Background: The hMSH2-hMSH6 heterodimer must coordinate mismatch binding with dual site adenosine nucleotide processing.
Results: An hMSH2-magnesium-ADP complex inhibits ATP hydrolysis by both the hMSH2 and hMSH6 subunits.
Conclusion: hMSH2 regulates adenosine nucleotide processing by the hMSH2-hMSH6 mismatch recognition heterodimer.
Significance: Understanding the molecular mechanism of hMSH2-hMSH6 function is crucial for elucidating the role of the mismatch repair pathway in human tumorigenesis.
Keywords: ADP, ATP, ATPases, Cancer Biology, Colon Cancer, DNA Repair
Abstract
The mechanics of hMSH2-hMSH6 ATP binding and hydrolysis are critical to several proposed mechanisms for mismatch repair (MMR), which in turn rely on the detailed coordination of ATP processing between the individual hMSH2 and hMSH6 subunits. Here we show that hMSH2-hMSH6 is strictly controlled by hMSH2 and magnesium in a complex with ADP (hMSH2(magnesium-ADP)-hMSH6). Destabilization of magnesium results in ADP release from hMSH2 that allows high affinity ATP binding by hMSH6, which then enhances ATP binding by hMSH2. Both subunits must be ATP-bound to efficiently form a stable hMSH2-hMSH6 hydrolysis-independent sliding clamp required for MMR. In the presence of magnesium, the ATP-bound sliding clamps remain on the DNA for ∼8 min. These results suggest a precise stepwise kinetic mechanism for hMSH2-hMSH6 functions that appears to mimic G protein switches, severely constrains models for MMR, and may partially explain the MSH2 allele frequency in Lynch syndrome or hereditary nonpolyposis colorectal cancer.
Introduction
Lynch syndrome or hereditary nonpolyposis colorectal cancer accounts for 1–3% of colorectal cancer cases and is caused by germ line mutations of the DNA mismatch repair (MMR)3 genes (1). The majority of mutations occur in MSH2 (∼40%) or MLH1 (∼50%) (2). MMR corrects mismatched nucleotides and insertion-deletion loops (IDLs) in DNA caused by polymerase errors, chemical modifications, and recombination between heterologous DNA sequences (3). MMR proteins also act as sensors to activate DNA damage checkpoints in response to alkylating agents and other DNA-damaging agents (4).
hMSH2 forms a heterodimer with hMSH6 that recognizes single base pair mismatches and small IDLs (5). A hallmark of MutS homologs (MSH) is the presence of highly conserved Walker A/B nucleotide binding domains (6). The ability of MSH proteins to bind and hydrolyze ATP (ATPase) is essential for MMR function and is stimulated by mismatched DNA (7–10). The ATPase cycle is composed of two major steps: γ-phosphate hydrolysis and the mismatch-dependent exchange of ADP for ATP (ADP → ATP exchange). ADP → ATP exchange is rate-limiting and provokes a conformational transition that results in the formation of a sliding clamp, which dissociates from the mismatch (8, 11). One possible model for MMR suggests that mismatch-dependent loading of multiple hydrolysis-independent sliding clamps activates downstream MMR components (8, 12–14), whereas another model suggests hydrolysis-dependent translocation to contact downstream MMR components (15–17). The management of ATP processing by the two MSH subunits is predicted to be significantly different for these two models.
A number of studies have detailed the coordination of mismatch binding and adenosine nucleotide processing by the individual Walker A/B domains of hMSH2 and hMSH6 (18–21). Structures of bacterial and human MSH revealed asymmetry in subunit structure and function, with one subunit, MSH6 in eukaryotes, making specific contact with the mismatch (22–24). Biochemical studies in bacteria and yeast have demonstrated asymmetric ATP processing between the two subunits as well. Importantly, Msh2 displays a high affinity for ADP, and Msh6 displays a high affinity for ATP (21, 25, 26). This difference in binding affinities translates into an asymmetry in ATP hydrolysis (18, 19, 27). However, the mechanism by which the individual hMSH2 and hMSH6 subunits coordinate ATP processing to regulate the conversion from mismatch binding to signaling downstream events in MMR has not been resolved (30).
Here we have examined hMSH2-hMSH6 subunit mutations of the Walker A domain to determine the mechanism of asymmetric ATP processing. We find that hMSH2 in a complex with magnesium and adenosine nucleotide regulates all aspects of ADP/ATP processing of the hMSH2-hMSH6 heterodimer. This central role of hMSH2 in the ATP processing may at least partially explain the allele frequency in Lynch syndrome or hereditary nonpolyposis colorectal cancer.
EXPERIMENTAL PROCEDURES
Proteins and DNA Substrates
hMSH2(K675A) and hMSH6(K1140A) mutations were generated by QuikChange site-directed mutagenesis (Stratagene). Wild-type and mutant hMSH2-hMSH6 heterodimers were expressed and purified as described previously (8). Biotinylated and digoxigenin-modified oligonucleotides were synthesized by Integrated DNA Technologies. Other oligonucleotides were synthesized by Sigma-Aldrich.
Adenosine Nucleotide Binding
ATP cross-linking experiments were performed in the absence of DNA with wild-type or mutant hMSH2-hMSH6 protein (200 nm). Samples were titrated with increasing concentrations (0–500 nm) of [γ-32P]ATP, [α-32P]ATP, [α-32P]ADP, or [γ-35S]ATP, as indicated, for 20 min at room temperature in a 10 μl reaction buffer containing 25 mm HEPES (pH 7.8), 110 mm NaCl, 1 mm DTT, 10% glycerol, and either 0 or 5 mm MgCl2, as indicated. The reactions were cross-linked on ice for 10 min at 120 mJ using a UV Stratalinker 1800 (Stratagene) and resolved on a 6% SDS-polyacrylamide gel. Bands were visualized using the Cyclone Storage Phosphor System (PerkinElmer Life Sciences) and analyzed by OptiQuantTM software. 0.7 μg of each protein was run on a separate gel and stained with Coomassie Blue to control for the amount of each protein loaded. ATPγS filter binding experiments were performed with 85 nm hMSH2-hMSH6 protein, 5 μm [γ-35S]ATP, 0.5–20 mm unlabeled ATPγS, and 0 or 2.5 mm MgCl2, as described previously (31). [γ-35S]ATP bound was measured in a Beckman LS 6500 Counter. ADP → ATP exchange assays were performed as described previously (31).
