Background: Non-steroidal anti-inflammatory drugs acting as γ-secretase modulators (GSMs) modulate γ-cleavage of amyloid precursor protein (APP).
Results: Modulation of γ-cleavage by GSMs in chimeric APP constructs showed that the transmembrane domain plays a pivotal role in determining drug sensitivity.
Conclusion: Modulatory effects on γ-cleavage appeared to be substrate sequence-specific.
Significance: γ-Secretase substrates may have differential response to GSMs.
Keywords: Amyloid, Amyloid Precursor Protein, Cd44, Notch, Presenilin
Abstract
A subset of non-steroidal anti-inflammatory drugs modulates the γ cleavage site in the amyloid precursor protein (APP) to selectively reduce production of Aβ42. It is unclear precisely how these γ-secretase modulators (GSMs) act to preferentially spare Aβ40 production as well as Notch processing and signaling. In an effort to determine the substrate requirements in NSAID/GSM activity, we determined the effects of sulindac sulfide and flurbiprofen on γ-cleavage of artificial constructs containing several γ-secretase substrates. Using FLAG-tagged constructs that expressed extracellularly truncated APP, Notch-1, or CD44, we found that these substrates have different sensitivities to sulindac sulfide. γ-Secretase cleavage of APP was altered by sulindac sulfide, but CD44 and Notch-1 were either insensitive or only minimally altered by this compound. Using chimeric APP constructs, we observed that the transmembrane domain (TMD) of APP played a pivotal role in determining drug sensitivity. Substituting the APP TMD with that of APLP2 retained the sensitivity to γ-cleavage modulation, but replacing TMDs from Notch-1 or ErbB4 rendered the resultant molecules insensitive to drug treatment. Specifically, the GXXXG motif within APP appeared to be critical to GSM activity. Consequently, the modulatory effects on γ-cleavage appears to be substrate-dependent. We hypothesize that the substrate present in the γ-secretase complex influences the conformation of the complex so that the binding site of GSMs is either stabilized or less favorable to influence the cleavage of the respective substrates.
Introduction
Alzheimer disease (AD),4 the most common neurodegenerative disease, is characterized by deposition of amyloid β-peptides (Aβ) in senile plaques in brain that are generated from the sequential cleavage of APP. First β-secretase, β-amyloid precursor protein site cleaving enzyme (BACE), cleaves APP in the extracellular domain and then γ-secretase cleavage (herein referred to as γ-cleavage) releases Aβ from the cell. This proteolytic cleavage occurs near the center of the TMD and is carried out by a protein complex that contains presenilin (PS), nicastrin, Aph-1, and Pen-2. γ-Secretase cleavage is labile, yielding Aβs with varying COOH termini, resulting in peptides from 34–43 amino acids long. Aβ42 is toxic to cells, aggregates rapidly, and is hypothesized to play a role in initiating AD pathogenesis. Inherited mutations in APP or PS that are thought to be causative in early onset AD lead to increased generation of Aβ and especially an increase in the ratio of Aβ42Aβ40 peptides that are generated.
There are currently no effective treatments for AD, and no therapy is available to prevent or delay its onset. One promising discovery was that certain NSAIDs reduce production of the Aβ42 isoform both in vitro and in vivo (1). Epidemiologic evidence suggests that some of these same NSAIDs reduce the risk of developing AD (2). Although the anti-inflammatory activity of these compounds may play a protective role, reducing Aβ42 generation could also slow the onset of disease symptoms. Chronic treatment of animals with NSAIDs led to significant reduction of amyloid pathology in the brain and improved memory function with a tight correlation between Aβ42-lowering activity of NSAIDs in cultured cells and in vivo efficacy (3–8). These studies suggested that NSAIDs could help delay or prevent AD by decreasing the production of the amyloidogenic Aβ42 species. Unlike other γ-secretase inhibitors, these NSAIDs do not block enzymatic activity. Rather, Aβ42 production is reduced, as shorter Aβ peptides such as Aβ37 and Aβ38 are elevated, with no changes in overall APP processing, indicating a modulation of γ-cleavage sites (1, 9–12). As a result, compounds with these general activities are now coined γ-secretase modulators (GSMs), which are heterogeneous in structure. Conversely, we have identified GSMs that have the opposite effect, namely increasing Aβ42 production (13). The ability of GSMs to reduce Aβ42 generation is altered by mutations in PS and can be recapitulated in highly purified preparations of human γ-secretase (14–16), supporting the hypothesis that this NSAID activity occurs within the PS-containing γ-secretase complex.
APP is only one of a burgeoning list of type 1 transmembrane proteins, now numbering over 80, that is cleaved by the PS-containing secretase complex (17). Other notable substrates include APP-like proteins (APLPs), Notch (18) and its ligands (19), ErbB4 (20), CD44 (21), p75 neurotrophin receptor (22), and LDL receptor-related protein (23). Thus, AD therapies based on non-selective γ-secretase inhibition may alter the cleavage of numerous substrates. Secretion of γ-cleaved peptides, analogous to Aβ, have been described from CD44 and Notch-1 (24, 25). Mass spectrometry of these peptides indicates similar heterogeneity in the C termini in their corresponding γ-cleavage sites. Whether these Aβ-like fragments have any physiological or pathophysiological function is not yet known. The lack of substrate specificity and variable cleavage sites that characterize γ-secretase activity suggest that the enzyme complex has a flexible structure.
As opposed to classic protease inhibitors, the remarkable feature about GSMs is that they shift cleavage with minimal effects on overall rate of cleavage, a property that likely explains the normal generation of Notch intracellular domain (NICD) and Amyloid intracellular domain (AICD) from Notch and APP, respectively. How GSMs accomplish this is an area of active investigation. GSMs can alter the conformation of PS-containing protease complexes and act non-competitively with peptidomimetic γ-secretase inhibitors (26–31). We recently reported that low potency GSMs and inverse GSMs can bind to APP and that mutation of the putative GSM binding site in APP dramatically reduced sensitivity to GSM activity (15). To encompass the recent reports that other GSMs may work differently and target the γ-secretase complex directly (30, 32), we designated the former as substrate-targeting GSMs (stGSMs) versus those that do not. Whether targeting γ-secretase directly, the substrate, or the interface of the substrate and γ-secretase complex, it is possible that the conformation of the γ-secretase complex itself might influence GSM activity. Indeed, the effect of GSM on γ-secretase cleavage can be modulated by changes to both the substrate or the γ-secretase complex, as shown by cells expressing either APP or PS1 mutations (11, 15, 28, 29). To investigate more thoroughly whether substrates determine the ability of GSMs to modulate γ-secretase activity, we examined the effect of two NSAIDs that show Aβ42-lowering GSM activity, sulindac sulfide and flurbiprofen, on γ-cleavage of CD44, Notch, and APLP2 and found that the TMD sequences have a marked influence on the ability of GSMs to modulate γ-cleavage.
