Background: A complete understanding of the role of βc ubiquitination in IL-5R biology is currently lacking.
Results: We identified three βc lysine residues that are required for JAK kinase binding to the receptor.
Conclusion: JAK kinase binding to βc via 3 lysines is essential for receptor ubiquitination.
Significance: We provide new mechanistic details regarding IL-5R biology, which is important for understanding eosinophil physiology.
Keywords: Eosinophils, Immunology, Jak Kinase, Receptor Modification, Signal Transduction, Ubiquitination, Cytokine Receptors, Interleukin 5, Allergic Inflammation, Asthma
Abstract
Eosinophils are multifunctional leukocytes implicated in the pathogenesis of numerous inflammatory diseases including allergic asthma and hypereosinophilic syndrome. Eosinophil physiology is critically dependent on IL-5 and the IL-5 receptor (IL-5R), composed of a ligand binding α chain (IL-5Rα), and a common β chain, βc. Previously, we demonstrated that the βc cytoplasmic tail is ubiquitinated and degraded by proteasomes following IL-5 stimulation. However, a complete understanding of the role of βc ubiquitination in IL-5R biology is currently lacking. By using a well established, stably transduced HEK293 cell model system, we show here that in the absence of ubiquitination, βc subcellular localization, IL-5-induced endocytosis, turnover, and IL-5R signaling were significantly impaired. Whereas ubiquitinated IL-5Rs internalized into trafficking endosomes for their degradation, ubiquitination-deficient IL-5Rs accumulated on the cell surface and displayed blunted signaling even after IL-5 stimulation. Importantly, we identified a cluster of three membrane-proximal βc lysine residues (Lys457, Lys461, and Lys467) whose presence was required for both JAK1/2 binding to βc and receptor ubiquitination. These findings establish that JAK kinase binding to βc requires the presence of three critical βc lysine residues, and this binding event is essential for receptor ubiquitination, endocytosis, and signaling.
Introduction
Allergic respiratory diseases are characterized in part by large numbers of proinflammatory eosinophils and their reactive products in the airways and blood (reviewed in Refs. 1 and 2). Interleukin-5 (IL-5) is the principal cytokine for eosinophil differentiation, activation, and prolonged survival. IL-5 initiates its biological effects by binding to the IL-5 receptor (IL-5R)3 composed of a ligand-specific IL-5 receptor α chain (IL-5Rα) and a shared signaling component, βc (3). Our previous studies revealed that IL-5 receptor signaling is partially terminated by ubiquitin/proteasome degradation of the βc cytoplasmic domain, followed by lysosomal degradation of the remaining truncated IL-5R complex (4). However, the molecular details underlying the role of βc ubiquitination in IL-5R down-regulation remain largely unknown.
In recent years, studies have shown that the 8-kDa ubiquitin (Ub) molecule is a “master regulator” of cellular physiology, whereby protein modification by this protein has the potential to regulate cellular processes as diverse as endocytosis, trafficking, cell cycle progression, signal transduction, inflammation, apoptosis, neural and muscular degeneration, and transcription (reviewed in Refs. 5–8). Modification of target proteins by Ub alters their molecular landscape resulting in changes that can affect their stability, interaction with other proteins, enzymatic activity, and even their subcellular localization (9–11). Different types of Ub-linkage topologies are associated with distinct biological functions (5–11). For example, poly-Ub chains formed through Ub Lys48 and Gly76 linkages are attached to substrates destined for proteasome degradation (7), whereas poly-Ub chains formed through Ub Lys29 or Lys63 have other nonproteolytic functions in cells, such as transcriptional regulation and membrane trafficking (8–11).
Pioneering work in the field of cellular trafficking has firmly established that Ub regulates protein transport between membrane compartments by serving as a sorting signal on protein cargo and by controlling the activity of trafficking machinery (12–15). As such, mono-Ub, multi-mono-Ub, and Lys63-linked poly-Ub chains appended to integral membrane proteins serve as regulated signals for internalization into the endocytic pathway. Indeed, studies with the growth hormone, epidermal growth factor (EGF), platelet-derived growth factor (PDGF), nerve growth factor (TrkA), and prolactin receptors have demonstrated an important regulatory role for ubiquitination in either receptor endocytosis and/or intracellular trafficking (9, 16–20).