ATP Hydrolysis
Steady-state ATPase activity was measured at multiple protein concentrations (15–100 nm) of hMSH2-hMSH6 by incubating with 16.5 nm [γ-32P]ATP in the presence of a 41-bp DNA oligonucleotide containing a central G/T mismatch, as described previously (31). For analysis of DNA-independent ATPase activity, protein concentration was increased to 200 nm. Experiments investigating the effect of free magnesium on ATPase activity were performed similarly. Free magnesium concentration was calculated using Maxchelator software (available from the Stanford University Web site) and adjusted taking into account ATP-magnesium complex formation and reported as indicated. Pre-steady-state measurements of ATP hydrolysis and phosphate (Pi) release were performed by stopped-flow (KinTek Corp., Austin, TX) experiments in which MDCC-labeled PBP and hMSH2-hMSH6 proteins were mixed with ATP, and the change in MDCC-PBP fluorescence upon binding Pi was monitored over time (lex = 425 nm, lem > 450 nm; buffer: 25 mm Hepes-NaOH, pH 7.5, 5 mm MgCl2, 110 mm NaCl, 1 mm DTT, 10% (v/v) glycerol). Five or more individual kinetic traces were averaged, and Pi concentration was determined from calibration curves, as described previously (32). The final concentrations were 0.2 mm hMSH2-hMSH6, 400 mm ATP, and 8 mm MDCC-PBP.
Surface Plasmon Resonance
A Biacore 3000TM was used to study real-time kinetics, magnesium-dependent nucleotide processing, and DNA dissociation with hMSH2-hMSH6. 41-bp DNA substrates containing a centrally located G/T base pair mismatch or G/C base pair were constructed as described previously (33) with the addition of covalently attached 5′-biotin. For double-blocked end studies, a 3′- digoxigenin was added. Purified DNA was attached to an SA Chip (GE Healthcare) coated with streptavidin. Constant flow experiments were performed in running buffer (RB) (25 mm HEPES, pH 7.8, 100 mm NaCl, 4% glycerol, 1 mm DTT, 0.005% surfactant, and 10 mm MgCl2 unless otherwise indicated). All free-end studies were performed by injecting binding buffer (RB + 30 nm indicated protein), followed by a RB wash to remove unbound protein from the DNA, and concluding with an ATP injection (RB + 250 μm ATP). The chip is regenerated following an injection of RB + 1 m NaCl.
Blocked end magnesium titration studies were carried out identically as above but with the following exceptions: an initial injection of RB + 50 nm anti-digoxigenin antibody (Roche Applied Science), no MgCl2 or 10 mm MgCl2 in RB or binding buffer, and varying levels of MgCl2 in the ATP buffer, as indicated. All plots and kinetic analysis were created using Origin 8.0TM and Microsoft ExcelTM software.
RESULTS
Adenosine Nucleotide Binding by hMSH2 and hMSH6 Subunits in the Absence of DNA
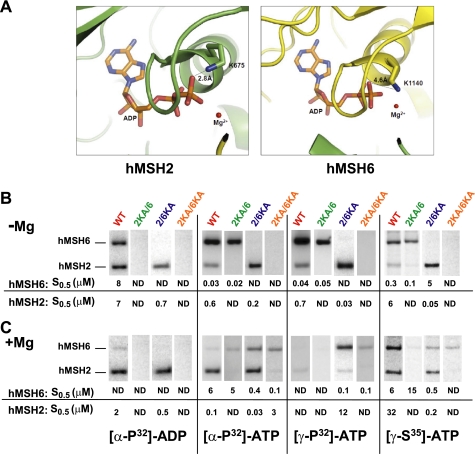
To address the individual contributions of hMSH2 and hMSH6, we constructed Lys to Ala substitution mutations within the highly conserved Walker A nucleotide binding domain (GXXXGK(S/T)) of each subunit (7). This Lys residue occupies slightly different positions in the hMSH2 and hMSH6 subunits (Fig. 1A), where the hMSH2(Lys-675) may form hydrogen bond interactions with the ADP hydrolysis product β-phosphate (Fig. 1A) (23). We purified wild type hMSH2-hMSH6 (WT), hMSH2(K675A)-hMSH6 (2KA/6), hMSH2-hMSH6(K1140A) (2/6KA), and hMSH2(K675A)-hMSH6(K1140A) (2KA/6KA) heterodimers and examined magnesium-dependent adenosine nucleotide cross-linking by the individual MSH subunits (Fig. 1, B and C) (12, 21, 31, 33). A relatively tight range for the S0.5 cross-linking affinity for each subunit at 95% confidence was observed for S0.5 between 0.01 and 10 μm (supplemental Table S1), although it is clear that only large differences in S0.5 should be considered significant. These calculations suggest that the S0.5 cross-linking affinity accurately reflects binding affinity for the individual subunits.
FIGURE 1.
Magnesium affects the affinity of hMSH6 for adenosine nucleotide. A, hMSH2(Lys-675) and hMSH6(Lys-1140) mapped to the human hMSH2-hMSH6 crystal structure (23). hMSH2 is shown in green. hMSH6 is in yellow. The distance between the Lys residue and β-phosphate is indicated. Typical hydrogen bond interaction distances occur between 1 and 3 Å. B and C, representative UV cross-linking reactions containing WT or Lys → Ala mutant heterodimers and 50 or 100 nm ATP, ADP, or ATPγS in the absence (B) or presence (C) of magnesium (5 mm). S0.5 values were calculated from a series of cross-linking reactions at different concentrations of nucleotide as described under “Experimental Procedures.” Each cross-link series was reproduced 2–6 times, and the average S0.5 is shown below. The range of S0.5 at 95% confidence is shown in supplemental Table S1.
In the absence of magnesium (non-hydrolytic conditions), a consistent pattern of [α-32P]ATP, [γ-32P]ATP, and [γ-35S]ATP cross-linking was observed that reliably predicted which subunit contained the Lys → Ala mutation (Fig. 1B). As shown previously, the hMSH6 subunit displayed a 10–100-fold increase in ATP affinity compared with the hMSH2 subunit (Fig. 1B) (21). In contrast, we note nearly equivalent cross-linking of the WT subunits to ADP in the absence of magnesium, although hMSH6 had more than 100-fold less affinity for ADP than ATP. No cross-linking was observed to either subunit of the 2KA/6 mutant protein (Fig. 1B), whereas ADP cross-linked significantly to the wild type hMSH2 subunit within the 2/6KA mutant protein (Fig. 1B).