EXPERIMENTAL PROCEDURES
Reagents
All compounds and reagents were from commercial venders as follows: Aβ1–22 (custom synthesis, American Peptide, Inc.); anti-mouse IgG-agarose beads (American Qualex); sulindac sulfide (Biomol); flurbiprofen (Cayman Chemical Co.); complete protease inhibitor pellet (Roche); and α-cyanohydroxycinnamic acid, M2-FLAG antibody, trifluoroacetic acid, acetonitrile, and phosphoramidon (Sigma-Aldrich). Monoclonal antibodies included 82E1 (IBL) specific to the cleaved N terminus of Aβ, Ab9 recognizing the N-terminal region of Aβ, and C-terminal specific antibodies to Aβ42 (2.1.3) and Aβ40 (13.1.1) (33). Synthetic APP-APLP2 Aβ-like peptides were custom-synthesized by Peptide 2.0, Inc. (Chantilly, VA).
cDNA Constructs
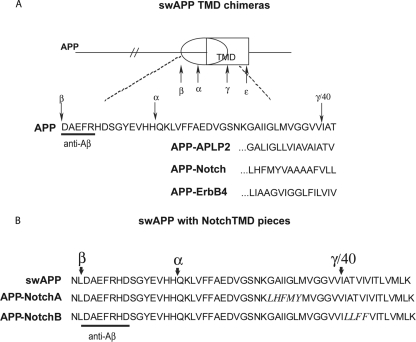
The FLAG epitope-tagged, extracellulary truncated Notch-1 construct (F-NEXT) and FLAG epitope-tagged extracellulary truncated CD44 construct (F-CD44ΔE) have been described previously (24, 25). In addition, we generated a FLAG epitope-tagged, extracellularly truncated APP construct (F-APPC83) by inserting the cDNA for the 83 C-terminal amino acids of APP into the HindIII and XhoI sites of pSecTag2HygB (Invitrogen). cDNAs encoding the APP-APLP2 and APP-Notch-1 chimera where the APP TMD sequences were substituted by the APLP2 and Notch TMD sequences, respectively, have been described previously (34). APP-ErbB4 was initially constructed by substitution of the ErbB4 TMD into wild-type APP695. These constructs were subcloned into Swedish APP695 (swAPP) in the retroviral vectors pLPCX or pLHCX (Clontech) to facilitate generation of stable cell lines. Schematics of these epitope-tagged and swAPP-chimeric constructs are shown in Fig. 2 and supplemental Fig. 1. Retroviral expression vectors for APP-Notch TMD-A and APP-Notch TMD-B were generated by site-directed mutagenesis with the Stratagene QuikChange PCR kit according to the manufacturer's instructions. Primers were designed to insert the appropriate sequences of Notch-1 and excise the corresponding sequence of APP. All DNA constructs were sequenced to assure accuracy.
FIGURE 2.
Transmembrane domain substitutions in APP. A, schematic diagram and partial sequences for chimeric molecules based on swAPP. The TMD of APP was removed, and TMDs from other γ-secretase substrates, APLP2, Notch-1, and ErbB4 were inserted. B, sequences of the Aβ-like region of chimeras composed of swAPP with substitutions of short segments from the Notch-1 TMD. Notch-1 sequences are shown in italics.
Cell Lines and Culture Conditions
HEK293 cells stably transfected with PS-1 wild-type, swAPP, and either F-NEXT or F-CD44ΔE cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml) in a 37 °C, humidified incubator with 5% CO2 with 400 μg/ml G418, 100 μg/ml hygomycin B, and 100 μg/ml Zeocin. HEK293 cells stably expressing F-APPC83 or F-NEXT were obtained following retroviral infection and selection for antibiotic resistance. Individual clones and pooled transfectants yielded similar expression levels and responses to GSM treatments. Therefore, to avoid clonal anomalies, only transfected pools were analyzed. CHO cells were grown in modification of Eagle's medium α with 10% serum, penicillin, and streptomycin as above. To generate stable lines, CHO cells were infected with retroviruses encoding APP-APLP2, APP-Notch-1, APP-ErbB4, APP-Notch TMD-A, or APP-Notch TMD-B and selected with puromycin or hygromycin B as appropriate and tested as pooled transfectants as above.
Immunoprecipitation and Mass Spectrometry MALDI-TOF
MALDI-TOF was performed on peptides immunoprecipitated from conditioned medium as described (35), with the following modifications. Complete protease inhibitor and phosphoramidon were added to collected media, and cellular debris was removed by centrifugation. For analysis of Aβ and chimeric Aβ peptides, a synthetic Aβ1–22 peptide was added to serve as an immunoprecipitation control and molecular weight standard. Immunoprecipitation was with anti-mouse IgG-agarose beads and a monoclonal antibody against Aβ1-x (Ab9 (13). For FLAG-tagged constructs, M2 anti-FLAG antibody was used for immunoprecipitation. The immunoprecipitated complexes were washed twice with Nonidet P40 buffer (50 mm Tris (pH 8), 150 mm NaCl and 1% Nonidet P40), once with 10 mm HEPES, and once with water. Extraction from the beads was with trifluoroacetic acid/water/acetonitrile 1:20:20 (v/v/v) saturated with α-cyanohydroxycinnamic acid. Spectra shown are representative of at least three experiments performed with triplicate samples. Treatment-induced changes were determined by calculating peak height versus the sum of heights of all examined peaks. When multiple N termini were detected, peptides with identical C termini (i.e. same predicted γ-cleavage site) were added together and normalized to total heights of all peaks. Means were compared by nonparametric one-way analysis of variance (Kruskal-Wallis Test) with Dunn's multiple comparisons post test. Results represent averages of all experiments ± S.D. repeated in three to seven experiments with two to three replicates in each experiment (*, p < 0.05; **, p < 0.01; !, p < 0.001 for treatment compared with vehicle control).
MALDI-TOF-TOF Analysis of FLAG-tagged Substrates
Samples were prepared as described for MALDI-TOF, except the final elusion was in trifluoroacetic acid:acetonitrile:water (1:20:20) without an energy-absorbing matrix. α-cyanohydroxycinnamic acid was added immediately before spotting on the MALDI-TOF-TOF target. Samples were analyzed by staff of the University of California San Diego Biomolecular Mass Spectrometry facility on an ABI 4800 MALDI-TOF-TOF.
