellular domain (NICD, ϵ cleavage at the S3 site) (4, 5). Other substrates are thought to be processed in an analogous manner, although most have not been characterized to the same level of detail as APP and Notch1.
One notable aspect of γ-secretase proteolysis is that the final cleavages that release Aβ, and the corresponding peptides from other substrates, occur at varying residues within the transmembrane domain. Thus, the length of Aβ produced from APP, or Aβ-like peptides, is variable. Moreover, the ratios of the various peptides vary between different substrates. For APP, the predominant Aβ produced is 40 amino acids in length, but smaller quantities of Aβ 1–42, Aβ 1–38, and other peptides can be detected in media from cells overexpressing APP or more physiological fluids such as human cerebral spinal fluid (6). Longer membrane-associated Aβ peptides (1–46, 1–47) can also be detected that are thought to represent an intermediate cleavage (ζ) occurring between the ϵ and γ sites (7, 8).
The factors regulating the levels and ratios of Aβ 1–42 has been an intense area of research because Aβ 1–42 is hypothesized to be the initiating molecule in Alzheimer's disease (1). A variety of mutations have been identified in APP that cause early-onset familial AD (9). Most of these mutations elevate the levels of Aβ 1–42 or increase the 42/40 ratio. However, some APP mutations that alter residues within the Aβ peptide have also been identified and increase the propensity of Aβ to form higher-order aggregates or amyloid fibrils. It is an ongoing question exactly how mutations in APP lead to shifts in Aβ fragment ratios produced by γ-secretase although various mechanisms have been proposed, including changes in the conformation of the enzyme complex (10, 11).
Different sites on APP have been identified that can regulate the levels of Aβ peptides or influence the length of Aβ peptides produced. Early studies have shown that γ-secretase displays loose specificity but that the site of cleavage was more dependent on the proximal portion of the transmembrane domain than on the distal portion (12, 13). Mutations in a GXXXG motif in the APP transmembrane domain have been reported to decrease Aβ 1–42 levels while at the same time increasing shorter Aβ fragments such as 34, 37, and 38 (14). The effect of these mutations has been ascribed to decreases in the dimerization of C99, suggesting that Aβ 1–42 levels can be regulated by the degree of C99 dimer formation. Another group has reported similar mutations in this region of C99 that also lead to shifts in Aβ cleavage away from 42 toward shorter fragments (15). The changes in Aβ cleavage resulting from mutation of this region of APP is reminiscent of the pharmacological effects of γ-secretase modulators (GSMs), a class of compounds that typically shift γ cleavage away from the 42 site toward shorter fragments (i.e. 39, 38, 37, 34, 33). We have reported recently that GSMs bind directly to the C99 substrate (termed substrate-targeting GSMs (stGSMs)) and that this interaction appears be responsible for their ability to modulate γ cleavage of Aβ (16). Richter et al. (17) have recently shown, using multiple biochemical methods, that GSMs can bind APP and Aβ, supporting our initial observation (16) although newer generations of GSMs reported to bind to Pen-2 (18) or PS1-N-terminal fragment (NTF) (19) do not appear to show such specificity.
Amino acids in the juxtamembrane region of APP and other substrates have been reported to regulate both γ and ϵ cleavage. Specifically, mutations at lysine 28 were shown to allow ϵ cleavage and ICD release to occur, whereas γ cleavage and Aβ production were abolished (20). These results suggest that amino acids in the JMD region of the substrate could influence proteolysis C-terminal to the JMD, in the center of the lipid bilayer. We became interested in this region of C99 because we have observed that two substrate-targeting GSM photoprobes (fenofibrate and flurbiprofen) bind and label this region (Fig. 1A). We focused on lysine 28 (relative to C99 and Aβ, Lys-624 relative to APP695) in particular because we have observed that Aβ42-lowering GSMs require a carboxylic acid for their activity and hypothesized that these compounds interacted with the positive charge of the lysine head group (16). We now report that mutations at Lys-28 facilitate additional γ-cleavages of APP. Specifically, they enable the production of novel short Aβ-like peptides with truncated carboxyl termini. The implication of these results for the proteolytic mechanism of γ-secretase and GSMs will be discussed.
EXPERIMENTAL PROCEDURES
DNA Plasmids
Construction of the pcDNA3.1-C99GVP and C99 juxtamembrane chimeras construct has been described previously (20). Point mutations within the luminal juxtamembrane domain (i.e. APP695-K28A, APP695-S26L, and APP695-K28S as well as C99GVP-G2S, C99GVP-S26L, C99GVP-N27S, and C99GVP-K28S) were generated using QuikChange (Stratagene) site-directed mutagenesis. All cDNAs were verified by sequencing. The Aβ and Aβ-like peptides generated from C99GVP and various mutant substrates were numbered with reference to the first N-terminal residue (Asp-1) of the Aβ peptide.