In the presence of magnesium (hydrolytic conditions), the pattern of WT cross-linking was substantially different (Fig. 1C). A significant decrease in hMSH6 cross-linking affinity was observed with both [α-32P]ATP and [γ-32P]ATP (Fig. 1C). On the contrary, the hMSH2 subunit cross-linked efficiently to [α-32P]ATP but not [γ-32P]ATP. These results are consistent with previous conclusions that the MSH2 and MSH6 subunits bind and hydrolyze ATP and that MSH2 retains the ADP hydrolysis product, whereas MSH6 releases it (21, 29). The decrease in hMSH6 cross-linking affinity reflects either the inability to rebind ATP after catalytic turnover or the inability to trap ATP associated with the heterodimer long enough for stable cross-linking. The wild type subunits within the 2KA/6 and 2/6KA heterodimers largely mimic the properties of the WT protein subunits. Remarkably, the 6KA subunit in both the 2/6KA and 2KA/6KA heterodimers displays nearly equivalent [α-32P]ATP and [γ-32P]ATP cross-linking affinity (Fig. 1C). These results suggest that the 6KA subunit may readily bind ATP in the presence of magnesium but remains deficient for ATP hydrolysis. We also noted that the stable ATP binding by hMSH6 appears to increase the affinity of hMSH2 for adenosine nucleotide ∼10-fold (compare hMSH2 [α-32P]ATP affinity in 2/6KA with or without magnesium; Fig. 1, B and C). Moreover, the corresponding elevated S0.5 for hMSH2 [γ-32P]ATP in the 2/6KA heterodimer demonstrates that the hMSH2 subunit remains capable of ATP hydrolysis and that the retained ADP (see [α-32P]ATP) largely blocks rebinding and cross-linking of [γ-32P]ATP. This hydrolysis effect on the S0.5 is an important consideration when evaluating adenosine nucleotide cross-linking experiments. Finally, the 2KA subunit does not cross-link adenosine nucleotide under any conditions, suggesting that it is incapable of binding and/or retaining ADP/ATP.
[γ-35S]ATP was used to examine ATP binding in the presence of magnesium under poorly hydrolyzable conditions. We verified that in the absence of magnesium [γ-35S]ATP binds with ∼10-fold weaker affinity to hMSH2-hMSH6 than 32P-labeled ATP (Fig. 1B; (21). It is important to be aware that during the 10-min cross-linking in the presence of magnesium, there is likely to be significant but slow hydrolysis of the [γ-35S]phosphate (kcat([γ-35S]ATP) ≤ 0.1 min−1) (34). For example, we observe a decrease in cross-linking affinity for both WT subunits compared with experiments in the absence of magnesium (Fig. 1, B and C). As shown above, hydrolysis may result in an inability to rebind and/or trap [γ-35S]ATP associated with the WT heterodimer, effectively decreasing the cross-linking affinity. Nevertheless, both WT subunits appear capable of binding [γ-35S]ATP simultaneously (Fig. 1C). These studies also confirm that the 6KA mutant subunit may bind [γ-35S]ATP in the presence of magnesium (Fig. 1C). Taking the cross-linking studies as a whole, we conclude that the Walker A lysines are required for ATP hydrolysis by both the hMSH2 and hMSH6 subunits, hMSH2(Lys-675) plays a role in binding/retaining ADP under all conditions, the hMSH6 subunit displays high affinity binding to ATP, and ATP binding by hMSH6 enhances ATP binding by hMSH2. We also observe that hMSH6 releases its ADP hydrolysis product and thereafter appears unable to cross-link adenosine nucleotide.
To further examine the effect of magnesium on ATP binding, we performed a filter-binding assay at 4 °C with [γ-35S]ATP, where any background hydrolysis is largely eliminated (supplemental Fig. S1A and Table 1) (34). In general, the [γ-35S]ATP filter binding (KD) and cross-link affinity (S0.5) in the presence or absence of magnesium appear largely equivalent (compare Fig. 1, B and C, and supplemental Table S1 with Table 1). We observed near saturation binding of the two possible Walker A/B sites by WT in the absence of magnesium (supplemental Fig. S1A). In contrast, the WT protein appears half-saturated in the presence of magnesium with an equivalent KD to the WT protein in the absence of magnesium (supplemental Fig. S1A and Table 1, see Bmax/ET). The 2/6KA mutant protein displays half-saturation independent of magnesium with a KD value nearly equivalent to that of the WT protein (supplemental Fig. S1A and Table 1). Interestingly, the 2KA/6 mutant protein also approaches half-saturation independent of magnesium (supplemental Fig. S1A), yet in the absence of magnesium, it displays a KD nearly equivalent to that of WT and 2KA/6, whereas in the presence of magnesium, the KD is increased 10-fold (supplemental Fig. S1A and Table 1). The saturation and KD of 2KA/6KA in the presence of magnesium mimic the 2KA/6 mutant protein in the presence of magnesium, whereas there is minimal [γ-35S]ATP- binding in the absence of magnesium. These results suggest combinatorial binding of [γ-35S]ATP by both subunits at less than saturating levels that is clearly affected by magnesium. To further determine the individual subunit contributions, we examined [γ-35S]ATP binding at different magnesium concentrations (supplemental Fig. S1B). The uniform binding by the 2/6KA protein across multiple magnesium concentrations is consistent with the cross-linking studies that show hMSH2 saturation at low magnesium that shifts to combinatorial subunit [γ-35S]ATP binding in the presence of magnesium (see Fig. 1, B and C). Similarly, the reduction in [γ-35S]ATP binding by WT and 2KA/6 with increasing concentrations of magnesium is consistent with decreasing hMSH6 [γ-35S]ATP binding as reflected in the cross-linking analysis, whereas the increased [γ-35S]ATP binding by 2KA/6KA reflects the ability of the hMSH6 subunit to bind ATP in the presence of magnesium. Taken as a whole, these results indicate that magnesium dramatically influences cross-linking affinity by MSH subunits that are ultimately manifest in altered equilibrium adenosine nucleotide binding by the MSH heterodimer.
TABLE 1.