Bicine-Urea Gel Analysis of γ-Secretase Products
For constructs with the swAPP backbone, cell lines were cultured and drug-treated as for mass spectrometry. Immunoprecipitates of γ-secretase products were prepared as described above. All immunoprecipitated material was eluted into the bicine-gel loading buffer at 95 °C. Bicine-urea gels were conducted as published previously (1). Proteins were electrophoretically transferred to nitrocellulose membrane and Western-blotted with anti-Aβ 82E1antibodies with subsequent quantification on a CCD imager (Syngene). For quantification, the results are expressed as averages ± S.E., normalized to control values. All experiments were repeated two to three times. Statistical analysis (two-tailed unpaired Student's t test) was performed with JMP software.
RESULTS
FLAG-tagged γ-Secretase Substrates
To determine whether the modulatory activity of GSMs on γ-secretase activity is limited to APP or rather occurs more generally on most γ-secretase substrates, we examined the pattern of γ-cleavage of CD44 and Notch-1 after drug treatment. FLAG epitope-tags were added to the sequences for extracellulary truncated CD44 (F-CD44ΔE), Notch-1 (F-NEXT), and APP (F-C83APP) to enable immunoprecipitation and MALDI-TOF analysis of secreted γ-cleaved fragments that are analogous to Aβ (referred to as F-NEXTβ, F-CD44β and F-Aβ (Refs. 24, 25 and supplemental Fig. 1). HEK cells stably coexpressing the swAPP and either F-CD44ΔE or F-NEXT were treated with sulindac sulfide (60 μm). We chose sulindac sulfide as the prototypic NSAID that has γ-secretase-modulating activity to reduce Aβ42 production.
If the binding site of GSMs is not specifically on APP alone, then this NSAIDs should also affect cleavage of the non-APP substrates. To ascertain that these cell lines were responsive to the dose of sulindac sulfide tested, we analyzed the MALDI-TOF spectra of Aβ peptides secreted from control and sulindac sulfide-treated cells coexpressing F-CD44ΔE and swAPP. We observed a robust increase in Aβ38 and a decrease in Aβ42 with sulindac sulfide as seen in our prior work (Fig. 1B and supplemental Fig. 2B). However, we detected essentially no changes in the profile of F-CD44β peptides with sulindac sulfide treatment as assessed by MALDI-TOF (Fig. 1A and supplemental Fig. 2A and Table 1). MALDI-TOF/TOF tandem mass spectrometry was performed to confirm the identity of the individual fragments (data not shown). Similarly, in HEK cells expressing F-NEXT derived from Notch-1 with or without swAPP, we could not detect any significant changes in peak heights of the respective F-NEXTβ peptides after sulindac sulfide treatment (data not shown). Therefore, both FLAG-tagged CD44 and Notch constructs failed to respond to γ-secretase modulation. In cells transfected with F-83-APP, we did detect an increase in Aβ38 levels as expected, although the reduction in Aβ42 was not significant (Fig. 1C). Similar results were seen regardless of whether the cells were singly or doubly transfected with the target construct together with APP (data not shown), indicating that the lack of changes in F-CD44β and F-NEXT is unlikely to be due to competing γ-substrates.
FIGURE 1.
Sulindac sulfide has no effect on γ-cleavage of F-CD44ΔE by MALDI-TOF. A, sulindac sulfide (60 μm) did not significantly shift the pattern of γ-secretase cleavage for F-CD44ΔE but shifted the γ-cleavage of Aβ generated from the same HEK cells coexpressing swAPP and F-CD44ΔE (B). This indicates appropriate drug penetration and activity in these cells. (*, p < 0.05; **, p < 0.01; !, p < 0.001). C, HEK293 cells stably expressing F-C83APP treated with sulindac sulfide showed the expected increase in peptides corresponding to γ-cleavage at Aβ38 although Aβ was fused to the FLAG epitope tag. Only a minimal trend in Aβ42 reduction was seen. γ-cleaved peptides from F-CD44ΔE and F-C83APP were immunoprecipitated with M2 anti-FLAG and analyzed by MALDI-TOF mass spectrometry. Peptides from F-CD44ΔE are denoted by the amino acids identified by their C termini as shown in supplemental Fig. 1 and Table 1. Aβ peptides were immunoprecipitated in B from culture media with anti-Aβ antibody (Ab9). Peptides are denoted by length in amino acids and standard Aβ numbering, with and analysis as described under “Experimental Procedures.” **, p < 0.01.
Effect of Chimeric APP-APLP2 Construct on GSM Sensitivity
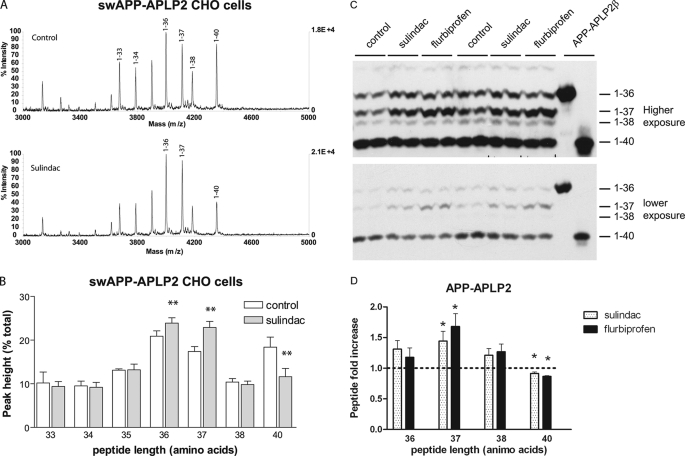
To confirm the preceding observations, we next investigated whether the sequence of the substrate TMD influences GSM modulatory activity. For these studies, the TMD of swAPP was substituted with sequences from APLP2, Notch-1, or ErbB4, all recognized γ-secretase substrates (Ref 36, Fig. 2A, and supplemental Tables 2–4 for masses and sequences). Using the swAPP backbone allowed immunoprecipitation of γ-cleaved products with Aβ N-terminal antibodies and easy detection by MALDI-TOF and immunoprecipitation/Western blotting. Aβ-like peptides from CHO cells stably transfected with APP-APLP2 chimera cDNA were strongly modulated by sulindac sulfide (Fig. 3A and B, and supplemental Table 2). The longest peptide, comprising 40 amino acids, was significantly reduced, with a concomitant increase in peptides of 36 and 37 amino acids in length. This pattern is highly reminiscent of the response in cells expressing intact APP. Because MALDI-TOF is only semi-quantitative, we performed Western blotting using bicine-urea gel from conditioned medium to validate the mass spectrometry results. Following immunoprecipitation/Western blotting, we observed four major species where the top and bottom bands comigrated with synthetic peptides that corresponded to the 1–36 and 1–40 of APP-APLP2 β peptides identified by MALDI-TOF (Fig. 3C). We therefore assigned the bands accordingly. Not unexpectedly, the intensity of the bands on Western blotting did not match the peak heights of the peptides seen by mass spectrometry. This is because the efficiency of ionization of the peptides embedded in the matrix is different for different peptides, a phenomenon well described for Aβ peptides, where Aβ42 is poorly ionized and, as a result, only a small peak is detected (37). Nevertheless, there was virtually complete concordance between the mass spectrometry and Western blotting data. APP-APLP2β 1–36 and 1–37 levels were increased by sulindac sulfide, whereas levels of 1–40 were decreased (Fig. 3D). As an additional confirmation, treatment of the cells with flurbiprofen (200 μm), another NSAID-based GSM that is structurally distinct from sulindac, also resulted in a very similar outcome by Western blotting (Fig. 3C). Therefore, these results showed that APP-APLP2 chimeric protein responded to GSM modulatory activity.
