Background: The conversion domain of the Prion protein, responsible for its switch into an abnormal form, is not clearly characterized.
Results: The H2H3 domain of the Prion protein can access two different conformations in mammalian cells, one of which is amyloid and resistant to proteinase K.
Conclusion: The H2H3 domain is a conformational switch.
Significance: The H2H3 domain may be the conversion domain of PrP.
Keywords: Amyloid, Prions, Protein Aggregation, Protein Chemistry, Protein Conformation, Protein Domains, Protein Misfolding
Abstract
The concept of prion is applied to protein modules that share the ability to switch between at least two conformational states and transmit one of these through intermolecular interaction and change of conformation. Although much progress has been achieved through the understanding of prions from organisms such as Saccharomyces cerevisiae, Podospora anserina, or Aplysia californica, the criteria that qualify a protein module as a prion are still unclear. In addition, the functionality of known prion domains fails to provide clues to understand the first identified prion, the mammalian infectious prion protein, PrP. To address these issues, we generated mammalian cellular models of expression of the C-terminal two helices of PrP, H2 and H3, which have been hypothesized, among other models, to hold the replication and conversion properties of the infectious PrP. We found that the H2H3 domain is an independent folding unit that undergoes glycosylations and glycosylphosphatidylinositol anchoring similar to full-length PrP. Surprisingly, in some conditions the normally folded H2H3 was able to systematically go through a conversion process and generate insoluble proteinase K-resistant aggregates. This structural switch involves the assembly of amyloid structures that bind thioflavin S and oligomers that are reactive to A11 antibody, which specifically detects protein oligomers from neurological disorders. Overall, we show that H2H3 is a conformational switch in a cellular context and is thus suggested to be a candidate for the conversion domain of PrP.
Introduction
Prion diseases are unique mammalian neurodegenerative diseases because their infectivity is mediated by the misfolding of an endogenous protein PrPC into an amyloid-structured conformer PrPSc (1). The β-sheet-enriched and aggregated PrPSc is able to template the conformational change of the α-helical PrPC, thus creating new infectious particles, which were at first characterized as partially resistant to proteinase K (PK)2 degradation (2–4). Prions are therefore defined as “infectious proteins” that are able to switch between at least two states, the prion state being able to replicate and propagate. This prion concept was successfully extended to a variety of proteins, mainly from yeast but also from fungi and marine mollusk, to explain non-Mendelian inheritance of phenotypes. In yeast, for example, the Sup35 and Ure2 proteins are considered to be the molecular determinants of the prion states [PSI+] and [URE3], respectively (5–7). Similarly, in the filamentous fungus Podospora anserina, the prion properties of Het-s protein explained the heterokaryon incompatibility phenotype of [Het-s] (8). More recently, Si et al. (9, 10) proposed a prion-based model for ApCPEB as a molecular mechanism of long term memory in Aplysia californica. In all of these cases, identification of a minimal prion-determining region was a strong asset for dissecting the role of the different domains in the proteins and for understanding their functions. Prion-determining regions appear to be an unstructured stretch of ∼70–200 amino acids that are, apart from Het-s, rich in glutamine/asparagine residues and have a bias against polar residues (9, 11–14). For instance, the prion domain of Sup35, called Sup35NM, has been characterized as sufficient for functional prion aggregation (15) and even for replication in mammalian cells, either in the cytosol (16) or attached to the membrane through a GPI anchor (17).
No definite region has been clearly identified as responsible for the change of conformation and the replication of PrP, dragging down attempts to understand both physiological and pathological function(s) of PrP and its subdomains. Although the putative replicative and templating prion domain of PrP is thought to be located in the globular C-terminal region, the unstructured N terminus (amino acids 23–89) is excluded from the infectious core of PrPSc (1). In 2007, Cobb et al. (18) proposed a model involving the last two helices of PrP, H2 and H3, as the major misfolding region, contrasting with other models centered on the misfolding of the central region (amino acids 89–175 or 116–164) (19, 20). More recent biophysical studies provided insights into brain-derived PK-resistant PrP and also implicated a conformational conversion of H2H3, which extends over the whole C-terminal region (amino acids ∼80–231) (21, 22). As shown by the apparent contradiction between these models, a consensus has yet to be found that should be based on in vivo studies. In favor of the H2H3 hypothesis, we have previously shown that (i) H2H3 can be independently generated in an Escherichia coli system and retain its native secondary and tertiary structure (23), (ii) H2H3 is able to fibrillize and reproduce the oligomerization pathways of PrP (23, 24), and (iii) in a cellular model of prion infection, polylysines are able to eliminate PK-resistant PrP through a direct interaction with H2H3 (25). These results led to the hypothesis that H2H3 could be the minimal region involved in the change of conformation of PrP.
In contrast to other prion-determining regions, H2H3 is natively structured. Unexpectedly, we observed that there was no natural propensity for the primary sequence of H2H3 to fold into helices and that the disulfide bond and the rest of the proteins are essential in its stabilization (23). It is not unusual for globular proteins to form amyloids (26). The metastable property of the structured H2H3 is consistent with amyloid misfolding of globular proteins under native conditions where environmental fluctuation may lead to local unfolding and cooperative restructuration (26). However, we need to establish conclusively whether, in vivo, H2H3 displays these alternative structures that are necessary, although not sufficient, to qualify it as a prion domain. Whether H2H3, when expressed in isolation in mammalian cells, retains its native structure and how the post-translational modifications may affect its stability are not known either.
Here, we generated cellular models expressing of GPI-anchored H2H3. Different H2H3- or H2H3Flag-expressing clones were selected and displayed membrane-anchored glycosylated H2H3. We then showed that the structured H2H3 could be induced to misfold into highly PK-resistant aggregate. Analysis of the aggregation process highlighted prion properties such as PK resistance, detergent insolubility, oligomers, and amyloid structures. The aggregation was confluence-dependent. We conclude that H2H3 presents a bistable structure in which the switch is controlled by the state of the cellular environment. Hence, this work supports a conformational switch function of H2H3 and casts new light onto the molecular bases of the pathogenesis of prion diseases and onto our fundamental understanding of the prion concept in general.