Antibodies and Aβ ELISAs
A rabbit polyclonal antibody against the last 20 amino acids of APP (CT20) was produced in house and used to detect expression of full-length APP, C99, C83, and AICD fragments. FLAG-tagged proteins were detected with anti-FLAG M2 antibody (Sigma). Two ELISAs to detect Aβ were used and have been described previously (22, 23). Briefly, amyloid-β peptides were captured by C-terminal-specific antibodies for Aβ40 (antibody 40.1) or Aβ42 (antibody 42.2) that were coated on Immulon 4 HBX ELISA plates (Thermo Scientific) at 25 μg ml−1 in PBS. Captured amyloid-β was then detected by an HRP-conjugated antibody reactive to the N-terminal epitope 1–16 of amyloid-β (antibody 9). Total Aβ was captured on antibody 9 ELISA plates and detected with 4G8-HRP (Covance). HRP was detected using TMB (KPL). Alternatively, Aβ40 and Aβ 42 in samples were captured onto 2G3 or 21F12 antibody-coated plates, respectively, and detected with a biotinylated 2H3 antibody (specific to Aβ 4–7). The fluorescence signal generated from a streptavidin/alkaline phosphatase conjugate (Roche) was measured with a CytoFluor microplate reader (Applied Biosystems). Synthetic Aβ40 or Aβ42 peptides (rPeptide, ultra pure, Hexafluoroisopropanol (HFIP)) were used to generate standard curves. Measurements were done in duplicate or triplicate.
Cell Culture and Transfection
Human embryonic kidney 293T (HEK 293T) cells or H4 neuroglioma cells (ATCC) were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 50 units/ml penicillin and streptomycin (37 °C, 5% CO2). Endotoxin-free (Qiagen) cDNA plasmids were transfected into 6- or 12-well tissue culture plates (Costar) using FuGENE6 reagent (Roche) according to the manufacturer's protocol. Cells and conditioned media were harvested 48 h posttransfection for analysis by ELISA or Western blot analysis. Complete protease inhibitor (Roche) was added to media and lysis buffers for cells.
Western Blot Characterization of APP Metabolites
After media collection, transfected or stable cells were harvested, washed with ice-cold PBS, and collected by centrifugation. Cells were lysed on ice with PBS containing 1% Triton X-100 containing protease inhibitor (Roche) for 20 min and cleared by centrifugation at 14,000 × g for 10 min. Protein concentration of the supernatant was determined using the BCA protein assay (Pierce), normalized, diluted as necessary, and mixed 1:4 in XT sample buffer (Bio-Rad). Samples were loaded onto Criterion XT Bis-Tris gels (4–12 or 12%) and separated using MES running buffer. Proteins were transferred with a modified protocol to facilitate transfer and retention of small APP CTF/Aβ/AICD fragments as well as full-length APP (24). Gels were transferred to either 0.2 μm nitrocellulose (Bio-Rad) or PVDF (Millipore, PSQ) in Tris-glycine buffer with 20% methanol at 0.2 A for 30 min followed by 0.5 A for 60 min. Membranes were rinsed in water following transfer, and then submerged for 5 min in PBS that had been heated to boiling in a microwave to increase Aβ and AICD detection via epitope retrieval (25). Membranes were blocked over night in 0.5% casein-PBS. Blots were probed with appropriate antibodies (82E1 (Immuno-Biological Laboratories) for Aβ and C99; 6E10 (Covance) for APP, C99, and Aβ; and CT20 (Sigma) for APP, C83, C99, and AICD). Two different secondary strategies were used. Secondary antibodies conjugated to HRP (Jackson ImmunoResearch Laboratories) were visualized with ECL Plus (GE Healthcare) or Supersignal West Femto (Pierce). Alternatively, secondary antibodies conjugated to near-infrared dyes (Alexa Fluor 680, Invitrogen; IR800, Licor) were visualized with the Licor Odyssey system.
Mass Spectrometry of Amyloid-β and AICD
Secreted Aβ and Aβ-like peptides were analyzed using immunoprecipitation followed by MALDI-TOF mass spectrometry (IP/MS) by minor modification of the technique described previously (16, 26). Aβ (1 to X) was immunoprecipitated from conditioned cell culture medium with antibody 9 (epitope Aβ 1–16) using sheep anti-mouse IgG magnetic Dynabeads (Invitrogen, catalog no. 112-01D). Synthetic β-amyloid (1–22) (Anaspec) was added as an immunoprecipitation control and mass standard. Proteins were eluted into 0.1% formic acid and 75% acetonitrile. Samples were mixed 1:1 with α-cyano-4-hydroxycinnamic acid matrix in methanol:acetonitrile:water (36:56:8%) and spotted on a ProteinChip Gold Array (Bio-Rad, catalog no. C55-30033). Samples were analyzed on a ProteinChip surface-enhanced laser desorption/ionization time-of-flight mass spectrometry PCS4000 (Bio-Rad). Typical settings were positive ion mode, laser power = 3000 nJ, 10 shots per position, calibrated using all-in-one peptide standard (Bio-Rad). Samples were analyzed in duplicate using separate spots. The same protocol was used to analyze AICD fragments (supplemental data), but IP was performed with anti-FLAG M2 magnetic beads (Sigma, catalog no. M8823). The theoretical average molecular weights of amyloid-β, Aβ-like, and AICD fragments were calculated using the ExPASy Compute pI/Mw tool.