FIGURE 3.
The γ-cleavage of the APLP2 transmembrane domain in the APP-APLP2 chimera is sensitive to sulindac sulfide. A, representative MALDI-TOF tracings of Aβ-like peptides secreted from CHO cells stably expressing the APP-APLP2 chimera. There was an increase in generation of shorter peptides and a decrease in detection of the longest 40-amino acid species after 60 μm sulindac sulfide treatment. Peaks are labeled according to the peptide length counted from the aspartate of the normal Aβ sequence. In the lower panel, only those peaks that responded to sulindac sulfide are identified. B, summary results from APP-APLP2 expressing CHO cells as from A (**, p < 0.01). C, bicine-urea gel analysis of immunoprecipitated Aβ-like peptides secreted from APP-APLP2 CHO cells. Photomicrograph of a representative Western blot analysis detected by anti-Aβ antibody (82E1). Short and longer exposures are shown to visualize all the different species. D, graphic summary of results from APP-APLP2-expressing CHO cells as from C. Sulindac sulfide and flurbiprofen increased the levels of shorter Aβ-like peptides (1–37) and reduced the longer species (1–42). *, p < 0.05 by Student's t test in comparison to control treatment (n = 3).
Effect of Chimeric APP-Notch Construct on GSM Sensitivity
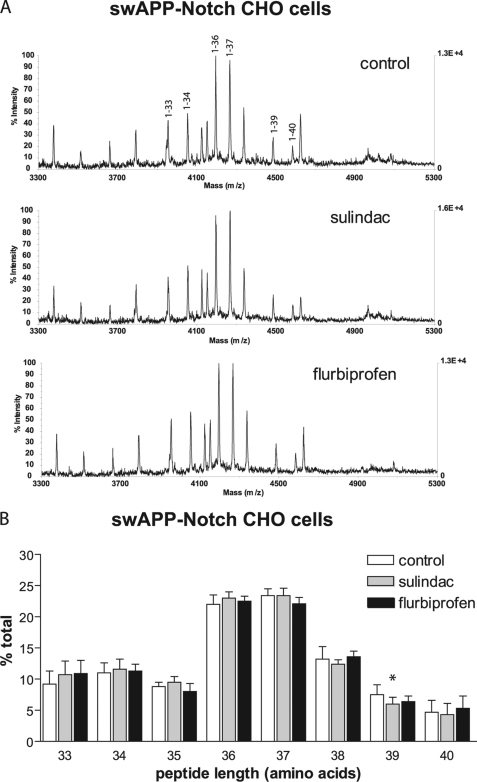
Because there is substantial sequence identity in the TMD between APLP2 and APP, it is perhaps not surprising that they responded to GSM in a similar fashion. We next analyzed the APP-Notch and APP-ErbB4 chimeric constructs, as their TMD sequences are more divergent from APP. Transfecting the APP-Notch chimeric construct stably in CHO cells (Fig. 4 and supplemental Table 3) or transiently in HEK cells (data not shown) showed essentially no changes other than a single peptide species containing 39 amino acids in length that was minimally reduced by sulindac sulfide treatment but not by flurbiprofen. Similarly, bicine-urea gel analysis after immunoprecipitation/Western blotting showed no significant changes in band intensity of the various species (data not shown). In this instance, however, we cannot positively correlate the bands on the bicine-urea gels to the MALDI-TOF-identified γ-cleaved species because we did not synthesize the corresponding APP-Notchβ peptides.
FIGURE 4.
Neither sulindac sulfide nor flurbiprofen altered γ-cleavage of the APP-Notch chimera containing the full TMD sequences of Notch-1. A, sample MALDI-TOF traces of Aβ-like peptides secreted from control- and NSAID-treated CHO cells stably expressing the APP-Notch chimera. Peaks are labeled according to the number of amino acids from the typical Aβ β-cleavage site. B, quantification of the peak heights from MALDI-TOF traces seen in A following treatment by sulindac sulfide and flurbiprofen on γ-secretase cleavage of APP-Notch in CHO cells. Neither drug altered the spectra significantly. A minor effect on the 39-amino acid peptide was significant. *, p < 0.05 in CHO but not reproducible in HEK cells (data not shown).
Effect of Chimeric APP-ErbB4 Construct on GSM Sensitivity
γ-Secretase cleavage of APP-ErbB4 yielded a major 38-amino acid Aβ-like species on MALDI-TOF spectra, corresponding to γ-cleavage after the -VIGGL. Interestingly, the peak heights of all the remaining peptide species were of minute sizes. Treatment of these cells with either sulindac sulfide or flurbiprofen did not alter the spectra of Aβ-like peptides (supplemental Fig. 3 and Table 4). However, given the diminutive size of most of these peaks, modest changes would likely not be detected.