MATERIALS AND METHODS
Chemicals and Antibodies
Peptide N-glycosidase F (PNGase F), phosphatidylinositol-specific phospholipase C (PIPLC), and M2 anti-FLAG antibody were purchased from Sigma-Aldrich.
Plasmids
pcDNA3 plasmid containing the MoPrP construct with 3F4 epitope was kindly provided by Vincent Béringue and colleagues. Mouse H2H3 (PrP amino acids 163–254 concatenated with signal peptide amino acids 1–24) and FLAG-tagged H2H3 (DYKDDDDK tag sequence inserted between signal peptide and H2H3) fragments were synthesized by GenScript. These constructs were then cloned into mammalian expression vector pcDNA3 using the blunt restriction site EcoRV. The correct ligation of the resulting constructs was assessed by sequencing.
Cell Culture
The rabbit kidney cell line RK13 (27, 28) and the mouse cholinergic septal neuronal cell line SN56 (29, 30) were grown in culture medium composed of OptiMem (Invitrogen), supplemented with 10% FCS and 1% penicillin-streptomycin. Clonal cell lines resulting from the transfection of RK13 cells, namely B5 (expressing H2H3), D3 (expressing H2H3Flag), F6 (expressing H2H3Flag), and E1 (expressing PrP) clones were maintained in the same conditions and passaged twice a week.
Confluence was assessed as in Ref. 31. Briefly, 100% confluence was defined when cells covered all the culture plate or flask. Before that point, the percentage of confluence was calculated as the ratio between the number of cells divided by the number in 100% confluent condition. After confluence, the number of added culture days was indicated.
Transfection
Transfection of RK13 with the different constructs was performed using Lipofectamine 2000 (Invitrogen), following the manufacturer's instructions. For transient transfection studies, the cells were recovered after 24 h and analyzed. Stable transfectants were selected using G418 (Invitrogen) at 0.3 mg/ml for 2 weeks. After cell sorting and cloning, medium containing G418 was replaced by normal medium.
Western Blot
The cells were removed from culture plates using nonenzymatic cell dissociation solution (Sigma-Aldrich) and lysed for 10 min at 4 °C (lysis buffer: 50 mm Tris-HCl, 0.5% sodium deoxycholate, 0.5% Triton X-100). Nuclei and cellular remnants were removed by centrifugation at 104 × g for 2 min. The amount of protein was determined and normalized to 200 μg/sample using bicinchoninic acid protein assay (Uptima). For PK digestion experiments, the samples were PK-treated (standard concentration: 3 μg/mg of total protein) for 30 min at 37 °C, 1 mm PMSF was added to block enzymatic reaction, and proteins were centrifuged at 2 × 104 × g for 1 h at 4 °C. The pellets were resuspended and heated for 5 min at 100 °C in Laemmli buffer. For other experiments, lysates (10–40 μg of total protein, as indicated) were not PK-digested but directly diluted in 2× Laemmli buffer.
The samples were subjected to SDS-12% polyacrylamide gel electrophoresis (CriterionXT precast gel; Bio-Rad) and electroblotted to nitrocellulose membranes in transfer buffer (12 mm Tris, 80 mm glycine, 10% isopropanol), at 70 V for 90 min. The membranes were blocked in 5% (w/v) fat-free milk, processed with the indicated antibody and anti-mouse horseradish-peroxidase-coupled secondary antibody (GE Healthcare). Revelation was performed using ECL kit (Millipore) by autoradiographic method.
PNGase F and PIPLC Treatment
For deglycosylation, proteins from cell lysates were denatured by heating in 2% SDS buffer and incubated at 37 °C overnight in buffer containing 0.5% SDS, 1% Triton X-100 with 2 units of PNGase F/100 μg of total proteins.
PIPLC treatment was performed on a single layer of live cells in 6-well plates in PBS (pH 7.4) by incubating at 37 °C for 2 h with 1 unit of PIPLC per ml. Supernatants were collected, and proteins were precipitated by adding four volumes of acetone at −20 °C. The samples were then stored at −20 °C for 2 h and centrifuged at 15,000 × g for 10 min. The pellets were washed in cold acetone, centrifuged again, resuspended in Laemmli buffer, and analyzed by Western blot.
Solubility Assay
F6 cells from a 6-well plate were recovered at the indicated confluence, extensively washed in cold PBS, pelleted, and resuspended in 200 μl of lysis buffer supplemented with protease inhibitors (Roche Applied Science). Protein concentrations were normalized at 2 mg/ml. After a short centrifugation at 104 × g for 2 min to remove nuclei and cellular remnants, supernatant was centrifuged at 2 × 104 × g for 2 h at 4 °C. Supernatants were diluted in 2× Laemmli buffer, and pellet was resuspended in 50 μl of 1× Laemmli buffer; both were heated for 5 min at 100 °C. 10 μl of supernatant and pellet samples were loaded in SDS-PAGE gel.
Flow Cytometry
Confluent monolayer cells were recovered with nonenzymatic cell dissociation solution (Sigma-Aldrich) treatment and extensively washed with cold PBS. E1 and F6 cells were respectively labeled with 3F4 and M2 antibodies (5 μg/ml) followed by fluorescein isothiocyanate-coupled secondary antibody (Dako) detection. We used mouse IgG2a and IgG1 (Dako) as isotype controls for 3F4 and M2 antibodies, respectively. For quantification of dead cells, propidium iodide (Molecular Probes) (1 μg/ml) was incubated with cells at 4 °C for 20 min, and cells were counted by flow cytometry. Small cellular remnants (<1 μm) were gated out. Flow cytometry was performed on a BD FACScan device. Cell sorting and cloning were performed on a inFlux Cell Sorter (Alliance Biosecure Foundation). Cells within the fluorescence gate were cloned in 96-well plates with 1 cell/well in culture medium. The data were analyzed with FlowJo version 8.8.6 software.