In Vitro γ-Secretase Assay
γ-Secretase activity was assayed as described previously in detail. Briefly, cells overexpressing the APP of interest were grown in 5- to 15-cm cell culture dishes to confluence and then treated overnight with the reversible γ-secretase IL-CHO (20 μm) to increase C99 substrate concentration. Microsomal membrane fractions were prepared and resuspended in 150 mm sodium citrate buffer (pH 6.8) for 2 h at 37 °C to induce γ-secretase activity. Samples were then analyzed by ELISA for Aβ production and IP/MS (Aβ or AICD) or standard Western blot analysis for total γ-secretase activity (ϵ cleavage and AICD production). Urea-SDS-PAGE was necessary to resolve AICD fragments (49–99 and 50–99) and was performed as described previously (27, 28).
RESULTS
Mutation of Lysine 28 Shifts the Site of Aβ Cleavage toward Smaller Peptides
We made a series of site-directed mutations at lysine 28 in APP and investigated their effects on Aβ production. Lysine was changed to alanine to remove the positive charge and replace it with a small, hydrophobic amino acid or glutamine, which is neutral but polar (29). These cDNA constructs were transfected into H4 neuroglioma cells, stable clones were isolated, and then the effect of these mutations on Aβ production was evaluated. Using sandwich ELISAs to measure Aβ levels, we observed typical levels and ratios of Aβ1–40 and Aβ1–42 from the APP wild-type clone. However, the APP K28Q and K28A mutants produced low or no detectable Aβ1–40 and Aβ1–42. Both APP K28Q and K28A produced similar levels of total Aβ (1-X) compared with wild-type APP (Fig. 1B). We subsequently used immunoprecipitation followed by mass spectrometry (IP/MS) to examine the full spectrum of Aβ peptides produced by the mutations and determine whether they shifted the γ-secretase cleavage site (26). In contrast to APP WT, the major species produced by APP K28Q was Aβ 1–33, followed by 1–40 (Fig. 1B). Smaller amounts of 1–37, 1–38, 1–39, 1–34, and 1–32 were also detected in the media. The APP K28A mutation predominantly produced Aβ 1–33 followed by Aβ 1–34, but no longer Aβ fragments were detectable (Fig. 1C).
Mutations of Lysine 28 Do Not Change the Site of ϵ-Cleavage
We next examined how these mutations affected release of the AICD fragment by ϵ-cleavage (Fig. 1A). In vitro γ-secretase assays were performed using membrane-enriched fractions from cells stably overexpressing APP K28Q, APP K28A, and APP V717F as a positive control known to alter ϵ cleavage. Samples from these assays were run on denaturing SDS/urea PAGE to examine production of AICD and specific cleavage sites (27). AICD 50–99 was the predominant cleavage fragment observed for both APP wild-type and K28A (Fig. 1D). In contrast, the familial Alzheimer's disease mutation APP V717F produced mainly AICD 49–99, followed by AICD 50–99, in agreement with previous reports (Fig. 1D) (28). AICD produced from membranes isolated from cells expressing a C-terminal FLAG-tagged C99 construct, with wild-type sequence or K28A, was analyzed via IP/MS. These data support the identification of 49–99 and 50–99 as the major AICD fragments as well as the observation that these mutations do not modulate the site of ϵ cleavage (supplemental Fig. 1).
C99-GVP Hybrid Substrates Also Shift Cleavage toward Production of Smaller Aβ Peptides
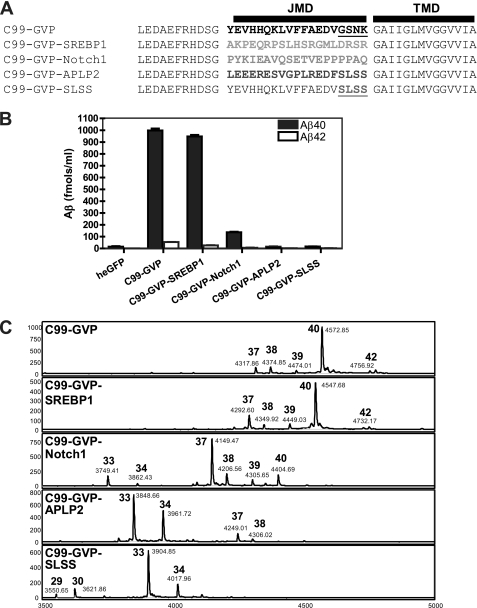
It has been reported previously that mutations in the JMD of APP could selectively abolish γ cleavage as assessed by Aβ production while maintaining ϵ cleavage on the basis of AICD production (20). Our current observation demonstrates that mutation of juxtamembrane lysine 28 shifts the site of γ cleavage rather than abolishing it. We therefore re-examined if the previous juxtamembrane mutations in APP had a similar effects on Aβ length. As shown previously, substitution of the JMD of APP C99 with the analogous region of other γ-secretase substrates produces hybrid substrates that are processed by γ-secretase and are cleaved at the ϵ site to produce AICD fragments (see Fig. 3A in Ref. 20). C99-GVP-SREBP1 produces similar levels and ratios of Aβ 1–40 and Aβ 1–42 compared with wild-type C99-GVP (Fig. 2B). In contrast, C99-GVP-Notch1 produces 6.5-fold less Aβ 1–40 than wild-type C99-GVP and no detectable Aβ 1–42. Both C99-GVP-APLP2 and C99-GVP-SLSS produce no detectable Aβ 1–40 or 1–42, despite equivalent expression levels determined by Western blot analysis (data not shown).