Mapping of Transmembrane Sequences in APP That Are Required for Modulation by GSM
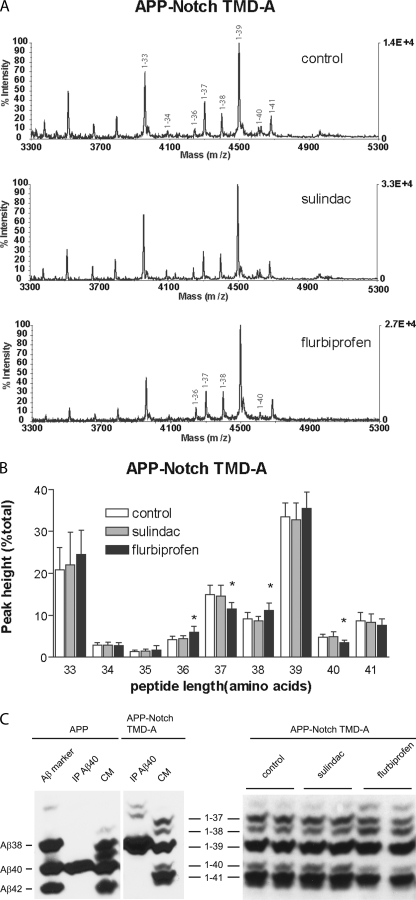
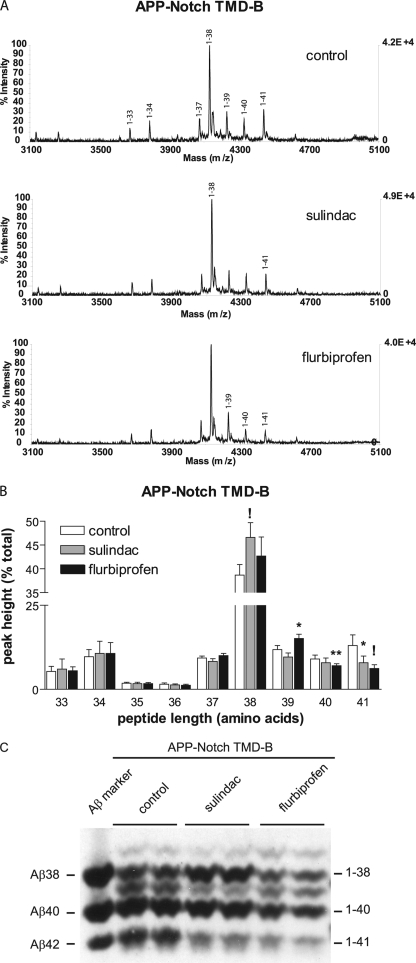
The preceding studies indicated that the TMD sequences have a major impact on the ability of GSM to modulate the levels of various Aβ-like peptides. To determine which residues of the APP TMD confer responsiveness to GSMs, we identified small TMD regions where GSM-sensitive APP and APLP2 sequences were similar to each other but divergent from Notch-1. Four to five amino acid domains of the Notch-1 TMD were substituted into the corresponding APP TMD. (Fig. 2B and supplemental Tables 5 and 6 list all the observed peaks and the corresponding sequences.) The APP-Notch TMD-A construct, which contains the first five residues of the Notch-1 TMD in place of six residues of APP, was chosen in large part because of the importance of the GXXXG motif identified as an APP dimerization motif (38). Interestingly, the MALDI-TOF trace showed a striking resemblance to intact APP (Fig. 5A). The major cleavage site was after the second valine (corresponding to Aβ40, now 39 amino acids in length). However, unlike APP, MALDI-TOF analyses showed that this construct was insensitive to sulindac sulfide treatment (Fig. 5, A and B). Treatment with flurbiprofen resulted in mild modulation of multiple peptide species that differed somewhat to wild-type APP, namely reduction of not only 1–40 but also the shorter 1–37 species. To further validate the mass spectrometry results, Western blotting following immunoprecipitation showed essentially similar results to the mass spectrometric analyses (Fig. 5C). Because the Aβ C terminus was preserved in the APP-Notch TMD-A construct, we could identify two of the bands in the Western blotting using Aβ40- and Aβ42-specific antibody. Fortuitously, this also allowed us to use Aβ ELISAs to quantify the Aβ-like peptides from the APP-Notch TMD-A construct. Indeed, consistent with the above findings, a careful dose response study showed no changes in levels of the Aβ-like peptides that end in Aβ40 and Aβ42 with increasing doses of either sulindac sulfide or flurbiprofen, all confirming that substituting the Notch sequences at the APP GXXXG motif rendered APP generally insensitive to GSM modulation (Fig. 6). A second chimera, APP-Notch TMD-B, this time with four amino acids of Notch-1 substituted at the Aβ42 γ-cleavage site (Fig. 2B), greatly altered the overall MALTI-TOF spectrum of the recovered Aβ peptides.1–38 appeared to represent the most abundant species, although by Western blotting, 1–38 was approximately as abundant as 1–40 (Figs. 5 and 7). This suggested that Aβ1–38 was ionized more efficiently than Aβ1–40. Although the pattern of Aβ peptides was altered, treatment with either sulindac or flurbiprofen nevertheless reduced the levels of the longest peptide (1–41) and variably altered the levels of the shorter peptides (Aβ 1–38 and 1–39), somewhat reminiscent of wild-type APP treated with GSMs. Taken together, our results showed conclusively that the sequences within the TMD of APP can influence the ability of GSMs to modulate the production of various Aβ peptides.
FIGURE 5.
Small regions of the Notch-1 transmembrane domain altered the pattern of APP γ-cleavage and sensitivity to NSAIDs. A, representative MALDI-TOF traces from a chimeric APP with a substitution of six amino acids at the beginning of the APP TMD with five residues from the Notch-1 sequence (APP-Notch TMD-A) stably expressed in CHO cells. The profile of γ-secretase cleavage products is similar to intact APP. The predominant peptide has 39 amino acids but represents the same valine γ-cleavage site as normal Aβ40. B, the APP-Notch TMD-A chimera had an altered NSAID response, eliminating the effect of sulindac sulfide and changing the pattern of peptides after flurbiprofen. Flurbiprofen increased the levels of a 36-amino acid peptide and decreased the peak for a 40-amino acid peptide. Peaks that underwent significant change are labeled in the panels for the appropriate drug treatment (*, p < 0.05). C, changes from MALDI-TOF were confirmed by immunoprecipitation and Western blot analyses. Bicine-urea gel analysis of immunoprecipitation (IP) of Aβ-like peptides secreted from APP-Notch TMD-A with Ab9 antibody was followed by blotting with 82E1 antibody. Because Aβ-like peptides from APP-Notch TMD-A retained the wild-type Aβ C termini, the secreted Aβ-like peptides can be unambiguously detected. In the left panel, immunoprecipitation of Aβ40 peptide was carried out from conditioned media (CM) of cells transfected with wild-type APP as well as APP-Notch TMD-A. Note that the Aβ-like 1–39 peptide has the same C terminus as Aβ40, as expected. Shown in the right panel is a representative Western blot analysis after immunoprecipitation of secreted Aβ40-like peptides from APP-Notch TMD-A cells following treatment with sulindac or flurbiprofen.
FIGURE 6.
There is no change in levels of Aβ-like peptides ending at Aβ40 and Aβ42 residues in APP-Notch TMD-A cells after sulindac or flurbiprofen treatment. Because the C termini of the Aβ-like peptides from APP-Notch TMD-A are identical to wild-type APP, this allowed quantification by Aβ ELISA where Aβ40 and Aβ42 corresponded to Aβ-like peptides 1–39 and 1–41, respectively. There are no changes in the levels of Aβ-like peptides after treatment with sulindac (B) or flurbiprofen (D) in APP-Notch TMD-A cells, but changes were detected in wild-type APP (A and C). The results are mean ± S.D. from three independent experiments.
FIGURE 7.