Immunofluorescent Microscopy
The cells were grown in Lab-Tek slides (Nunc) following the cell culture procedure. For immunofluorescent labeling of PrP or H2H3Flag, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100, unless stated otherwise. 3F4 and M2 monoclonal antibodies were used to detect PrP and H2H3Flag, respectively. Overconfluent (100% + 4d) F6 cells were more sensitive to the immunofluorescent labeling protocol, and many dissociated from the slide. A11 antibody (Invitrogen) (32) detecting protein oligomers implicated in different neurodegenerative disorders was also tested. Fluorescent Alexa Fluor 488- or 568-conjugated secondary antibodies (Molecular Probes) were used for detection. Thioflavin S (ThS) staining followed the protocol of Kimura et al. (33). Briefly, when applicable, after secondary antibody incubation, the cells were washed three times with G1T (1% gelatin, 0.5 mg/ml bovine serum albumin, 150 mm NaCl, 50 mm HEPES, 0.1% Tween 20), stained in 0.001% ThS for 20 min, and washed three times with G1T. The coverslips were mounted on slides using Vectashield mounting medium with DAPI (Vector Labs). Image acquisition was performed on a Leica DM6000B fluorescence microscope or on a Leica confocal SPE with a 40× ACS APO objective and a 1.15 numerical overture. Bright field microscopy observations were taken from Zeiss Axiovert S100 microscope, with a 10× objective.
Circular Dichroism
The CD experiment was performed in a Jasco 810 device with a 0.01-cm circular quartz cuvette. Recombinant murine H2H3 was used at 100 μm in 5 mm citrate buffer adjusted to pH 3.5, at 25 °C. The spectrum was recorded from 260 to 195 nm and averaged from four scans collected at 200 nm·min−1. The plot was normalized by molarity for the ellipticity.
RESULTS
The C-terminal Two Helices H2 and H3 Can Be Stably Transfected in Mammalian Cells
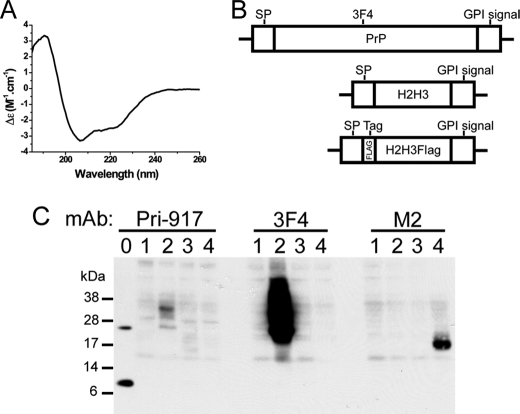
We previously showed by NMR that recombinant ovine H2H3 naturally acquires the same structure as the native H2H3 in full-length PrP (23). We first verified whether the murine sequence is also able to generate an α-helical recombinant H2H3. As shown by circular dichroism, mouse H2H3 displays a typical α-helix spectrum characterized by local minima at 209 and 220 nm (Fig. 1A), consistently with the in vitro results on ovine H2H3. To check whether H2H3 can be independently expressed in mammalian cells with processing and post-translational modifications similar to PrPC, we built a cellular expression model of a GPI-anchored H2H3 in a rabbit kidney cell line RK13, which does not express endogenous PrP and allows prion replication when expressing PrP (28).
FIGURE 1.
Transfection of H2H3 constructs. A, CD spectrum of recombinant mouse H2H3. The local minima at 209 and 220 nm are characteristic of α-helix structure. B, schematic representation of the three constructs: full-length mouse PrP containing the 3F4 epitope, H2H3 with signal peptide (SP) and GPI anchor propeptide (GPI signal) from PrP, and H2H3Flag with the FLAG tag (DYKDDDDK) inserted between signal peptide and H2H3. C, transient expression of transfected RK13 with the three constructs, analyzed by Western blot using different monoclonal antibodies. Lane 0, recombinant PrP and H2H3; lane 1, empty vector pcDNA3-transfected cells; lane 2, PrP-transfected cells; lane 3, H2H3-transfected cells; lane 4, H2H3Flag-transfected cells. 10 μg of proteins from cell lysate were loaded.
RK13 cells were transfected with four different constructs, including the empty plasmid pcDNA3 as a control (Fig. 1B). The inserts coding for H2H3 and an N-terminally FLAG-tagged H2H3 included the N-terminal signal peptide sequence and the C-terminal propeptide sequence of full-length PrP (GPI signal) (Fig. 1B), allowing correct localization and anchoring to the membrane through a glycosylphosphatidylinositol anchor. As a control, the sequence of mouse full-length PrP was used, containing the 3F4 epitope at position amino acids 109–112 (Fig. 1B) (34, 35).
After 24 h of transfection, the cells were recovered and analyzed by Western blot for transient expression of the different constructs (Fig. 1C). Despite our efforts in testing a large panel of antibodies, we could not find an efficient one specifically recognizing the C terminus of PrP. We thus used Pri-917 monoclonal antibody (36), which we found was able to detect H2H3. As shown in Fig. 1C (left panel, lane 0), Pri-917 detects the mixture of recombinant full-length PrP and H2H3. We were able to detect a band around 17 kDa in lane 3 corresponding to the H2H3-transfected cells. When probed with 3F4 monoclonal antibody, the typical glycoforms of full-length PrP was detected only in PrP-transfected cells, with a molecular mass ranging from 19–21 kDa to up to ∼40 kDa (Fig. 1C, center panel, lane 2). The signal was much stronger than with Pri-917, indicating a very low sensitivity of the latter. Because we could not get a clear signal for H2H3Flag with Pri-917 antibody (Fig. 1C, left panel, lane 4), we used the M2 monoclonal antibody raised against the FLAG tag and showed the expression of a ∼18-kDa protein only in H2H3Flag-transfected cells (Fig. 1C, right panel, lane 4), which corresponded to a glycosylated form of H2H3Flag as shown in subsequent deglycosylation experiments (Fig. 2A) and when compared with the theoretical molecular mass of the protein, which is 9–10 kDa. In each case, cells transfected with the empty vector did not show any specific signal (Fig. 1C, lanes 1).