FIGURE 2.
Substitution of alternative substrates for the juxtamembrane domain of C99 produces hybrid substrates that are processed by γ-secretase but shift the size of Aβ produced. A, sequences of the substrates that were substituted for the JMD of C99-GVP. These hybrids are cleaved at the ϵ site by γ-secretase and signal to the nucleus (20). B, ELISA analysis of Aβ 40 and 42 production by hybrid C99-GVP constructs. C99-GCP-SREBP1 produces similar amounts of 40 and 42 compared with the wild type. The Notch1, APLP2, and SLSS constructs have greatly impaired Aβ 40/42 production. C, IP/MALDI-TOF MS analysis of Aβ (1-X) production in cells transfected with hybrid C99-GVP reveals that all constructs produce Aβ species but show greatly altered patterns of the final C-terminal cleavage site of γ-secretase. In general, the γ site is shifted to produce shorter Aβ fragments. The largest shift toward smaller Aβ species is seen with C99-GVP-APLP2 and C99-GVP-SLSS (which are the four amino acid residues from APLP2 that changed from the GSNK sequence in wild-type C99). The molecular weights of hybrid Aβ fragments were calculated to include their representative mutations and the dipeptide LE, which is retained on the N terminus after signal peptide cleavage of C99 (20, 21).
Again, we used IP/MS to determine whether alternative γ cleavage sites were generated. IP/MS analysis of conditioned media from HEK293T cells transfected with C99-GVP-SREBP1 revealed Aβ 40 as the predominant species, similar to C99-GVP (Fig. 2C). For C99-GVP-Notch1, Aβ 37 was the major peptide detected. The C99-GVP-APLP2 construct produced even smaller Aβ species with Aβ 33 > Aβ 34 > Aβ 37 > Aβ 38, whereas C99-GVP-SLSS had the greatest shift in Aβ size. Aβ 33 was the major peak detected, but we also observed small amounts of Aβ 29 and Aβ 30, which are the smallest Aβ fragments we have observed. Taken together, these results explain why “normal” Aβ 40 and 42 fragments were not detected in the initial report, wherein the ELISA assay relied on capture of Aβ species ending at 40 or 42, followed by detection with an N-terminally-directed antibody. Substitution of amino acids in the JMD region of APP can dramatically shift the final cleavage site of γ-secretase and, consequently, the length of Aβ secreted.
Lysine 28 Plays a Critical Role in Determining the Final Size of Aβ Produced by γ-Secretase
The dramatic shifts in Aβ observed with the C99-GVP-SLSS mutant highlight the importance the GSNK region play in the final site of γ-secretase cleavage (Figs. 1 and 2). We next examined point mutations in GSNK to dissect the contribution of each residue to Aβ production. These constructs were transiently expressed in HEK 293T cells and produced levels of total Aβ (1-X) that were comparable with WT levels (GSNK sequence) and inhibited by treatment with the γ-secretase inhibitor DAPT (Fig. 3). When Aβ X-40 levels were measured, three constructs were significantly decreased: GLNK, GSNS, and SLSS, consistent with our previous report (see Fig. 5, B and C in Ref. 20). Levels of Aβ X-40 were decreased by 25% in the GLNK construct. For GSNS and SLSS, Aβ X-40 levels were decreased to a greater extent and were nearly undetectable (Fig. 3). This result suggests that mutation of S26L can also affect the length of Aβ but to a lesser extent than mutation of K28S.
FIGURE 3.
Lysine 28 is a critical determinate of the final size of secreted Aβ. Point mutations (shown in red) were made in the GSNK (25 to 28) region of the juxtamembrane region of C99-GVP by swapping the corresponding amino acid from the APLP2 region (SLSS, Fig. 2A). All mutants produced similar levels of total Aβ (1-X) compared with the wild-type (GSNK) control. Two point mutations showed decreased Aβ 40 (X-40), despite making comparable total amounts of Aβ. GSNS had the greatest Aβ 40 decrease and was similar to SLSS. Both total Aβ and Aβ 40 were completely inhibited by treatment with the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester DAPT (20 μm, not shown). As illustrated in Fig. 2, a decrease in Aβ 40 in this assay is a surrogate marker for a shift in Aβ toward smaller Aβ species. This result suggests that Lys-28 is the major residue regulating Aβ length in this region. The same transfected cells were used for the collection of conditioned media for the detection of 1-X and X-40 within each N (3), providing an internal control. All values are percentages (mean ± S.D.) of signals from the wild-type GSNK construct (set to 100%). One-way analysis of variance with post hoc Tukey's t test; ***, p < 0.001; *, p < 0.05. n.s., not significant.