Small regions of the Notch-1 transmembrane domain inserted in the Aβ42 γ-cleavage site altered the pattern of APP γ-cleavage and sensitivity to NSAIDs. A, representative MALDI-TOF traces from CHO cells stably expressing APP-Notch TMD-B resulted in a γ-secretase cleavage profile that is similar to intact APP, although a peptide of 42 amino acid residues was not detected. However, the predominant Aβ peptide has 38 rather than 40 amino acids. Sulindac sulfide increased the peptide 38 amino acids. Flurbiprofen reduced the levels of 40- and 41-amino acid peptides and increased the Aβ39 peptide. Peaks that underwent significant change are labeled in the panels for the appropriate drug treatment. B, quantification of the MALDI-TOF traces shown in A (*, p < 0.05; **, p < 0.01; !, p < 0.001). C, representative immunoprecipitation/Western blot analysis of APP-Notch TMD-B cells treated with sulindac and flurbiprofen. Conditioned media were collected and Aβ peptides were immunoprecipitated and analyzed on bicine-urea gel by Western blotting and generally confirmed the changes seen by MALDI-TOF.
DISCUSSION
It is now established that a subset of NSAIDs in high doses act as GSMs modulating the γ-secretase cleavage of APP by reducing the amyloidogenic Aβ42 peptides. Despite the potential of GSMs in Alzheimer's disease therapeutics, there is a paucity of information about the substrate within the γ-secretase complex that GSMs likely interact with to modulate γ-secretase activity. The studies reported here were undertaken to determine the impact of the substrate sequence on the modulatory effect of GSMs. We found that the substrate sequence does indeed alter the ability of two NSAIDs, sulindac sulfide and flurbiprofen, to modulate the γ-cleavage site. FLAG-tagged constructs based on three different γ-secretase substrates, APP, CD44, and Notch-1, showed differential responses to sulindac sulfide. Specifically, sulindac sulfide modulated the pattern of γ-cleavage products from F-C83APP, partially resembling that seen with full-length APP, but did not shift γ-cleavage in cells expressing the F-CD44 or F-NEXT construct. The notion that the TMD sequence of the substrate plays a pivotal role in determining GSM sensitivity was confirmed by the analyses of chimeric APP molecules with different TMD substitutions. In particular, when the TMD of APP was replaced by APLP2, a markedly similar modulation of γ-secretase activity as compared with wild-type APP was observed. In contrast, APP containing the TMD of Notch-1 or ErbB4 was not susceptible to GSM modulation. Indeed, when regions containing only four to five amino acids of the Notch-1 TMD replaced comparable amino acid residues in APP, there were large effects in both drug sensitivity and the overall pattern of γ-cleavage products detected. Consequently, although the γ-secretase complex appears to be rather non-discriminant as to the primary sequence of its substrate, our studies showed that there are fine details in the γ-cleavage pattern and response to GSM modulation that are sequence-specific.
Using the FLAG-tagged artificial substrates engineered to allow MALDI-TOF identification of secreted γ-secretase products, we determined that APP, CD44, and Notch-1 have varied sensitivities to sulindac sulfide. In the F-C83APP construct, the mass spectrometry profile showed cleavages that were similar to wild-type APP and somewhat similar modulation of the Aβ peptides following drug treatment. However, the effects seen in F-CD44 and F-NEXT constructs were largely negative, suggesting that different γ-secretase substrates do not respond in a similar same manner to NSAID treatment.
Results from the APP chimeric molecules reinforced the studies with the FLAG-tagged constructs in that the TMD of the substrates appeared to significantly alter the effects of sulindac sulfide and flurbiprofen on γ-cleavage. When the TMD of APP was replaced by that from APLP2, Notch-1, or ErbB4, the responsiveness of γ-secretase to NSAIDs changed substantially. The TMD of APLP2, which shows considerable sequence homology to APP (75% identical), was sensitive to sulindac sulfide in a fashion highly reminiscent of APP. Namely, smaller species were increased, and production of the largest peptide was reduced. In contrast, the TMD of ErbB4 or Notch-1 inserted into APP prevented NSAID modulation of γ-cleavage. Further analyses showed that even small substitutions in the APP TMD were able to change both the overall γ-cleavage pattern and the sensitivity to NSAID. TMD substitution near the luminal face of APP (APP-Notch TMD-A) yielded a cleavage pattern that was similar to Aβ. The major peak (1–39) corresponded to the -MVGGVV C terminus that characterizes Aβ40 in wild-type APP, with other cleavages in a relatively normal distribution. Surprisingly, this construct was resistant to modulation by sulindac sulfide. This result highlights the importance of the GXXXG motif in generating full-length Aβ peptides first defined by the Multaup laboratory and also consistent with the results reported by Kukar et al. using different GSMs (38–40). The second small TMD substitution (APP-Notch TMD-B) had a different cleavage pattern, with the largest peak corresponding to the Aβ38 C terminus -MVGG, yet it was responsive to both drugs tested. The former result is not surprising because single amino acid mutations, both artificial and those occurring in familial Alzheimer's disease, have been shown to have profound effects on the Aβ42/Aβ40 ratio and the overall profile of Aβ as observed by MALDI-TOF (41–44). In addition, fluorescence resonance energy transfer studies indicate that mutations and drugs that cause the same shift in cleavage site preference cause similar changes in PS conformation (28). In summary, these results appear to reinforce the notion that the domain in APP that is required for GSM activity is at or near the GXXXG motif and not at the C terminus of Aβ where one might have anticipated, as it is where the alteration of γ-cleavage actually takes place.
A study by Okochi et al. found that γ-cleavage of a Notch-1-based truncated construct was shifted by treatments with NSAIDs in a manner analogous to that seen in APP by bicine-urea gels (45). They reported that sulindac sulfide decreased generation of an Nβ (cognate Notch Aβ) fragment terminating in -AAAAFV, whereas we observed that cleavage at this same site in the APP-Notch chimeric construct (1–40) was unchanged by either sulindac sulfide or flurbiprofen. The discrepancy between the two studies could be due to the different Notch-1 constructs and analysis methods used. Our APP-Notch construct was an artificial chimera, whereas Okochi et al. (45 used a truncated Notch construct. Further, we assessed primarily by MALDI-TOF, whereas the changes in secreted Nβ peptides were assessed by gel electrophoresis alone by Okochi et al. (45). Mass spectrometry is only semiquantitative, and given the difference in systems, our study could have missed the subtle alterations reported by Okochi et al. (45) using a more authentic substrate. However, our study was more directed toward overall substrate specificity of GSMs, and in this regard, the findings are all internally consistent.