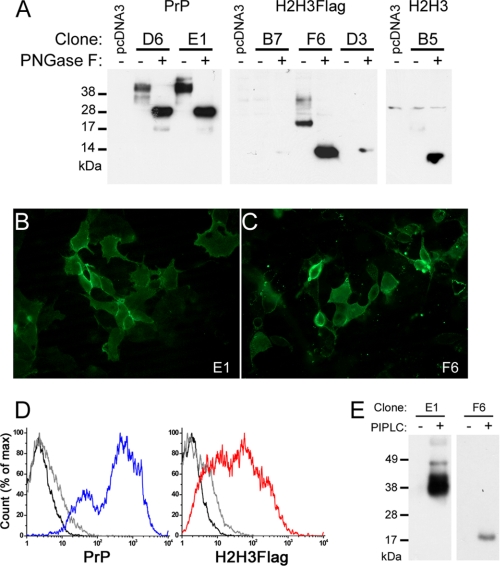
FIGURE 2.
Cell biology of selected PrP, H2H3, and H2H3Flag clones. A, study of the glycosylation of the selected clones. 10 μg of proteins from cell lysate (40 μg for B5), treated (+) or not (−) by PNGase F. D6 and E1 clones expressing PrP, B7, F6, and D3 clones expressing H2H3Flag, B5 clone expressing H2H3 were tested using 3F4, M2, and Pri-917 antibodies, respectively. B and C, fluorescence microscopy on nonpermeabilized cells. B, E1 cells displayed surface labeling of 3F4 antibody coupled with Alexa Fluor 488 secondary antibody. C, F6 cells displayed surface labeling of M2 antibody coupled with Alexa Fluor 488 secondary antibody. D, flow cytometry measurement of surface labeled PrP (blue) or H2H3Flag (red). In each plot, unlabeled control cells (black line) and isotype control labeled cells (gray line) were used. E, PrP and H2H3Flag are GPI-anchored. Western blot analysis of acetone-precipitated supernatant of PIPLC-treated (+) or untreated (−) E1 and F6 cells, using 3F4 and M2 antibodies, respectively.
To select stable clonal cell lines, we used a cell sorting strategy. MoPrP RK13, H2H3Flag RK13, and H2H3 RK13 cell surfaces were respectively labeled with 3F4, M2, Pri-917 antibodies and then with Alexa 488-coupled secondary antibody. The selected population corresponded to the uppermost 0.16, 0.055, and 0.037% fluorescent cells of MoPrP RK13, H2H3Flag RK13, and H2H3 RK13 cultures, respectively (supplemental Fig. S1A). As shown in supplemental Fig. S1B, clones with a detectable level of expression could be found, although probing with Pri-917 required more effort to reveal acceptable signal. Overall, we were able to generate, through a cell sorting selection and cloning strategy, clonal cell lines stably expressing different levels of expression for the three constructs of interest at the cell surface.
H2H3 Harbors Post-translational Modifications Similar to PrPC
In PrPC, the glycosylation process and the addition of a GPI anchor is associated with a stabilized membrane-bound α-helical structure of the H2H3 domain (37, 38). To explore the processing of H2H3, we first assessed the glycosylation pattern by PNGase F treatment. As expected, full-length PrP in both D6 and E1 clones was glycosylated (Fig. 2A) and, after PNGase treatment, revealed the unglycosylated PrP at ∼28 kDa and a faint signal at 19 kDa, corresponding to the truncated C-terminal C2 fragment (39). Similarly, for the 3 H2H3Flag clones B7, F6 and D3, removal of glycosylation by PNGase F allowed the detection of a single band at ∼14 kDa, slightly above recombinant H2H3 migration band caused by the presence of the FLAG tag (Fig. 2A and supplemental S1C). The three selected clones displayed different expression levels. For the F6 clone, which has the highest expression level, we could also detect higher molecular mass bands up to ∼33 kDa, which could correspond to other glycoforms (Fig. 2A). They were, however, present at a low proportion, giving rise to an original pattern, although similar to full-length PrP: no detectable unglycosylated, a strong intermediately glycosylated, and minor hyperglycosylated forms. We compared the expression of all selected clones, after PNGase F treatment, using the only common antibody Pri-917 and found similar level of expression for E1 and F6 and a slightly weaker expression for D6 and B5 (supplemental Fig. S1C).
These data prompted us to verify whether the protein could be ultimately localized on the outer plasma membrane. Using both immunofluorescent microscopy on nonpermeabilized cells and flow cytometry on surface-labeled cells (Fig. 2, B–D), we demonstrated that PrP-expressing E1 clone and H2H3Flag-expressing F6 clone displayed surface fluorescent labeling of their respective protein. Furthermore, Western blot analysis of the protein content in the supernatant of E1 or F6 cells with or without phospholipase C treatment (PIPLC), which released GPI-anchored proteins in the medium, indicated that at least a proportion of PrP in E1 and H2H3Flag in F6 were anchored on the plasma membrane through a GPI anchor (Fig. 2E), consistent with the localization results.