FIGURE 5.
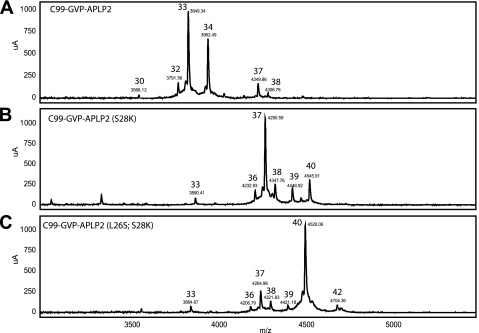
Serine 26 influences Aβ size to a lesser extent than Lysine 28. Rescue experiments, analogous to the ones performed for C99-GVP Notch1 (Fig. 4), were carried out for C99-GVP-APLP2. A, Aβ33 and 34 are the major species produced from C99-GVP-APLP2 following IP/MS analysis. B, following mutation of S28K in C99-GVP-APLP2, Aβ37 and 40 now predominant, demonstrating a shift toward longer species. C, double mutation (L26S, S28K) of C99-GVP-APLP2 is necessary to completely shift cleavage toward Aβ40 as the major detectable species. Mass spectra show the mass-to-charge ratio (m/z) on the x axis. The y axis is signal intensity (μA, arbitrary units).
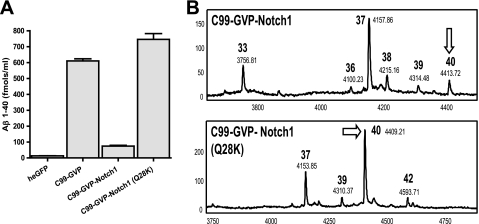
If Lys-28 is a key determinant of the final cleavage site of Aβ, we should be able to reverse the shift in cleavage that we have observed in these hybrid constructs. To test this hypothesis, we mutated glutamine 28 in C99-GVP-Notch1 to lysine. Although Aβ 37 is the major Aβ species in C99-GVP-Notch1, mutation of Glu to Lys eliminates the production of shorter fragments, i.e. Aβ 33, and shifts cleavage back to Aβ 40, with levels comparable with the wild type (Fig. 4). Next, we made the same mutation (S28K) in C99-GVP-APLP2 and observed the appearance of Aβ 40 but not at wild-type levels (Fig. 5). Double mutation (L26S, S28K) of C99-GVP-APLP2 produced a substrate that was predominantly cleaved at Aβ 40.
FIGURE 4.
Mutation of Q28K in C99-GVP Notch1 restores production of Aβ40 to wild-type C99 levels. A, the contribution of glutamine 28 to the shift in Aβ in the hybrid C99-GVP Notch1 protein was examined by mutating this residue to lysine as found in WT C99. Compared with C99-GVP Notch1, the Q28K mutation produced predominantly Aβ 40 as measured by ELISA or immunoprecipitation/MALDI-TOF MS analysis (B) of Aβ species.
DISCUSSION
The regulation of γ-secretase cleavage at the γ and ϵ sites has been an intense area of investigation. Inhibition of γ-secretase cleavage of APP to reduce Aβ is being pursued as a therapeutic strategy for AD. However, it is now widely accepted that inhibition of ϵ cleavage, which reduces levels of ICDs, can have serious side effects resulting from impaired nuclear signaling of the numerous γ-secretase substrates (30, 31). Toxicity from impaired Notch signaling is the best characterized example of this phenomenon (32, 33). The discontinuation of the phase III clinical trial for the gamma-secretase inhibitor semagacestat (LY-450139) may have been related to mechanism-based toxicity. However, no data has been published. Collectively, these findings highlight the need to understand regulation of γ and ϵ cleavage specificity to help design inhibitors that are selective for APP or preferentially inhibit or modulate γ cleavage.
In this study, we examined the role of specific amino acid residues in the JMD region of APP on the specificity of γ and ϵ cleavage by γ-secretase. We focused on lysine 28 in APP because it was part of the GSM binding region we identified previously (16), and we and others have shown that this region of APP can influence γ-secretase production of Aβ peptides (12, 20). Data from a previous study suggested that mutations in this JMD region of APP selectively inhibited γ cleavage (Aβ production) without effects on ϵ cleavage (AICD production) (20). Our current findings reveal that these mutations do permit γ cleavage but actually shift the final site to favor production of shorter Aβ species. This observation was not made earlier because the original assays, and most commercial Aβ ELISAs, were specific to Aβ 1–40 or 1–42 by virtue of the C terminus-specific capture antibodies employed. This assay strategy does not detect production of any smaller fragments (Aβ 1–41 down to 1–24).