ATP binding
| Protein | With magnesium |
Without magnesium |
||
|---|---|---|---|---|
| KD | Bmax/ET | KD | Bmax/ET | |
| μm | μm | |||
| WT | 0.6 | 1.6 | 0.8 | 2.1 |
| 2KA/6 | 6.0 | 0.2 | 0.6 | 1.5 |
| 2/6KA | 0.7 | 2.0 | 1.1 | 1.4 |
| 2KA/6KA | 3.4 | 0.4 | 86.7 | 0.004 |
The hMSH2-Magnesium Complex Controls Steady-state ATPase Activity
In the absence of DNA, the WT and Lys → Ala heterodimers display a low level steady-state ATPase activity. Notably, the 2KA/6 mutant appears consistently 5-fold more active than the ot her heterodimers (supplemental Fig. S2A and Table 2). In pre-steady-state ATPase assays, WT displays an initial rapid burst of hydrolysis followed by a slower linear rate (supplemental Fig. S2B and Table 2). The 2KA/6 mutant displays a nearly identical burst followed by a slow linear rate, indicating that hMSH6 is fully active and can continue to hydrolyze ATP in the absence of nucleotide binding to hMSH2 (supplemental Fig. S2B and Table 2). However, the 2/6KA mutant lacks the initial burst and only displays a slow linear hydrolysis rate (supplemental Fig. S2B and Table 2). These data confirm earlier findings from yeast studies that, in the absence of mismatched DNA, the MSH6 subunit is responsible for a rapid burst of ATP hydrolysis, whereas the MSH2 subunit has a slower ATPase activity (29). The similarity between the WT and the 2/6KA steady-state rates (supplemental Fig. S2A) indicates that WT catalytic turnover is limited by slow hMSH2 activity. This observation, coupled with the cross-linking studies and the high steady-state ATPase rate exhibited by the 2KA/6 mutant, wherein hMSH2 is nucleotide-free, suggests that adenosine nucleotide-bound hMSH2 acts as an allosteric regulator that blocks catalytic turnover by hMSH6 following its initial burst of ATP hydrolysis, most likely by blocking hMSH6 ATP binding.
TABLE 2.
ATPase activities
| Protein | Steady-state ATPase (no DNA) |
Steady-state ATPase (G/T) |
Pre-steady-state ATPase (no DNA) |
|||
|---|---|---|---|---|---|---|
| Km | kcat | Km | kcat | Burst ratea | Steady stateb | |
| μm | min−1 | μm | min−1 | s−1 | s−1 | |
| WT | 43.2 | 0.3 | 61.7 | 19.7 | 0.74 | 0.03 |
| 2KA/6 | 132.7 | 1.6 | 177.8 | 2.6 | 1.02 | 0.04 |
| 2/6KA | 47.5 | 0.3 | 80.6 | 1.4 | NDc | 0.04 |
| 2KA/6KA | 80.7 | 0.2 | 30.5 | 0.1 | ND | ND |
a Data were fit with an exponential equation.
b Data were fit with a linear equation.
c ND, not determinable.
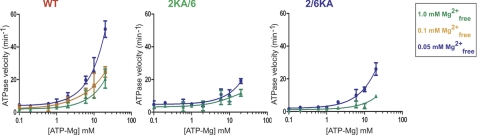
The effect of magnesium on ATP processing by the WT and Lys → Ala heterodimers appeared to indicate that magnesium might also act as a catalytic regulator. To examine the effect of magnesium on hMSH2-hMSH6 steady-state ATPase, we adjusted the magnesium and ATP in the reaction to fix the free magnesium at a wide range of ATP-magnesium concentrations (Fig. 2). If magnesium functions as a catalytic regulator (e.g. by controlling the rate-limiting step of hMSH2(ADP) release and/or rebinding of ATP by hMSH6), then lower free magnesium should reduce protein-bound magnesium at equilibrium, which would then allow unregulated rounds of ATP binding and hydrolysis. We found that the WT ATPase rate increased dramatically at low free magnesium concentrations, ultimately reaching an ATPase velocity ∼50 times greater than in the presence of excess free magnesium (Fig. 2). The 2KA/6 mutant displayed higher basal ATPase activity, consistent with reduced down-regulation of the hMSH6 ATPase by hMSH2, that increased ∼6-fold with lower free magnesium. In contrast, the 2/6KA increased ∼30-fold at low free magnesium concentrations. The simplest interpretation of these results is that low free magnesium enhances release of ADP by hMSH2, which we have previously demonstrated provides the largest barrier to MSH ATPase activity (8, 29). These observations contrast with studies of E. coli MutS that suggest that elevated magnesium stimulates ADP release (20), although the differential effects of magnesium on the MutS homodimer subunits may not be discernible. Our findings are comparable with those of kinesin and G-proteins, where reduced magnesium stimulates nucleotide diphosphate release (30, 35, 36). Because hMSH6 does not retain ADP following hydrolysis and because the 2KA mutant does not bind ATP under any conditions (see Fig. 1C), the increase in 2KA/6 ATPase at low free magnesium suggests that magnesium bound/retained by one or both of the hMSH2 and hMSH6 subunits may act as at least a partial allosteric regulator of the hMSH6 ATPase.
FIGURE 2.
The steady-state ATPase activity of hMSH2-hMSH6 is inhibited by excess free magnesium. Steady-state ATP hydrolysis in the absence of DNA was performed at various concentrations of ATP and at 1.0 mm (green), 0.1 mm (yellow), and 0.05 mm (blue) free magnesium as indicated (36). Data points represent an average of three independent experiments with S.D. (error bars) for each point (sometimes within the symbol).
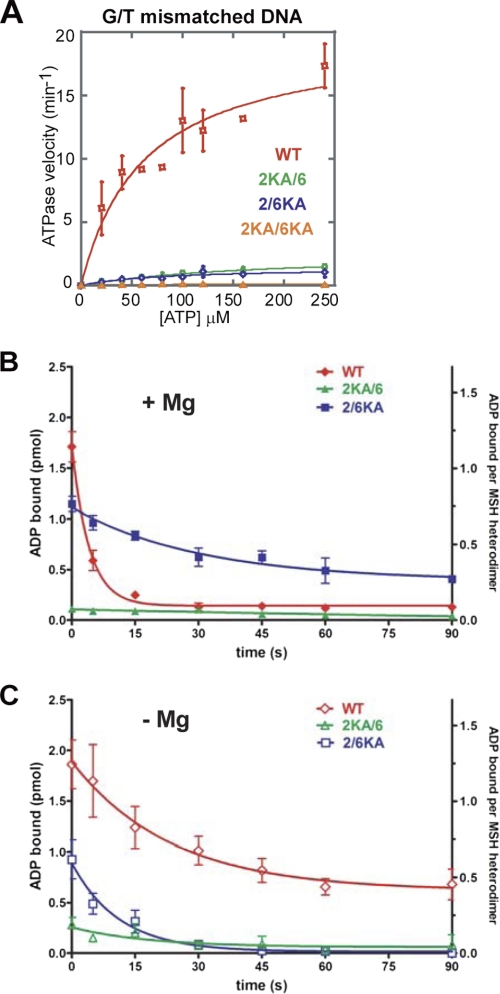
To further test the hypothesis that magnesium regulates ADP release by hMSH2, we examined the mismatch-independent release of ADP from hMSH2-hMSH6 following the addition of excess ATP in the presence or absence of magnesium. The WT and 2/6KA proteins bound ADP in the presence of magnesium, whereas the 2KA/6 mutant was severely deficient (supplemental Fig. S3; see results at time 0). These results are consistent with preferential ADP binding by the hMSH2 subunit, as indicated by the cross-linking studies (Fig. 1C). In the absence of magnesium, there was an increase in ADP binding by the WT protein that reflects the increased binding by the hMSH6 subunit (supplemental Fig. S3). ADP binding to WT in the presence of magnesium is quite stable (supplemental Fig. S3 and Table 3). However, in the absence of magnesium, we observe faster and nearly complete release of ADP (koff = 0.016 s−1 compared with 0.007 s−1) (Table 3). ADP release from the 2/6KA mutant also increases in the absence of magnesium (koff = 0.014 s−1 compared with 0.006 s−1; Table 3). These results combined with the free magnesium ATPase studies suggest that magnesium controls the release of ADP by hMSH2 and subsequent catalytic turnover.