H2H3 Switches to a PK-resistant Form under Specific Conditions
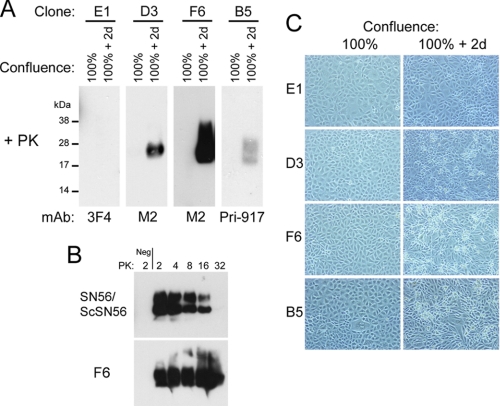
Evaluation of the resistance of H2H3 to PK digestion showed that at 100% confluence + 2 days, B5 (expressing H2H3 without the FLAG tag), D3 and F6 cells generated a PK-resistant form of H2H3 (Fig. 3A), hereafter called H2H3res, whereas before and at 100% confluence, H2H3 remained PK-sensitive. As a control, in the same conditions, PrP-expressing E1 cells did not show this phenomenon (Fig. 3A). Thus generation of PK-resistant species was specific to H2H3 and was not due to the FLAG tag.
FIGURE 3.
PK resistance of H2H3 and H2H3Flag. A, spontaneous PK resistance of H2H3 and H2H3Flag. Cell lysates from 100% confluent and 100% confluent + 2 days cultures were PK-digested. D3, F6, and B5 displayed PK-resistant proteins, in contrast to E1 cells. B, comparative PK resistance assay between Chandler-infected SN56 cells and overconfluent F6 cells. Increasing concentrations of PK (μg/mg of total proteins) were used, and samples were analyzed by Western blot using Saf83 antibody for ScSN56 and M2 for F6 cells. Negative controls (Neg) included uninfected SN56 cells and 100% confluent F6 cells. C, bright field observation of E1, D3, F6, and B5 cells at 100% confluence and 100% confluence + 2 days (2d), with a 10× objective. Only E1 cells at 100% confluence + 2 days did not show visible refringent dead cells.
When compared with classical scrapie prion from infected SN56 cells, H2H3res from F6 cells displayed a much higher resistance to PK digestion (Fig. 3B). As negative controls for PK digestion, uninfected SN56 and 100% confluent F6 cells were used, whereas Chandler-infected SN56 (ScSN56) and overconfluent (100% + 2 days) F6 cells were used as PK-resistant samples (Fig. 3B).
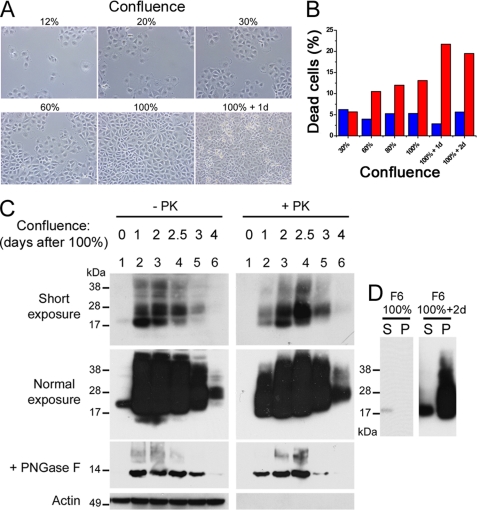
For B5, D3 and F6 clones, at 100% confluence + 2 days, refringent cells began to appear in the culture in contrast to the PrP-expressing E1 clone (Figs. 3C and 4, A and B). Quantification of the ratio of dead cells in F6 cultures indicated an increase up to ∼25% (Fig. 4B). H2H3res was generated in live cells, because with the extensive wash with PBS, most of the dead cells were not analyzed in the PK resistance Western blot assay. We then analyzed the electrophoretic profile of H2H3 as a function of confluence (Fig. 4C). F6 cells were plated at day 0 by serial dilution and incubated until day 7. The confluence state was assessed by microscopy. Both PK-resistant H2H3 and total H2H3 were analyzed by Western blot using M2 antibody probing. Compared with 100% confluence (Fig. 4C, normal exposure, lane 1), we observed a dramatic increase of total H2H3 after 1 day (Fig. 4C, normal exposure, lane 2). At this stage, a new higher molecular band appeared at ∼25–28 kDa (Fig. 4C, short exposure, lanes 2–6), which we found to be another glycoform as demonstrated by PNGase F treatment (Fig. 4C). Higher bands at ∼34 and ∼38–45 kDa could also be observed, probably corresponding to dimers (Fig. 4C, short exposure, lanes 2–6). As cells went further in their confluence state, we observed a shift from the lower glycoform to the high glycoform (Fig. 4C, short exposure, lanes 2–6), as well as a decrease in the total amount of H2H3. The 100% confluent F6 cells displayed no trace of H2H3res, indicating that at this stage, H2H3 was PK-sensitive (Fig. 4C, lanes 1). However, beginning at confluence 100% + 1 day, large amounts of H2H3res were detected. We also observed a shift from the lower glycoform to the higher one in PK-digested samples starting earlier than without PK digestion. A solubility assay showed that the lower glycoform was associated with detergent-soluble H2H3, whereas the higher glycoform was only found in the pellet of overconfluent F6 cells (Fig. 4D). In 100% confluent cells, no insoluble H2H3 fraction could be found in these conditions, in agreement with the PK resistance assay (Fig. 4D).
FIGURE 4.
Characterization of the aggregation correlated to confluence. A, different confluence states of F6 cells observed by bright field microscopy. B, flow cytometry quantification of propidium iodide-positive cells (dead cells) in E1 (blue) and F6 (red) cultures as a function of confluence. C, analysis of the electrophoretic profile of H2H3Flag, probed by M2 antibody, as a function of confluence. F6 were plated at day 0 by serial one half-dilution and analyzed at day 6, with (+) or without (−) PK digestion, loading 200 μg or 10 μg of total proteins, respectively. Top panels, two autoradiographic exposures were performed. Middle panels, aliquots of the samples were treated with PNGase F to assess the glycosylation status of the protein. Loaded amount of proteins corresponded to 1/10th of the upper panels load. Bottom panels, actin was used a normalization control. For lane 4, the date of confluence was intermediate between lanes 3 and 5. D, solubility assay. F6 cell lysates from 100% confluent and 100% confluent + 2 days cultures were separated as a detergent-soluble fraction (S) and pellet fraction (P) and probed by M2 antibody in Western blot.