Although the endocytic program has long been regarded as a critical mechanism of receptor down-regulation, through the degradation of signaling receptors and their ligands, it is now appreciated that signaling processes persist throughout the endocytic route, and that intracellular endocytic stations (“signaling endosomes”) actually contribute to the sorting of signals in time and space (21–26). Indeed, emerging evidence connects endocytosis to complex cellular programs such as cell motility, cell fate determination, mitosis, and apoptosis (21–26). As such, several studies exploring the connection between endocytosis and receptor tyrosine phosphorylation have been reported for the EGF, transforming growth factor-β (TGF-β), acetylcholine receptor, protease-activated receptor 1 (PAR1), and TrkA receptors (27–30). For the IL-5R, we have shown previously that inhibition of both clathrin-dependent and clathrin-independent endocytosis blocks the association of pSTAT5 and pMAPK with the IL-5R, indicating that entry of activated IL-5Rs into the endocytic pathway provides a mechanism for receptor interaction with signaling molecules (31). Therefore, because ubiquitination plays such an important role in receptor endocytosis and signal transduction, we set out to investigate the interplay between the βc ubiquitination axis and IL-5R endocytic trafficking and signaling. Here, we identify a cluster of three key lysine residues located in the βc membrane-proximal region that are required for JAK1/2 binding to the receptor, an event that appears to be necessary for βc ubiquitination and signaling.
EXPERIMENTAL PROCEDURES
Cell Culture, Materials, and Inhibitors
The human embryonic kidney cell line, HEK293 (purchased from ATCC), was maintained in DMEM supplemented with 10% FBS and 10 μg/ml of gentamicin. The human erythroleukemic cell line, TF-1-F11, was previously described (4), and endogenously expresses the IL-5, IL-3, and GM-CSF receptors. Recombinant human IL-5 was previously described (4). Cycloheximide (used at 10 μg/ml) was purchased from Calbiochem. Monensin (used at 40 μm) was from Sigma. Cy3-IL-5 was prepared as described in Ref. 31 and used at 50 ng/ml. On-TARGET Plus siRNA targeting human JAK2 (catalog number L-003146-00), and siGENOME SMART pool siRNAs JAK1 (catalog number M-003145-02), and siGenome negative control pool siRNA (catalog number D-001206-13) were purchased from Thermo Scientific. Antibodies used for immunofluorescence microscopy and flow cytometry were purchased from the following companies: anti-βc (clone 3D7, BD Biosciences; H-300, Santa Cruz Biotechnology), anti-IL-5Rα (R&D Systems), anti-transferrin receptor (BD Biosciences), anti-Rab11 (Cell Signaling), anti-LAMP1 (BD Biosciences), PE- and APC-conjugated rat anti-mouse IgG1 antibodies were purchased from BD Biosciences, and FITC-conjugated donkey anti-goat IgG was from Santa Cruz Biotechnology.
Construction of βc Ub-deficient Mutants and Generation of Stable Cell Lines
WT βc cDNA (GenBankTM accession number AAA18171) cloned in pCMV-Script (Stratagene) was used as a template for site-directed mutagenesis of βc cytoplasmic lysines 1–16 to arginine using the multisite site-directed mutagenesis kit (Stratagene) in a stepwise process with specific mutagenesis primers listed under supplemental Table S1. All βc Lys to Arg mutations were fully sequenced to confirm sequence modifications. The mutated βc K16R cDNA was then cloned into Virapower pLentivector (Invitrogen). During this stepwise mutagenesis process, we collected βc KtoR mutants with fewer mutations and generated our library of βc mutants listed in Fig. 7. The βc ΔBox 1 mutant (in pCMV Script) was constructed by designing primers corresponding to WT βc cDNA, which deleted the amino acid sequence, WEEKIPNPSKSH.
FIGURE 7.
Summary of βc Lys to Arg mutants used to identify lysine residues necessary for βc ubiquitination. A, schematic illustration of our panel of βc KtoR mutants depicting intracellular βc lysines (asterisks) and their substitution to arginine (dots). In addition, βc ΔBox 1 mutant was generated by deleting the Box 1 motif in the βc cytoplasmic domain using specific deletion primers. All βc constructs (WT and mutants) were cloned in pCMV-Script and stably transfected into IL-5Rα-expressing HEK293 cells. Dual-receptor expressing cell lines were selected in the presence of Geneticin (0.85 mg/ml) and blasticidin (6 μg/ml). IL-5-induced ubiquitination was evaluated as described in the legend to Fig. 8. Results are summarized to the right of the illustrations. @, βc K(4–6) is ubiquitinated prior to and after IL-5 stimulation. TM, transmembrane domain. White boxes denote Box 1 and 2 motifs. B, amino acid sequence of the first 20 amino acids (450–470) in the βc cytoplasmic domain. The Box 1 motif, which was deleted in the βc ΔBox 1 mutant, is underlined. The 3 critical lysine residues are marked with an asterisk.