TABLE 3.
Nucleotide exchange
| Protein | No DNA |
G/T |
||
|---|---|---|---|---|
| koff with magnesium | koff without magnesium | koff with magnesium | koff without magnesium | |
| s−1 | s−1 | s−1 | s−1 | |
| WT | 0.007 | 0.016 | 0.24 | 0.04 |
| 2KA/6 | NDa | ND | ND | ND |
| 2/6KA | 0.006 | 0.014 | 0.03 | 0.09 |
| 2KA/6KA | NDb | ND | ND | ND |
a ND, not determinable.
b ND, not done.
Mismatch-provoked ATP Processing
We examined the subunit contribution to the mismatch-stimulated ATPase activity of hMSH2-hMSH6 (Fig. 3A and Table 2). As reported previously (8), mismatched DNA significantly stimulates the WT ATPase activity (kcat = 0.3 min−1, kcat(DNA) = 19.7 min−1; Fig. 3A and Table 2). However, mismatched DNA failed to stimulate the ATPase activity of 2KA/6 and 2/6KA mutant proteins (Fig. 3A and Table 2). These results suggest that the ability to appropriately bind and/or hydrolyze ATP by both subunits is necessary for the efficient mismatch-stimulated ATPase. We next examined mismatch-dependent ADP → ATP exchange by WT and Lys → Ala mutants (Fig. 3B and Table 3). In the presence of magnesium, mismatched DNA provoked rapid ADP → ATP exchange by the WT protein (koff = 0.24 s−1). In contrast, the 2/6KA mutant performed ADP → ATP exchange ∼3-fold more slowly (koff = 0.09 s−1), and the 2KA/6 mutant protein did not appreciably bind or exchange ADP (Fig. 3B and Table 3). Our cross-linking studies show that the hMSH6 subunit is nucleotide-free in the WT protein, whereas the hMSH6 subunit in the 2/6KA mutant binds ATP (see Fig. 1C). These observations suggested that an ATP-bound hMSH6 subunit might inhibit the release of ADP by hMSH2. To test this hypothesis, we performed a similar ADP → ATP exchange experiment in the absence of magnesium, where the hMSH6 subunit within the 2/6KA mutant does not bind ATP, whereas the hMSH6 subunit of the WT protein readily binds ATP (see Fig. 1B). We found that ADP is completely released from the 2/6KA mutant, whereas ADP release by the WT protein is dramatically reduced (Fig. 3C). These results are consistent with the conclusion that hMSH6 must be nucleotide-free prior to interaction with a mismatch for efficient ADP → ATP exchange by hMSH2 to occur.
FIGURE 3.
Mismatch-provoked hydrolysis and ADP → ATP exchange of hMSH2-hMSH6. A, steady-state ATP hydrolysis in the presence of a 41-bp oligonucleotide containing a central G/T mismatch was performed using 200 nm WT, 2KA/6, 2/6KA, or 2KA/6KA protein and [γ-32P]ATP. Following incubation at 37 °C for 30 min, the amount of γ-32P released was determined as described previously (8). Data points represent an average of three independent experiments with S.D. and were fit with the Michaelis-Menten equation. B and C, WT and Lys → Ala mutants were prebound to [3H]ADP. The release of ADP upon the addition of a 41-bp G/T mismatched DNA (100 nm) and ATP (1 mm) in the presence (B) or absence (C) of magnesium (5 mm) was measured using filter binding as described previously (8). Data points represent the average of three independent experiments with S.D. (error bars) and were fit to a single exponential decay.
hMSH2 and Magnesium Control the Formation of a Stable Sliding Clamp
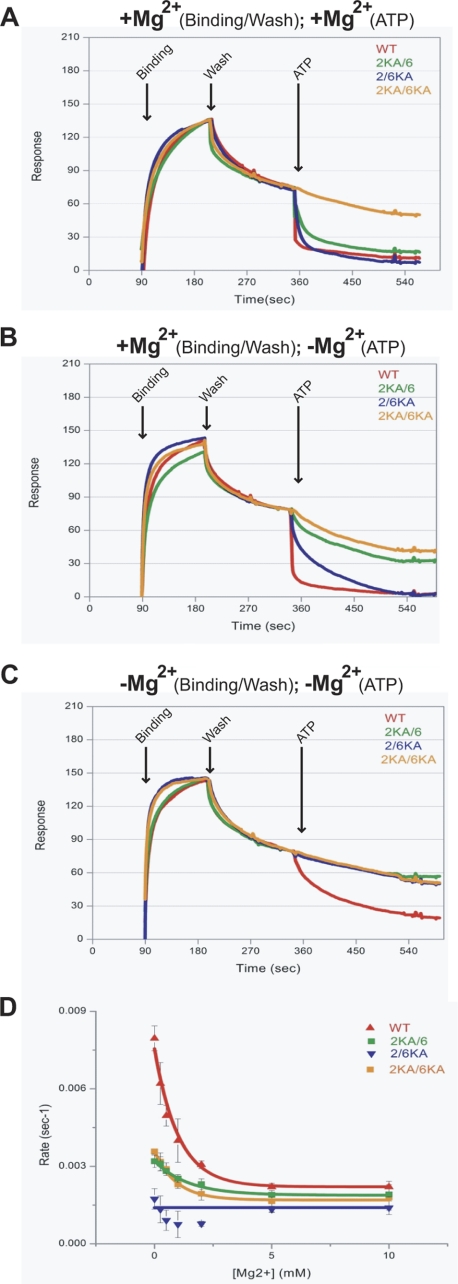
Mismatch-provoked ADP → ATP exchange induces the formation of an hMSH2-hMSH6 sliding clamp that is capable of bidirectional diffusion along DNA (11, 21). To determine whether the distinct adenosine nucleotide processing properties of hMSH2 and hMSH6 are important for clamp formation, we employed surface plasmon resonance and examined ATP-dependent dissociation of the heterodimers from mismatched DNA (Fig. 4A and supplemental Table S2). Importantly, the WT and mutant proteins display similar kon and koff, consistent with nearly identical mismatch binding/dissociation (KD; Fig. 4A and supplemental Table S2). In the presence of magnesium, the addition of ATP resulted in rapid dissociation of the WT protein consistent with the formation of a sliding clamp that is released from the open end of the DNA (Fig. 4A and supplemental Table S2). In contrast, the 2KA/6KA mutant fails to dissociate and displays a slow koff expected for equilibrium mismatch dissociation. This result suggests that the 2KA/6KA is incapable of forming a sliding clamp. The 2KA/6 and the 2/6KA mutant proteins were capable of dissociating from the mismatch, although at significantly slower rates compared with WT (Fig. 4A and supplemental Table S2).