H2H3 Forms Oligomers and Amyloid Aggregates
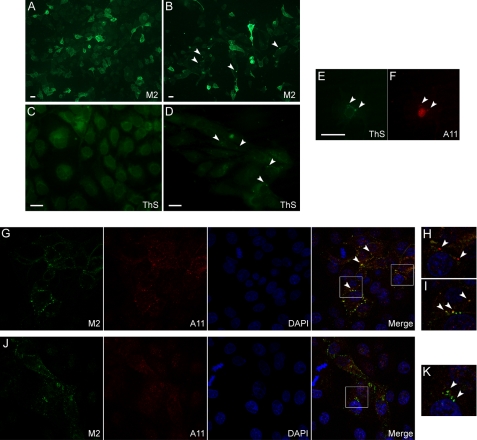
We wondered whether PK resistance would be associated with macroscopic aggregation of H2H3. M2-labeled overconfluent F6 cells (confluence 100% + 4 days) displayed heterogeneous fluorescent aggregates (Fig. 5B, arrows), which were not found in 100% confluent F6 cells (Fig. 5A). Similarly, ThS labeling showed specific appearance of puncta in overconfluent F6 cells (Fig. 5D), in contrast to the diffuse background fluorescence of confluent F6 cells (Fig. 5C).
FIGURE 5.
H2H3Flag oligomerization and amyloidogenesis. A–F, epifluorescence microscopy of 100% confluent (A and C) or 100% confluent + 4 days (B, D, E, and F) permeabilized F6 cells. Scale bars, 10 μm. A and B, M2 labeling of H2H3Flag localization showed the appearance of heterogeneous punctuate assemblies in overconfluent cells (arrows), in contrast to confluent cells. C and D, specific puncta-shaped thioflavin S staining in overconfluent cells (arrows). E and F, colabeling of overconfluent F6 cells with ThS (E) and A11 antibody (F). The arrows indicate assemblies that are positive for both staining. G–K, confocal microscopy of 100% confluent F6 cells, colabeled with M2 (green) and A11 (red) antibodies. G–I, in some cells, punctuate A11 labeling was observed, partially associated (G and I, arrows) or not (H, arrows) with M2 labeling. J and K, in general, A11 labeling was diffuse, and no colocalization was found with M2 (K, arrows).
To qualify the structure of H2H3res aggregates, we performed a double labeling with ThS, which is specific of amyloid structure, and A11 anti-oligomers antibody (32), which specifically targets oligomers from amyloid proteins associated with neurodegenerative diseases. Although subconfluent to confluent (100% confluence) F6 cells only showed some diffuse immunoreactivity for A11 antibody in the nuclei (supplemental Fig. S2), overconfluent (100% confluence + 4 days) cells sometimes displayed a heterogeneous punctuate pattern of A11 labeling (Fig. 5F) associated with ThS fluorescence in the cytoplasm (Fig. 5E). ThS fluorescence could not be detected before 100% confluence + 4 days.
To better visualize the initial steps of aggregation, we observed confluent F6 cells colabeled with M2 and A11 antibodies by confocal microscopy (Fig. 5, G–K). In some fields, we could detect punctuate A11 reactivity, indicating the presence of oligomers (Fig. 5G). Some A11-positive aggregates were not associated with H2H3Flag (Fig. 5H, arrows), but most of them partially colocalized with M2 green fluorescence (Fig. 5, G and I, arrows), indicating oligomerizations of H2H3Flag found at the junction of two cells. In general, we would find M2-positive cells with diffuse low reactivity for A11 (Fig. 5J). H2H3Flag was essentially found at the membrane, as expected (Fig. 5J), but did not colocalize with A11 (Fig. 5K).
DISCUSSION
In this study, we developed new cellular models to gain insights into the mechanisms of H2H3 structural switch as the molecular basis supporting PrP change of conformation. Expressing the H2H3 domain (amino acids 163–231) in RK13 cells leads to the N-glycosylation and GPI attachment of this region. Both glycosylation and GPI anchor are post-translational processes that require trafficking through the endoplasmic reticulum and Golgi apparatus (40). The presence of these post-translational modifications constitutes an indirect proof that the expressed H2H3 was correctly folded and localized according to its signal peptide. This is consistent with H2H3 being an independent folding domain, as proven by NMR studies of the isolated recombinant H2H3 domain (23). This observation is not trivial because H2H3 in PrP is structurally closely embedded in the rest of the globular domain, comprising S1-H1-S2.
In a search for alternative conformational states of H2H3, we found a correlation between cell confluence and the aggregation state of H2H3. After reaching confluence, different independent H2H3 clones systematically and spontaneously generated PK-resistant proteins, whereas full-length PrP-expressing E1 clone did not show this phenomenon. Here we wish to stress that this phenomenon was observed for three independent clones expressing either H2H3Flag or H2H3, thus discarding any concern that the results could be due to clone-specific properties unrelated to H2H3 expression. Furthermore, because we used clones that express different levels of H2H3, either comparable with or lower than PrP expression in E1 cells, generation of PK-resistant species is not simply due to an overexpression of H2H3.
The H2H3 aggregates are highly PK-resistant when compared with a standard Chandler prion strain replicated in permissive SN56 cells and form punctuate assemblies in cells, as visualized by fluorescent microscopy. After PK treatment, the molecular mass of H2H3 and H2H3Flag did not shift, and the FLAG tag remained detectable, suggesting that the structure of the assembly can even protect a region not directly involved in the resistant core. This observation may relate to the PK resistance and solvent inaccessibility of the whole C-terminal region (amino acids 90–231) in PrPSc (1, 21).