The temporal sequence of γ and ϵ cleavage has been an ongoing question in the field since the fragments were identified (Fig. 1A) (34). Our finding that N-terminal mutations outside the transmembrane domain of APP can cause dramatic shifts (up to seven residues) in the final site of the γ cut, without any significant change at the ϵ site, support the idea that ϵ cleavage occurs first, followed by sequential proteolysis, as has been proposed by a number of groups (8, 35, 36). Although we favor this sequential cleavage model of γ-secretase activity, one unexplained phenomenon is the C-terminal heterogeneity in the final site of γ cleavage to produce Aβ and analogous peptides from other substrates (5). Some early-onset AD mutations in APP and PS1/2 clearly alter the amounts and lengths of secreted Aβ (3). However, the precise mechanisms that regulate when γ-secretase cleavage terminates and Aβ is released are not well characterized.
Examination of amino acids that are conserved across γ-secretase substrates provides some insight into the sequence of cleavage events. All known γ-secretase substrates are type-I single transmembrane proteins with an enrichment of lysine and arginine residues at the inner (cytoplasmic) membrane boundary toward the C terminus (3). Multiple positively charged amino acids are frequently enriched on the end of helices buried in the lipid bilayer and serve as a stop-transfer sequences and membrane anchors (37). For APP, the C-terminal triplet lysine residue is essential for membrane incorporation and γ-secretase processing (12, 38). In contrast, membrane anchoring residues, such as lysine and arginine, at the JMD boundary of the transmembrane domain of substrates is highly variable (3, 39). On the basis of this requirement, we propose that γ-secretase must first cleave its substrate near the carboxyl terminal membrane-cytoplasmic interface (the ϵ site) to release the constraint imposed by the multiresidue anchor sequence, allowing further proteolysis of the remaining membrane stub to proceed, as was first proposed by Ihara and colleagues (10). This would explain why nearly all γ-secretase substrates are cleaved at a similar site near the carboxyl terminal inner membrane leaflet (ϵ cleavage), whereas the subsequent site of intramembrane γ cleavage varies widely (3). In this model, γ-secretase remains stationary in the membrane, whereas the substrate is “pulled” into the active site. Our data from mutations to C99, as well as C99-Notch, suggest that a lysine at position 28 can anchor the N terminus, thus limiting substrate access to the active site and preventing further processing into shorter peptide fragments. Interestingly, lysine anchors have some vertical flexibility, and this may explain why, although Aβ 1–40 is the major fragment, smaller peptides are also generated to a lesser extent (40). Mutation of K28A would remove this constraint, increase hydrophobicity, and allow further entry of the substrate into the γ-secretase active site, leading to the production of shorter Aβ fragments. Our results and the current understanding of membrane protein dynamics support this model, but further experiments are needed to rigorously test this hypothesis in more detail (41).
The influence of mutations in the transmembrane domain of APP on γ and ϵ cleavage is well documented (28, 42–44). In an accompanying manuscript, Sagi et al, (49) show that the sequence of the transmembrane domain also alters the ability of GSMs to modulate γ cleavage. They identify a five-amino-acid region in the APP transmembrane domain adjacent to Lys-28 that greatly influences GSM activity. This work, combined with our previous finding that some GSMs bind to APP in the same region, suggest that GSM activity is substrate-dependent. The precise mechanism of GSM activity is still debated, and different mechanisms have been proposed, including allosteric modulation of PS1 (45), binding to Pen-2 (18), or binding to a portion of the Aβ domain within APP (16, 17). Our current observation that mutation of Lys-28 can mimic the effect of GSMs (shifting the site of γ cleavage) supports but does not prove that these compounds work by binding to APP. We propose that this residue interacts with the negative carboxylic acid moiety that is found in most GSMs and is critical for the ability to lower Aβ42 (46). It has recently been reported that some point mutations in the GSM binding site on APP are still modulated by GSMs, although to varying degrees (47). This is not surprising, on the basis of our current results. The GSM binding site reported previously (16) and the critical APP sequence that influences GSM activity are at least five amino acids in length and require larger mutations to completely attenuate GSM-induced changes in γ cleavage (49). More detailed studies are ongoing to further refine the minimum mutations in the APP sequence necessary to fully attenuate GSM activity.
In summary, an unappreciated fact demonstrated in this study is the profound impact N-terminal JMD residues can have on the ultimate size of Aβ peptides generated from cleavage inside the membrane (12, 14, 20). Our results are compatible with the sequential proteolytic model of γ-secretase activity but reveal a new level of complexity. The secreted γ site product (Aβ40/42) is not solely determined by the initial ϵ-cut (Aβ 48/49) but is also critically regulated by amino acids such as lysine 28 in C99 at the membrane interface. Finally, our results suggest how GSMs, such as some NSAIDs, can bind this region of APP to modulate Aβ species without inhibition of AICD production.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants AG20206 (to T. E. G.), AG29886 (to E. H. K.), AG032362 (to T. L. K.), and NS069289 (to T. L. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1, Tables 1–11, and references.