FIGURE 4.
Efficient sliding clamp formation requires ATP binding by both hMSH2 and hMSH6 subunits. Shown are surface plasmon resonance (Biacore) studies of WT and Lys → Ala heterodimer dissociation from a mismatched oligonucleotide in the presence of ATP (250 μm). A, protein was bound to the mismatch, washed, and dissociated from the mismatch with ATP in the presence of magnesium (10 mm). B, protein was bound to the mismatch and washed in the presence of magnesium (10 mm) and dissociated from the mismatch with ATP in the absence of magnesium. C, protein was bound, washed, and dissociated from the mismatch with ATP in the absence of magnesium. D, rates of ATP-induced dissociation of WT and Lys → Ala mutant protein from a double-blocked end mismatched oligonucleotide in the presence of increasing concentrations of magnesium.
We then examined the effect of magnesium on ATP-provoked hMSH2-hMSH6 mismatch dissociation (Fig. 4, B and C). When magnesium was included during binding but excluded during the addition of ATP, the WT protein again dissociated rapidly from DNA, the 2/6KA mutant dissociated more slowly than in the presence of magnesium, and the 2KA/6 dissociation was inhibited severely enough that it resembled the 2KA/6KA. We interpret these results as follows. When magnesium is present at any stage in the surface plasmon resonance system (Fig. 4, A and B), the 6KA subunit retains at least some ATP binding activity, and the 2/6KA can attain a sliding clamp form capable of dissociating from mismatched DNA. In contrast, the 2KA subunit does not retain sufficient ATP binding affinity when magnesium is excluded during the ATP dissociation step, and the 2KA/6 heterodimer exhibits seriously defective sliding clamp formation. To further test these conclusions, we omitted magnesium from both the binding and dissociation steps to produce a condition where all combinations of the Lys → Ala heterodimers have lost their affinity of ATP (see Fig. 1B). The absence of magnesium did not affect mismatch binding for any of the proteins (Fig. 4C). However, ATP-provoked dissociation by WT occurred more slowly, and none of the Lys → Ala mutant proteins dissociated from the mismatched DNA (Fig. 4C). In general, the ATP-dependent dissociation from mismatched DNA appeared to mimic the magnesium-dependent ability of both MSH subunits to bind and cross-link ATP (compare Fig. 4, A–C, with Fig. 1, B and C). These results are consistent with the conclusion that both the hMSH2 and hMSH6 subunits must be bound by ATP to efficiently dissociate from the mismatched DNA.
To determine whether the proteins formed sliding clamps during ATP-provoked dissociation, we blocked the open end of the mismatched DNA with digoxigenin-anti-digoxigenin (Fig. 4D) (37). These experiments were performed as for Fig. 4B except that the magnesium concentration was varied with the addition of ATP. At high concentrations of magnesium (5 mm), the WT protein is stable for ∼8 min on the double-blocked end mismatched DNA (koff = 0.002 s−1). This result indicates that the WT protein sliding clamps are stably trapped on the double-blocked end mismatched DNA (11). However, as magnesium concentrations were decreased, the koff increased significantly (koff = 0.008 s−1), consistent with direct dissociation of the WT protein from the double-blocked end mismatched DNA (Fig. 4D). We observe little change in dissociation kinetics for the mutant proteins (Fig. 4D). Taken together with the cross-linking data, these results suggest that that magnesium plays an essential role in coordinating the appropriate ATP processing that ultimately results in stable sliding clamps.
DISCUSSION
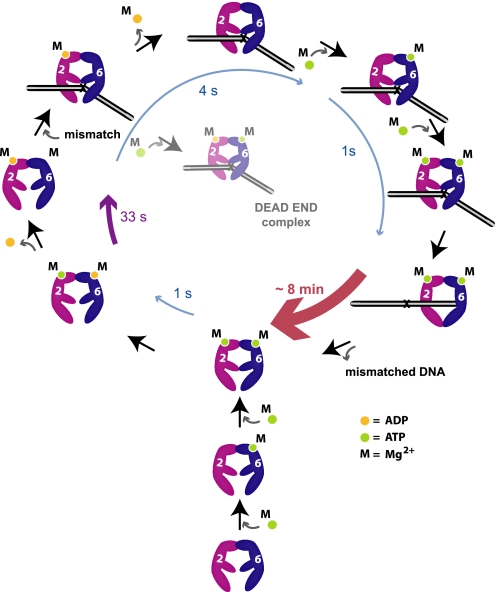
The hMSH2-hMSH6 ATPase is limited by ADP release, which is stimulated by mismatch binding (8, 30). Subsequent ATP binding by hMSH2-hMSH6 produces a sliding clamp (8, 30). Several models have been proposed for MMR (16). Our studies detail the kinetic steps associated with ADP/ATP processing by the individual subunits of human hMSH2-hMSH6 (Fig. 5) and appear uniquely consistent with the molecular switch model for MMR (Fig. 5) (8, 11).
FIGURE 5.