It is remarkable that, up to confluence, H2H3- or H2H3Flag-expressing clones do not show aggregation, which proves the existence of a process of conversion for H2H3 in a given condition and not only a nonspecific native aggregation occurring at all times. This conclusion is supported by the results showing that H2H3 is stably folded, glycosylated, and normally brought to the membrane before showing any sign of aggregation. When we searched for the early aggregation events of 100% confluent F6 cells, we found colocalizations at cell junctions of M2 and A11 antibodies detections observed by confocal microscopy, suggesting that H2H3Flag oligomerization occurs first at the membrane vicinity. We could not see this kind of colocalization in all cells, suggesting local favorable conditions for oligomerization. Reactivity to A11 antibody, described by Kayed et al. (32) as being able to specifically detect oligomeric misfolded forms of proteins and peptides implicated in neurological disorders, associated with ThS dye binding, thus providing a structural characterization of H2H3 aggregation in the cellular context. Overall, these data suggest a picture of the structural switch process for H2H3 in a cellular context. To explain the cellular dynamics of amyloid aggregation, we speculate that when confluence is reached, an increased local concentration and stability of glycosylated H2H3 caused by membrane contacts and junction trigger or enhance the oligomerization and aggregation of membrane-bound H2H3. Endocytosis and endosomal pathways would further distribute them inside cells, as observed with ThS fluorescence. Subsequent cell death may be due to signaling pathways triggered by H2H3 or by some toxicity of H2H3 aggregates. Indeed, the cell culture conditions are not harsh, because control RK13 and PrP-expressing E1 clone do not display aberrant cell death when overconfluent. Moreover, H2H3-expressing cells that are analyzed in this study are live cells because washing steps remove most of the dead cells. Thus the observed properties, such as acquisition of PK resistance or amyloid structure, are not cell death-induced artifacts but rather, cell death may be a consequence of H2H3 conversion.
In contrast to yeast prions, H2H3 aggregation may arise from a natively structured protein and not only unstructured glutamine/arginine-rich domains (26). However, systematically generating PK-resistant aggregates in specific cellular conditions is quite unusual, even for proteins with prion properties. For instance, when yeast prions such as Sup35 or Ure2 are overexpressed, the rate of spontaneous apparition of the prion state is increased by several orders of magnitude (7, 11, 12, 15, 41) but still remains low compared with the systematic conversion of H2H3 when confluence is reached. This indicates that, compared with yeast prions, H2H3 may have a higher propensity to undergo a structural change. This is supported by our previous work in vitro showing that the absence of S1-H1-S2 favors fibrillization of H2H3, which occurs more easily than in full-length PrP (23). Interestingly, it is usual for recombinant proteins such as Sup35, Ure2, or PrP to produce amyloid fibrils in mild denaturing conditions, but fibrillization from recombinant H2H3 does not require these facilitating conditions (23): this is consistent with the spontaneous generation of misfolded H2H3 reported here and implies that the cellular environment is able per se to produce sufficient energetic and thermodynamic fluctuations for H2H3 initial conversion. We thus propose that the separation of S1-H1-S2 from the H2H3 bundle in PrP is a prerequisite for misfolding as suggested in Ref. 42 and that exposing or isolating H2H3 is a necessary step in mammalian cells for misfolding and for generation of aggregated PK-resistant species.
Further developments of this work include the study of transmissibility of the H2H3 aggregates from cell to cell, because a prion domain must be able to template the formation of new misfolded proteins. However, we have not managed to get results about this issue because it is technically difficult to distinguish between transmissible and spontaneous conformational change of H2H3 in our cellular model. We thus performed a heterologous transmissibility study using an in vivo transgenic mice model overexpressing PrP. A study of the potential infectivity of PK-resistant H2H3 aggregates inoculated to PrP-overexpressing transgenic mice is still in progress.
In conclusion, the independently expressed H2H3 domain is natively folded as it would within full-length PrP, harboring similar post-translational modifications. A specific cellular event linked to confluence triggers a change of conformation of H2H3 into an aggregated, insoluble, and amyloid form. Prion-based mechanisms, whether pathological or functional, require the misfolding of a specific domain. When such a domain is natively structured, the molecular basis of the prion phenomenon is related to the existence of two stable conformational folds, which is unique for prions but not unusual in general. This study demonstrates in a cellular context that the H2H3 domain of PrP follows such a model and is able to access two different structures. We thus provide evidence that H2H3 is a plausible candidate in vivo for being the prion domain of PrP.
Supplementary Material
Acknowledgments
We thank Vincent Béringue and Jérôme Chapuis for providing RK13 cells and MoPrP plasmid construct; Bruce Wainer for providing SN56 cell line; Annalisa Pastore, Hubert Laude, and Vincent Béringue for comments; Ludmilla Sissoeff, Frédéric Auvray, and Fabien Aubry for technical assistance in confocal microscopy and cell sorting; and Kausik Si for technical advice on ThS staining.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PK
- proteinase K
- PNGase F
- peptide N-glycosidase F
- PIPLC
- phosphatidylinositol-specific phospholipase C
- GPI
- glycosylphosphatidylinositol
- ThS
- thioflavin S.