- Aβ
- amyloid β
- APP
- amyloid β (A4) precursor protein
- AICD
- amyloid precursor protein intracellular domain
- AD
- Alzheimer's disease
- GSM
- γ-secretase modulator
- JMD
- juxtamembrane domain
- IP
- immunoprecipitation.
REFERENCES
- 1. Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 2. Thinakaran G., Koo E. H. (2008) J. Biol. Chem. 283, 29615–29619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beel A. J., Sanders C. R. (2008) Cell. Mol. Life Sci. 65, 1311–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Okochi M., Steiner H., Fukumori A., Tanii H., Tomita T., Tanaka T., Iwatsubo T., Kudo T., Takeda M., Haass C. (2002) EMBO J. 21, 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okochi M., Fukumori A., Jiang J., Itoh N., Kimura R., Steiner H., Haass C., Tagami S., Takeda M. (2006) J. Biol. Chem. 281, 7890–7898 [DOI] [PubMed] [Google Scholar]
- 6. Ford M. J., Cantone J. L., Polson C., Toyn J. H., Meredith J. E., Drexler D. M. (2008) J. Neurosci. Methods 168, 465–474 [DOI] [PubMed] [Google Scholar]
- 7. Zhao G., Mao G., Tan J., Dong Y., Cui M. Z., Kim S. H., Xu X. (2004) J. Biol. Chem. 279, 50647–50650 [DOI] [PubMed] [Google Scholar]
- 8. Takami M., Nagashima Y., Sano Y., Ishihara S., Morishima-Kawashima M., Funamoto S., Ihara Y. (2009) J. Neurosci. 29, 13042–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brouwers N., Sleegers K., Van Broeckhoven C. (2008) Ann. Med. 40, 562–583 [DOI] [PubMed] [Google Scholar]
- 10. Sato T., Dohmae N., Qi Y., Kakuda N., Misonou H., Mitsumori R., Maruyama H., Koo E. H., Haass C., Takio K., Morishima-Kawashima M., Ishiura S., Ihara Y. (2003) J. Biol. Chem. 278, 24294–24301 [DOI] [PubMed] [Google Scholar]
- 11. Uemura K., Lill C. M., Li X., Peters J. A., Ivanov A., Fan Z., DeStrooper B., Bacskai B. J., Hyman B. T., Berezovska O. (2009) PLoS ONE 4, e7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murphy M. P., Hickman L. J., Eckman C. B., Uljon S. N., Wang R., Golde T. E. (1999) J. Biol. Chem. 274, 11914–11923 [DOI] [PubMed] [Google Scholar]
- 13. Lichtenthaler S. F., Wang R., Grimm H., Uljon S. N., Masters C. L., Beyreuther K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munter L. M., Voigt P., Harmeier A., Kaden D., Gottschalk K. E., Weise C., Pipkorn R., Schaefer M., Langosch D., Multhaup G. (2007) EMBO J. 26, 1702–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kienlen-Campard P., Tasiaux B., Van Hees J., Li M., Huysseune S., Sato T., Fei J. Z., Aimoto S., Courtoy P. J., Smith S. O., Constantinescu S. N., Octave J. N. (2008) J. Biol. Chem. 283, 7733–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kukar T. L., Ladd T. B., Bann M. A., Fraering P. C., Narlawar R., Maharvi G. M., Healy B., Chapman R., Welzel A. T., Price R. W., Moore B., Rangachari V., Cusack B., Eriksen J., Jansen-West K., Verbeeck C., Yager D., Eckman C., Ye W., Sagi S., Cottrell B. A., Torpey J., Rosenberry T. L., Fauq A., Wolfe M. S., Schmidt B., Walsh D. M., Koo E. H., Golde T. E. (2008) Nature 453, 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richter L., Munter L. M., Ness J., Hildebrand P. W., Dasari M., Unterreitmeier S., Bulic B., Beyermann M., Gust R., Reif B., Weggen S., Langosch D., Multhaup G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14597–14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kounnas M. Z., Danks A. M., Cheng S., Tyree C., Ackerman E., Zhang X., Ahn K., Nguyen P., Comer D., Mao L., Yu C., Pleynet D., Digregorio P. J., Velicelebi G., Stauderman K. A., Comer W. T., Mobley W. C., Li Y. M., Sisodia S. S., Tanzi R. E., Wagner S. L. (2010) Neuron 67, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohki Y., Higo T., Uemura K., Shimada N., Osawa S., Berezovska O., Yokoshima S., Fukuyama T., Tomita T., Iwatsubo T. (2011) EMBO J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ren Z., Schenk D., Basi G. S., Shapiro I. P. (2007) J. Biol. Chem. 282, 35350–35360 [DOI] [PubMed] [Google Scholar]
- 21. Lichtenthaler S. F., Multhaup G., Masters C. L., Beyreuther K. (1999) FEBS Lett. 453, 288–292 [DOI] [PubMed] [Google Scholar]