Taken together, our results indicate that NSAIDs do exhibit substrate specificity in their ability to modulate γ-cleavage (27, 40). Further, our findings add to the observations reported in the accompanying manuscript by Kukar et al. showing that the juxtamembrane APP sequence can have profound influence on the length of Aβ peptides (47). Nevertheless, the precise mechanisms of action of GSMs remain unresolved. Given the dramatically increased potency of second- and third-generation GSMs (31, 46), substrate binding is unlikely to account for overall GSM activity (30). Because γ-secretase cleaves multiple substrates at multiple positions, we hypothesize that with certain substrates, the γ-secretase complex assumes a structure that allows GSMs to interact with the complex to stabilize certain conformations over others and, in so doing, modulate γ-cleavage. However, whether γ-cleavage modulation holds promise in Alzheimer's disease therapeutics will only become clear when potent compounds with optimized activity reach the clinic for testing.
Supplementary Material
Acknowledgments
We thank Karen Jansen for construction of the APP-ErbB4 construct; Drs. Christian Haass, Sven Lammich, and Masayasu Okochi for providing the F-NEXT constructs and cell lines; and Drs. Dennis Selkoe and Jimin Zhang for providing the APP-Notch and APP-APLP2 transmembrane domain chimera constructs. We also thank Ann Shih and Barbara Cottrell for technical and laboratory assistance, and Dr. Elizabeth Komives for providing mass spectrometry expertise and a critical review.
This work was supported, in whole or in part, by National Institutes of Health Grants AG 20206 (to T. E. G. and E. H. K.) and AG005131–22 (to S. A. S.). This work was also supported by an American Federation for Aging Research/Ellison Medical Foundation fellowship (to S. A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1–6.
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- Aβ
- amyloid β
- TMD
- transmembrane domain
- NSAID
- non-steroidal anti-inflammatory drug
- GSM
- γ-secretase modulator
- PS
- presenilin
- APLP
- amyloid precursor-like protein
- F-APPC83
- FLAG epitope-tagged extracellularly truncated amyloid precursor protein
- F-Aβ
- γ-secretase cleaved fragment from F-APPC83
- F-CD44ΔE
- FLAG epitope-tagged extracellularly truncated CD44
- F-CD44β
- γ-secretase cleaved fragment from F-CD44ΔE
- F-NEXT
- FLAG epitope-tagged extracellularly truncated Notch-1
- F-NEXTβ
- γ-secretase cleaved fragment from F-NEXT
- swAPP
- Swedish mutation of human amyloid precursor protein.
REFERENCES
- 1. Weggen S., Eriksen J. L., Das P., Sagi S. A., Wang R., Pietrzik C. U., Findlay K. A., Smith T. E., Murphy M. P., Bulter T., Kang D. E., Marquez-Sterling N., Golde T. E., Koo E. H. (2001) Nature 414, 212–216 [DOI] [PubMed] [Google Scholar]
- 2. Szekely C. A., Thorne J. E., Zandi P. P., Ek M., Messias E., Breitner J. C., Goodman S. N. (2004) Neuroepidemiology 23, 159–169 [DOI] [PubMed] [Google Scholar]
- 3. Lim G. P., Yang F., Chu T., Gahtan E., Ubeda O., Beech W., Overmier J. B., Hsiao-Ashec K., Frautschy S. A., Cole G. M. (2001) Neurobiol. Aging 22, 983–991 [DOI] [PubMed] [Google Scholar]
- 4. Lim G. P., Yang F., Chu T., Chen P., Beech W., Teter B., Tran T., Ubeda O., Ashe K. H., Frautschy S. A., Cole G. M. (2000) J. Neurosci. 20, 5709–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jantzen P. T., Connor K. E., DiCarlo G., Wenk G. L., Wallace J. L., Rojiani A. M., Coppola D., Morgan D., Gordon M. N. (2002) J. Neurosci. 22, 2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Groen T., Kadish I. (2005) Brain Res. Brain Res. Rev. 48, 370–378 [DOI] [PubMed] [Google Scholar]
- 7. McKee A. C., Carreras I., Hossain L., Ryu H., Klein W. L., Oddo S., LaFerla F. M., Jenkins B. G., Kowall N. W., Dedeoglu A. (2008) Brain Res. 1207, 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Townsend K. P., Praticò D. (2005) FASEB J. 19, 1592–1601 [DOI] [PubMed] [Google Scholar]
- 9. Sagi S. A., Weggen S., Eriksen J., Golde T. E., Koo E. H. (2003) J. Biol. Chem. 278, 31825–31830 [DOI] [PubMed] [Google Scholar]
- 10. Eriksen J. L., Sagi S. A., Smith T. E., Weggen S., Das P., McLendon D. C., Ozols V. V., Jessing K. W., Zavitz K. H., Koo E. H., Golde T. E. (2003) J. Clin. Invest. 112, 440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weggen S., Eriksen J. L., Sagi S. A., Pietrzik C. U., Ozols V., Fauq A., Golde T. E., Koo E. H. (2003) J. Biol. Chem. 278, 31831–31837 [DOI] [PubMed] [Google Scholar]
- 12. Beher D. (2008) Curr. Top Med. Chem. 8, 34–37 [DOI] [PubMed] [Google Scholar]
- 13. Kukar T., Murphy M. P., Eriksen J. L., Sagi S. A., Weggen S., Smith T. E., Ladd T., Khan M. A., Kache R., Beard J., Dodson M., Merit S., Ozols V. V., Anastasiadis P. Z., Das P., Fauq A., Koo E. H., Golde T. E. (2005) Nat. Med. 11, 545–550 [DOI] [PubMed] [Google Scholar]
- 14. Fraering P. C., Ye W., Strub J. M., Dolios G., LaVoie M. J., Ostaszewski B. L., van Dorsselaer A., Wang R., Selkoe D. J., Wolfe M. S. (2004) Biochemistry 43, 9774–9789 [DOI] [PubMed] [Google Scholar]
- 15. Czirr E., Cottrell B. A., Leuchtenberger S., Kukar T., Ladd T. B., Esselmann H., Paul S., Schubenel R., Torpey J. W., Pietrzik C. U., Golde T. E., Wiltfang J., Baumann K., Koo E. H., Weggen S. (2008) J. Biol. Chem. 283, 17049–17054 [DOI] [PubMed] [Google Scholar]
- 16. Hahn S., Brüning T., Ness J., Czirr E., Baches S., Gijsen H., Korth C., Pietrzik C. U., Bulic B., Weggen S. (2011) J. Neurochem. 116, 385–395 [DOI] [PubMed] [Google Scholar]