REFERENCES
- 1. Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeArmond S. J., McKinley M. P., Barry R. A., Braunfeld M. B., McColloch J. R., Prusiner S. B. (1985) Cell 41, 221–235 [DOI] [PubMed] [Google Scholar]
- 3. Bolton D. C., McKinley M. P., Prusiner S. B. (1982) Science 218, 1309–1311 [DOI] [PubMed] [Google Scholar]
- 4. Meyer R. K., McKinley M. P., Bowman K. A., Braunfeld M. B., Barry R. A., Prusiner S. B. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 2310–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patino M. M., Liu J. J., Glover J. R., Lindquist S. (1996) Science 273, 622–626 [DOI] [PubMed] [Google Scholar]
- 6. Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. (1996) EMBO J. 15, 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- 7. Wickner R. B. (1994) Science 264, 566–569 [DOI] [PubMed] [Google Scholar]
- 8. Coustou V., Deleu C., Saupe S., Begueret J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9773–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Si K., Lindquist S., Kandel E. R. (2003) Cell 115, 879–891 [DOI] [PubMed] [Google Scholar]
- 10. Si K., Choi Y. B., White-Grindley E., Majumdar A., Kandel E. R. (2010) Cell 140, 421–435 [DOI] [PubMed] [Google Scholar]
- 11. Uptain S. M., Lindquist S. (2002) Annu. Rev. Microbiol. 56, 703–741 [DOI] [PubMed] [Google Scholar]
- 12. Masison D. C., Maddelein M. L., Wickner R. B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Osherovich L. Z., Cox B. S., Tuite M. F., Weissman J. S. (2004) PLoS Biol. 2, E86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Balguerie A., Dos Reis S., Ritter C., Chaignepain S., Coulary-Salin B., Forge V., Bathany K., Lascu I., Schmitter J. M., Riek R., Saupe S. J. (2003) EMBO J. 22, 2071–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Derkatch I. L., Chernoff Y. O., Kushnirov V. V., Inge-Vechtomov S. G., Liebman S. W. (1996) Genetics 144, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krammer C., Kryndushkin D., Suhre M. H., Kremmer E., Hofmann A., Pfeifer A., Scheibel T., Wickner R. B., Schätzl H. M., Vorberg I. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 462–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Speare J. O., Offerdahl D. K., Hasenkrug A., Carmody A. B., Baron G. S. (2010) EMBO J. 29, 782–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cobb N. J., Sönnichsen F. D., McHaourab H., Surewicz W. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18946–18951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Govaerts C., Wille H., Prusiner S. B., Cohen F. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeMarco M. L., Daggett V. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2293–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smirnovas V., Baron G. S., Offerdahl D. K., Raymond G. J., Caughey B., Surewicz W. K. (2011) Nat. Struct. Mol. Biol. 18, 504–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baron G. S., Hughson A. G., Raymond G. J., Offerdahl D. K., Barton K. A., Raymond L. D., Dorward D. W., Caughey B. (2011) Biochemistry 50, 4479–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adrover M., Pauwels K., Prigent S., de Chiara C., Xu Z., Chapuis C., Pastore A., Rezaei H. (2010) J. Biol. Chem. 285, 21004–21012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chakroun N., Prigent S., Dreiss C. A., Noinville S., Chapuis C., Fraternali F., Rezaei H. (2010) FASEB J. 24, 3222–3231 [DOI] [PubMed] [Google Scholar]
- 25. Xu Z., Adrover M., Pastore A., Prigent S., Mouthon F., Comoy E., Rezaei H., Deslys J. P. (2011) FASEB J., in press [DOI] [PubMed] [Google Scholar]
- 26. Chiti F., Dobson C. M. (2009) Nat. Chem. Biol. 5, 15–22 [DOI] [PubMed] [Google Scholar]
- 27. Christofinis G. J., Beale A. J. (1968) J. Pathol. Bacteriol. 95, 377–381 [DOI] [PubMed] [Google Scholar]
- 28. Vilette D., Andreoletti O., Archer F., Madelaine M. F., Vilotte J. L., Lehmann S., Laude H.(2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4055–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hammond D. N., Lee H. J., Tonsgard J. H., Wainer B. H. (1990) Brain Res. 512, 190–200 [DOI] [PubMed] [Google Scholar]
- 30. Magalhães A. C., Baron G. S., Lee K. S., Steele-Mortimer O., Dorward D., Prado M. A., Caughey B. (2005) J. Neurosci. 25, 5207–5216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Su H. W., Yeh H. H., Wang S. W., Shen M. R., Chen T. L., Kiela P. R., Ghishan F. K., Tang M. J. (2007) J. Biol. Chem. 282, 9883–9894 [DOI] [PubMed] [Google Scholar]
- 32. Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G.(2003) Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 33. Kimura Y., Koitabashi S., Fujita T. (2003) Cell Struct. Funct. 28, 187–193 [DOI] [PubMed] [Google Scholar]
- 34. Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., Diringer H. (1987) J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kascsak R. (2010) J. Biol. Chem. 285, le5, author reply le6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Demart S., Fournier J. G., Creminon C., Frobert Y., Lamoury F., Marce D., Lasmézas C., Dormont D., Grassi J., Deslys J. P. (1999) Biochem. Biophys. Res. Commun. 265, 652–657 [DOI] [PubMed] [Google Scholar]
- 37. Lowe J. B., Marth J. D. (2003) Annu. Rev. Biochem. 72, 643–691 [DOI] [PubMed] [Google Scholar]
- 38. Cancellotti E., Barron R. M., Bishop M. T., Hart P., Wiseman F., Manson J. C. (2007) Biochim. Biophys. Acta 1772, 673–680 [DOI] [PubMed] [Google Scholar]
- 39. Chen S. G., Teplow D. B., Parchi P., Teller J. K., Gambetti P., Autilio-Gambetti L. (1995) J. Biol. Chem. 270, 19173–19180 [DOI] [PubMed] [Google Scholar]
- 40. Helenius A., Aebi M. (2001) Science 291, 2364–2369 [DOI] [PubMed] [Google Scholar]
- 41. Chernoff Y. O., Derkach I. L., Inge-Vechtomov S. G. (1993) Curr. Genet. 24, 268–270 [DOI] [PubMed] [Google Scholar]
- 42. Eghiaian F., Daubenfeld T., Quenet Y., van Audenhaege M., Bouin A. P., van der Rest G., Grosclaude J., Rezaei H.(2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.