An illustration of the kinetic events associated with hMSH2 and hMSH6 subunits during ADP/ATP processing for MMR. Bottom and lower left of cycle, hMSH6 rapidly binds ATP (green dot) followed by rapid binding of ATP to hMSH2. hMSH6 rapidly hydrolyzes ATP and releases the resultant ADP (yellow dot). hMSH2 hydrolyzes ATP more slowly yet retains ADP. This process results in the major steady-state species hMSH2(ADP)-hMSH6(apo). The likely disposition of magnesium (M) for each subunit is shown. Remaining cycle, upon mismatch interaction, hMSH2 rapidly releases ADP (4 s). Under conditions where hMSH6 is bound to ATP at the mismatch prior to ADP release, a dead-end complex is formed that cannot undergo nucleotide exchange. hMSH6 rapidly binds ATP, which enhances ATP binding by hMSH2 (1 s). Binding of ATP by both hMSH2 and hMSH6 results in the formation of a stable sliding clamp (∼8 min) that dissociates from the mismatch to execute the downstream steps of MMR. Dissociation from the DNA containing the mismatch and ATP hydrolysis completes the hMSH2-hMSH6 ATPase cycle.
Nucleotide-free hMSH2-hMSH6 is likely to be rare in the absence of mismatched DNA because the MSH6 subunit binds and hydrolyzes ATP rapidly (Fig. 5) (21). Moreover, our cross-linking studies suggest that ATP-bound hMSH6 significantly stimulates ATP binding by MSH2 (Fig. 1C). Pre-steady-state ATPase analysis indicates that hMSH6 hydrolyzes ATP rapidly (1 s−1), whereas hMSH2 hydrolyzes ATP more slowly (0.03 s−1), similar to yeast Msh2-Msh6 (29). As observed with the yeast proteins, we find that the hMSH6-ADP complex is short lived (21). In contrast, hMSH2 remains tightly associated with ADP (hMSH2(ADP); Fig. 5).
The steady state ATPase of hMSH2-hMSH6 in solution matches the slow rate of hMSH2 hydrolysis. These results are consistent with the conclusion that ADP release by hMSH2 also limits the hMSH6 ATPase. However, the kinetic difference in ATP hydrolysis by hMSH2 (0.03 s−1) and hMSH6 (1 s−1) would seem to suggest that the hMSH6 subunit might hydrolyze >30 ATP before the hMSH2(ADP) complex is established. The simplest solution for this conundrum is to propose that any adenosine nucleotide-bound form of hMSH2 is capable of suppressing hMSH6 ATP hydrolysis. Indeed, the significant increase in the hMSH2 cross-linking affinity (S0.5) when hMSH6 is constrained to an ATP-bound state (see 2/6KA cross-linking studies) suggests that both subunits are likely to bind ATP prior to hydrolysis by hMSH6. A subsequent two-step hydrolysis process that includes release of the ADP hydrolysis product by hMSH6 would ultimately result in hMSH2(ADP) and hMSH6 in the apo state, the predominant form observed in cross-linking studies here and by Mazur et al. (21).
The observation that ATP hydrolysis substantially increases with reduced free magnesium suggests that magnesium plays an essential role in maintaining the hMSH2(ADP) complex. A similar mechanism has been reported for the monomeric kinesin K1F1A, whose ATPase activity is also inhibited by excess free magnesium (36). Control of nucleotide diphosphate (NDP) release by magnesium mimics features of G protein nucleotide exchange, as we previously proposed (30). Our observations contrast studies with the E. coli MutS that suggest that magnesium enhances ADP release (20). The modest increase in 2KA/6 ATPase activity at low free magnesium appears to suggest that the hMSH6 subunit may also retain magnesium following catalytic turnover that then inhibits subsequent catalytic cycles. Together, these observations underline a central role for magnesium in the control of hMSH2-hMSH6 ATPase activity (Fig. 5).
Following mismatch binding, ADP release takes ∼4 s and probably involves disruption of magnesium at the Walker A/B site (Table 3 and Fig. 5) (36). Release of ADP and magnesium by hMSH2 relieves the nucleotide binding inhibition of hMSH6, which rapidly binds ATP. This order of events appears different from yeast Msh2-Msh6, where Msh6 is thought to bind ATP after binding to the mismatch, leading to the release of ADP by Msh2 (21). As noted above, our results indicate that under conditions where hMSH6 binds and cross-links stably to ATP, the nucleotide cross-linking affinity of hMSH2 increases (see 2/6KA in Fig. 1C). We also noted that the hMSH2 subunit within the 2/6KA heterodimer hydrolyzes ATP and remains ADP-bound (see 2/6KA in Fig. 1C). However, mismatch-provoked ADP release by 2/6KA when the mutant hMSH6 subunit may bind but not hydrolyze ATP is significantly inhibited and probably forms a dead-end complex (see 2/6KA, +Mg in Figs. 1C and 3B; see Fig. 5). In contrast, ADP release by hMSH2 is relatively rapid and complete when the hMSH6 subunit is incapable of binding adenosine nucleotide (see 2/6KA, −Mg in Figs. 1B and 3C) or is nucleotide-free (see WT, +Mg in Fig. 1C and Fig. 3B). Together, these results suggest that hMSH6 must be in the apo state for efficient mismatch-dependent release of ADP by hMSH2 (Fig. 5).
ATP binding by hMSH2-hMSH6 results in dissociation from the mismatch and the formation of a sliding clamp that takes ∼1 s (Fig. 4A, supplemental Table S2, and Fig. 5) (11, 38). The process of mismatch binding, ADP → ATP exchange, and sliding clamp formation (5 s) appears remarkably similar to T. aquaticus MutS kinetics (3 s) deduced by single-molecule FRET studies (Fig. 5) (37).
The efficient formation of a stable sliding clamp requires ATP binding by both hMSH2 and hMSH6 subunits (Fig. 4, A–C). These observations are consistent with similar studies of the yeast Msh2-Msh6 (21) and E. coli MutS (20). Because hydrolysis is suppressed (29), the hMSH2-hMSH6 sliding clamp remains in a dual ATP-bound state as well as stably associated with the mismatched DNA for ∼8 min, a length of time nearly equivalent to a complete MMR excision reaction (29). This kinetic stability appears remarkably similar to the ∼10-min stability of T. aquaticus MutS sliding clamps on mismatched DNA (37). Moreover, magnesium plays an essential role in maintaining the stability of the hMSH2-hMSH6 sliding clamp because its absence increases direct dissociation ∼5-fold. Finally, the central role of the hMSH2(magnesium-ADP) complex in the regulation of hMSH2-hMSH6 functions may be an additional factor that explains the prevalence of MSH2 mutations in Lynch syndrome or hereditary nonpolyposis colorectal cancer (28).