- 17. Haapasalo A., Kovacs D. M. (2011) J. Alzheimer's Dis. 25, 3–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mumm J. S., Kopan R. (2000) Dev. Biol. 228, 151–165 [DOI] [PubMed] [Google Scholar]
- 19. Ikeuchi T., Sisodia S. S. (2003) J. Biol. Chem. 278, 7751–7754 [DOI] [PubMed] [Google Scholar]
- 20. Vidal G. A., Naresh A., Marrero L., Jones F. E. (2005) J. Biol. Chem. 280, 19777–19783 [DOI] [PubMed] [Google Scholar]
- 21. Murakami D., Okamoto I., Nagano O., Kawano Y., Tomita T., Iwatsubo T., De Strooper B., Yumoto E., Saya H. (2003) Oncogene 22, 1511–1516 [DOI] [PubMed] [Google Scholar]
- 22. Jung K. M., Tan S., Landman N., Petrova K., Murray S., Lewis R., Kim P. K., Kim D. S., Ryu S. H., Chao M. V., Kim T. W. (2003) J. Biol. Chem. 278, 42161–42169 [DOI] [PubMed] [Google Scholar]
- 23. May P., Reddy Y. K., Herz J. (2002) J. Biol. Chem. 277, 18736–18743 [DOI] [PubMed] [Google Scholar]
- 24. Lammich S., Okochi M., Takeda M., Kaether C., Capell A., Zimmer A. K., Edbauer D., Walter J., Steiner H., Haass C. (2002) J. Biol. Chem. 277, 44754–44759 [DOI] [PubMed] [Google Scholar]
- 25. Okochi M., Steiner H., Fukumori A., Tanii H., Tomita T., Tanaka T., Iwatsubo T., Kudo T., Takeda M., Haass C. (2002) EMBO J. 21, 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takahashi Y., Hayashi I., Tominari Y., Rikimaru K., Morohashi Y., Kan T., Natsugari H., Fukuyama T., Tomita T., Iwatsubo T. (2003) J. Biol. Chem. 278, 18664–18670 [DOI] [PubMed] [Google Scholar]
- 27. Beher D., Clarke E. E., Wrigley J. D., Martin A. C., Nadin A., Churcher I., Shearman M. S. (2004) J. Biol. Chem. 279, 43419–43426 [DOI] [PubMed] [Google Scholar]
- 28. Uemura K., Lill C. M., Li X., Peters J. A., Ivanov A., Fan Z., DeStrooper B., Bacskai B. J., Hyman B. T., Berezovska O. (2009) PLoS ONE 4, e7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uemura K., Farner K. C., Hashimoto T., Nasser-Ghodsi N., Wolfe M. S., Koo E. H., Hyman B. T., Berezovska O. (2010) Nat. Commun. 1, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kounnas M. Z., Danks A. M., Cheng S., Tyree C., Ackerman E., Zhang X., Ahn K., Nguyen P., Comer D., Mao L., Yu C., Pleynet D., Digregorio P. J., Velicelebi G., Stauderman K. A., Comer W. T., Mobley W. C., Li Y. M., Sisodia S. S., Tanzi R. E., Wagner S. L. (2010) Neuron 67, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oehlrich D., Berthelot D. J., Gijsen H. J. (2011) J. Med. Chem. 54, 669–698 [DOI] [PubMed] [Google Scholar]
- 32. Tomita T., Ohki Y., Yokoshima S., Higo T., Shimada N., Koizumi H., Osawa S., Fukuyama T., Iwatsubo T. (2010) 40th Annual Meeting of the Society for Neuroscience, San Diego, CA, November 16, 2010, Program 627.3, Society for Neuroscience, Washington, D.C [Google Scholar]
- 33. McGowan E., Pickford F., Kim J., Onstead L., Eriksen J., Yu C., Skipper L., Murphy M. P., Beard J., Das P., Jansen K., Delucia M., Lin W. L., Dolios G., Wang R., Eckman C. B., Dickson D. W., Hutton M., Hardy J., Golde T. (2005) Neuron 47, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang J., Ye W., Wang R., Wolfe M. S., Greenberg B. D., Selkoe D. J. (2002) J. Biol. Chem. 277, 15069–15075 [DOI] [PubMed] [Google Scholar]
- 35. Uljon S. N., Mazzarelli L., Chait B. T., Wang R. (2000) Methods Mol. Biol. 146, 439–452 [DOI] [PubMed] [Google Scholar]
- 36. De Strooper B. (2010) Physiol. Rev. 90, 465–494 [DOI] [PubMed] [Google Scholar]
- 37. Wang R., Sweeney D., Gandy S. E., Sisodia S. S. (1996) J. Biol. Chem. 271, 31894–31902 [DOI] [PubMed] [Google Scholar]
- 38. Munter L. M., Voigt P., Harmeier A., Kaden D., Gottschalk K. E., Weise C., Pipkorn R., Schaefer M., Langosch D., Multhaup G. (2007) EMBO J. 26, 1702–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Richter L., Munter L. M., Ness J., Hildebrand P. W., Dasari M., Unterreitmeier S., Bulic B., Beyermann M., Gust R., Reif B., Weggen S., Langosch D., Multhaup G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14597–14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kukar T. L., Ladd T. B., Bann M. A., Fraering P. C., Narlawar R., Maharvi G. M., Healy B., Chapman R., Welzel A. T., Price R. W., Moore B., Rangachari V., Cusack B., Eriksen J., Jansen-West K., Verbeeck C., Yager D., Eckman C., Ye W., Sagi S., Cottrell B. A., Torpey J., Rosenberry T. L., Fauq A., Wolfe M. S., Schmidt B., Walsh D. M., Koo E. H., Golde T. E. (2008) Nature 453, 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lichtenthaler S. F., Ida N., Multhaup G., Masters C. L., Beyreuther K. (1997) Biochemistry 36, 15396–15403 [DOI] [PubMed] [Google Scholar]
- 42. Lichtenthaler S. F., Wang R., Grimm H., Uljon S. N., Masters C. L., Beyreuther K. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murphy M. P., Hickman L. J., Eckman C. B., Uljon S. N., Wang R., Golde T. E. (1999) J. Biol. Chem. 274, 11914–11923 [DOI] [PubMed] [Google Scholar]
- 44. Tesco G., Ginestroni A., Hiltunen M., Kim M., Dolios G., Hyman B. T., Wang R., Berezovska O., Tanzi R. E. (2005) J. Neurochem. 95, 446–456 [DOI] [PubMed] [Google Scholar]
- 45. Okochi M., Fukumori A., Jiang J., Itoh N., Kimura R., Steiner H., Haass C., Tagami S., Takeda M. (2006) J. Biol. Chem. 281, 7890–7898 [DOI] [PubMed] [Google Scholar]
- 46. Zettl H., Weggen S., Schneider P., Schneider G. (2010) Trends Pharmacol. Sci. 31, 402–410 [DOI] [PubMed] [Google Scholar]
- 47. Kukar T. L., Ladd T. B., Robertson P., Pintchovski S. A., Moore B., Bann M. A., Ren Z., Jansen-West K., Malphrus K., Eggert S., Maruyama H., Cottrel B. A., Das P., Basi G. S., Koo E. H., Golde T. E. (2011) J. Biol. Chem. 286, 39804–39812 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.