- 22. Johnson-Wood K., Lee M., Motter R., Hu K., Gordon G., Barbour R., Khan K., Gordon M., Tan H., Games D., Lieberburg I., Schenk D., Seubert P., McConlogue L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1550–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kukar T., Murphy M. P., Eriksen J. L., Sagi S. A., Weggen S., Smith T. E., Ladd T., Khan M. A., Kache R., Beard J., Dodson M., Merit S., Ozols V. V., Anastasiadis P. Z., Das P., Fauq A., Koo E. H., Golde T. E. (2005) Nat. Med. 11, 545–550 [DOI] [PubMed] [Google Scholar]
- 24. Otter T., King S. M., Witman G. B. (1987) Anal. Biochem. 162, 370–377 [DOI] [PubMed] [Google Scholar]
- 25. Rosen R. F., Tomidokoro Y., Ghiso J. A., Walker L. C. (2010) J. Vis. Exp. doi: 10.3791/1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang R., Sweeney D., Gandy S. E., Sisodia S. S. (1996) J. Biol. Chem. 271, 31894–31902 [DOI] [PubMed] [Google Scholar]
- 27. Kawooya J. K., Emmons T. L., Gonzalez-DeWhitt P. A., Camp M. C., D'Andrea S. C. (2003) Anal. Biochem. 323, 103–113 [DOI] [PubMed] [Google Scholar]
- 28. Kakuda N., Funamoto S., Yagishita S., Takami M., Osawa S., Dohmae N., Ihara Y. (2006) J. Biol. Chem. 281, 14776–14786 [DOI] [PubMed] [Google Scholar]
- 29. Morrison K. L., Weiss G. A. (2001) Curr. Opin. Chem. Biol. 5, 302–307 [DOI] [PubMed] [Google Scholar]
- 30. Barten D. M., Meredith J. E., Jr., Zaczek R., Houston J. G., Albright C. F. (2006) Drugs in R&D 7, 87. [DOI] [PubMed] [Google Scholar]
- 31. Golde T. E., Kukar T. L. (2009) Science 324, 603–604 [DOI] [PubMed] [Google Scholar]
- 32. van Es J. H., van Gijn M. E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D. J., Radtke F., Clevers H. (2005) Nature 435, 959–963 [DOI] [PubMed] [Google Scholar]
- 33. Kopan R., Ilagan M. X. G. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dillen K., Annaert W. (2006) in International Review of Cytology (Kwang W. J. ed), pp. 215–300, Academic Press, Washington, D. C: [DOI] [PubMed] [Google Scholar]
- 35. Fukumori A., Fluhrer R., Steiner H., Haass C. (2010) J. Neurosci. 30, 7853–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu X. (2009) J. Alzheimer's Dis. 16, 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. von Heijne G., Gavel Y. (1988) Eur. J. Biochem. 174, 671–678 [DOI] [PubMed] [Google Scholar]
- 38. Usami M., Yamao-Harigaya W., Maruyama K. (1993) J. Neurochem. 61, 239–246 [DOI] [PubMed] [Google Scholar]
- 39. Killian J. A., von Heijne G. (2000) Trends Biochem. Sci. 25, 429–434 [DOI] [PubMed] [Google Scholar]
- 40. Strandberg E., Killian J. A. (2003) FEBS Lett. 544, 69–73 [DOI] [PubMed] [Google Scholar]
- 41. Holt A., Killian J. A. (2010) European Biophysics Journal 39, 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Jonghe C., Esselens C., Kumar-Singh S., Craessaerts K., Serneels S., Checler F., Annaert W., Van Broeckhoven C., De Strooper B. (2001) Hum. Mol. Genet. 10, 1665–1671 [DOI] [PubMed] [Google Scholar]
- 43. Hecimovic S., Wang J., Dolios G., Martinez M., Wang R., Goate A. M. (2004) Neurobiol. Dis. 17, 205–218 [DOI] [PubMed] [Google Scholar]
- 44. De Strooper B. (2010) Physiol. Rev. 90, 465–494 [DOI] [PubMed] [Google Scholar]
- 45. Lleó A., Berezovska O., Herl L., Raju S., Deng A., Bacskai B. J., Frosch M. P., Irizarry M., Hyman B. T. (2004) Nat. Med. 10, 1065–1066 [DOI] [PubMed] [Google Scholar]
- 46. Narlawar R., Baumann K., Czech C., Schmidt B. (2007) Bioorg. Med. Chem. Lett. 17, 5428–5431 [DOI] [PubMed] [Google Scholar]
- 47. Page R. M., Gutsmiedl A., Fukumori A., Winkler E., Haass C., Steiner H. (2010) J. Biol. Chem. 285, 17798–17810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao G., Cui M. Z., Mao G., Dong Y., Tan J., Sun L., Xu X. (2005) J. Biol. Chem. 280, 37689–37697 [DOI] [PubMed] [Google Scholar]
- 49. Sagi S. A., Lessard C. B., Winden K. D., Maruyama H., Koo J. C., Weggen S., Kukar T. L., Golde T. E., Koo E. H. (2011) J. Biol. Chem. 286, 39794–39803 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.