Supplementary Material
Acknowledgments
We thank Ravindra Amunugama for assistance with the preparation and analysis of structural data shown in Fig. 1A and Sarah Javaid for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants CA115783 (to C. D. H.) and CA67007 (to R. F.). This work was also supported by National Science Foundation Grant MCB 1022203 (to M. M. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- MMR
- mismatch repair
- IDL
- insertion-deletion loop
- MSH
- MutS homolog(s)
- hMSH2 and hMSH6
- human MSH2 and MSH6, respectively
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- 6KA
- hMSH6(K1140A)
- 2KA
- hMSH2(K675A)
- 2KA/6
- hMSH2(K675A)-hMSH6
- 2/6KA
- hMSH2-hMSH6(K1140A)
- 2KA/6KA
- hMSH2(K675A)-hMSH6(K1140A)
- HDCC
- N-(2-(1-maleimidyl)ethyl)-7-(diethylamino)coumarin-3-carboxamide
- Bmax
- maximum binding saturation
- ET
- total enzyme.
REFERENCES
- 1. de la Chapelle A. (2005) Fam. Cancer 4, 233–237 [DOI] [PubMed] [Google Scholar]
- 2. Peltomäki P. (2005) Fam. Cancer 4, 227–232 [DOI] [PubMed] [Google Scholar]
- 3. Kolodner R. D., Marsischky G. T. (1999) Curr. Opin. Genet. Dev. 9, 89–96 [DOI] [PubMed] [Google Scholar]
- 4. Jiricny J. (2006) Nat. Rev. Mol. Cell. Biol. 7, 335–346 [DOI] [PubMed] [Google Scholar]
- 5. Acharya S., Wilson T., Gradia S., Kane M. F., Guerrette S., Marsischky G. T., Kolodner R., Fishel R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13629–13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haber L. T., Pang P. P., Sobell D. I., Mankovich J. A., Walker G. C. (1988) J. Bacteriol. 170, 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haber L. T., Walker G. C. (1991) EMBO J. 10, 2707–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gradia S., Acharya S., Fishel R. (1997) Cell 91, 995–1005 [DOI] [PubMed] [Google Scholar]
- 9. Blackwell L. J., Bjornson K. P., Modrich P. (1998) J. Biol. Chem. 273, 32049–32054 [DOI] [PubMed] [Google Scholar]
- 10. Alani E., Sokolsky T., Studamire B., Miret J. J., Lahue R. S. (1997) Mol. Cell Biol. 17, 2436–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gradia S., Subramanian D., Wilson T., Acharya S., Makhov A., Griffith J., Fishel R. (1999) Mol. Cell 3, 255–261 [DOI] [PubMed] [Google Scholar]
- 12. Heinen C. D., Wilson T., Mazurek A., Berardini M., Butz C., Fishel R. (2002) Cancer Cell 1, 469–478 [DOI] [PubMed] [Google Scholar]
- 13. Jiang J., Bai L., Surtees J. A., Gemici Z., Wang M. D., Alani E. (2005) Mol. Cell 20, 771–781 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., Li G. M. (2005) Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 15. Blackwell L. J., Martik D., Bjornson K. P., Bjornson E. S., Modrich P. (1998) J. Biol. Chem. 273, 32055–32062 [DOI] [PubMed] [Google Scholar]
- 16. Kolodner R. D., Mendillo M. L., Putnam C. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12953–12954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Modrich P., Lahue R. (1996) Annu. Rev. Biochem. 65, 101–133 [DOI] [PubMed] [Google Scholar]
- 18. Antony E., Hingorani M. M. (2004) Biochemistry 43, 13115–13128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Antony E., Khubchandani S., Chen S., Hingorani M. M. (2006) DNA Repair 5, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lebbink J. H., Fish A., Reumer A., Natrajan G., Winterwerp H. H., Sixma T. K. (2010) J. Biol. Chem. 285, 13131–13141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mazur D. J., Mendillo M. L., Kolodner R. D. (2006) Mol. Cell 22, 39–49 [DOI] [PubMed] [Google Scholar]
- 22. Lamers M. H., Perrakis A., Enzlin J. H., Winterwerp H. H., de Wind N., Sixma T. K. (2000) Nature 407, 711–717 [DOI] [PubMed] [Google Scholar]
- 23. Warren J. J., Pohlhaus T. J., Changela A., Iyer R. R., Modrich P. L., Beese L. S. (2007) Mol. Cell 26, 579–592 [DOI] [PubMed] [Google Scholar]
- 24. Obmolova G., Ban C., Hsieh P., Yang W. (2000) Nature 407, 703–710 [DOI] [PubMed] [Google Scholar]
- 25. Antony E., Hingorani M. M. (2003) Biochemistry 42, 7682–7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bjornson K. P., Modrich P. (2003) J. Biol. Chem. 278, 18557–18562 [DOI] [PubMed] [Google Scholar]
- 27. Drotschmann K., Yang W., Kunkel T. A. (2002) DNA Repair 1, 743–753 [DOI] [PubMed] [Google Scholar]
- 28. Peltomäki P., Vasen H. F. (1997) Gastroenterology 113, 1146–1158 [DOI] [PubMed] [Google Scholar]
- 29. Wang H., Hays J. B. (2002) J. Biol. Chem. 277, 26136–26142 [DOI] [PubMed] [Google Scholar]
- 30. Fishel R. (1998) Genes Dev. 12, 2096–2101 [DOI] [PubMed] [Google Scholar]
- 31. Cyr J. L., Heinen C. D. (2008) J. Biol. Chem. 283, 31641–31648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jacobs-Palmer E., Hingorani M. M. (2007) J. Mol. Biol. 366, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iaccarino I., Marra G., Palombo F., Jiricny J. (1998) EMBO J. 17, 2677–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Javaid S., Manohar M., Punja N., Mooney A., Ottesen J. J., Poirier M. G., Fishel R. (2009) Mol. Cell. 36, 1086–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng J. Q., Jiang W., Hackney D. D. (1998) Biochemistry 37, 5288–5295 [DOI] [PubMed] [Google Scholar]
- 36. Nitta R., Okada Y., Hirokawa N. (2008) Nat. Struct. Mol. Biol. 15, 1067–1075 [DOI] [PubMed] [Google Scholar]
- 37. Jeong C., Cho W. K., Song K. M., Cook C., Yoon T. Y., Ban C., Fishel R., Lee J. B. (2011) Nat. Struct. Mol. Biol. 18, 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mendillo M. L., Mazur D. J., Kolodner R. D. (2005) J. Biol. Chem. 280, 22245–22257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.