WT βc and IL-5Rα cDNAs (31) were cloned into a Virapower pLentivector (Invitrogen), and constructs were fully sequenced. Lentiviral particles containing each receptor chain were made following the manufacturer's instructions, concentrated using Amicon Ultrafiltration tubes (Millipore), and titered in HeLa cells for stable transductions. Initially, we established a parental cell line by stably transducing the HEK293 cell line with IL-5Rα only. Stably transduced cells were selected in the presence of 6 μg/ml of blasticidin, and surviving cells were sorted by flow cytometry. This stable cell line was used for transduction with WT βc and βc K16R viral particles, as well as all βc KtoR pCMV Script constructs. All pLenti-transduced cell lines were sorted by flow cytometry for confirmation of dual receptor expression.
Immunofluorescence Microscopy
Stably transduced HEK293 cells expressing either WT IL-5Rs or βc K16R with IL-5Rα were plated on poly-d-lysine-coated coverslips for 24 to 48 h prior to IL-5 stimulation (10 ng/ml). Freshly isolated human eosinophils were isolated as previously described (31) and cultured overnight in serum-containing RPMI in the absence of IL-5. The following day, cells (25,000 cells per slide) were fixed with 4% formaldehyde for 20 min, cytospun (700 rpm for 5 min), and processed for antibody labeling as described below for all cells: cells were permeabilized with 0.2% Triton, blocked with 10% normal goat serum, and incubated with 1:50 dilution mouse/anti-human-βc (BD) mAb, 1:100 rabbit/anti-human βc (H300), 1:25 goat/anti-human-5Rα polyclonal (R&D Systems), 1:50 mouse/anti-human TFR receptor mAb, 1:25 rabbit/anti-human Rab11, or 1:25 dilution of mouse/anti-human LAMP-1 antibodies (BD Biosciences) for 2 h at RT. After washing unbound antibodies (3 times with 1× PBS), cells were incubated with 1:1,000 dilution donkey anti-goat IgG/Alexa-488 (for anti-5Rα polyclonal Abs), 1:1,000 donkey anti-mouse IgG/Alexa-594 (for anti-βc mAb), and 1:1000 donkey anti-rabbit IgG/Alexa-594 (for βc H-300) or goat anti-rabbit IgG/Alexa-488 (for anti-Rab11), and 1:1,000 goat anti-mouse IgG/Alexa-488 (for anti-TFR-R and anti-LAMP1). After washing unbound secondary Abs (3 times with PBS), cells were mounted with Prolong Gold anti-fade reagent (which contains DAPI counterstain, Molecular Probes).
Deconvolution fluorescence microscopy was performed on a Zeiss AxioImager Z1 epifluorescence microscope with a Plan Apochromat ×63/1.4 numerical aperture oil-immersion objective. Images were acquired with a Zeiss Axiocam CCD camera using Axiovision software. Z-series of optical sections were performed at 0.24-μm increments, followed by deconvolution analysis with AxioVision software. For all images, only middle Z-stack sections are shown to display the intracellular compartment. In addition, for co-localization assays, images were further analyzed with AV4 MOD Widefield Multi-unmixing software (Zeiss) to eliminate any cross-talk (bleed-through) between the 488- and 594-nm channels. The scale bar on the images is 10 μm. Cells showing positive co-localization had three or more co-localized vesicles per cell.
Cy3-IL-5 Binding and Endocytosis Assay
Cy3-labeled IL-5 (10 or 50 ng/ml) was added to chilled HEK293 cells for 20 min for cell surface binding. Cells were washed extensively to remove unbound ligand and either processed for microscopy (supplemental Fig. S3B), flow cytometry analysis (supplemental Fig. S3A), or moved to 37 °C to initiate endocytosis for the desired time point (supplemental Fig. S2); endocytosis was stopped by washing with cold PBS. For these assays, cells were not acid washed to detect the remaining Cy3-IL-5 signal on the cell surface after endocytosis.
Cycloheximide Chase Assay
WT βc and βc K16R expressing cells were pretreated with 10 μg/ml of cycloheximide for 1 h as described in Ref. 32. Cells were then stimulated with 10 ng/ml of IL-5 in the presence of cycloheximide and harvested (chased) at various time points shown in the graph. Cells were lysed with RIPA buffer (1% Nonidet P-40, 1% Triton, 0.1% SDS, 0.15 m NaCl, 0.01 m sodium phosphate, pH 7.2, 0.2 mm EDTA, 5 mm N-ethylmalemide, 1 mm sodium vanadate, 1 mm PMSF, 1 μg/ml of leupeptin, 1 μg/ml of pepstatin) for 20 min on ice. Fifty μg of lysate was resolved by lauryl dodecyl sulfate-PAGE, followed by immunoblot (IB) analysis using anti-βc and anti-actin antibodies (Sigma). Degradation curves were generated by measuring βc band densities from the IB images using AlphaView software (Alpha Innotech) and normalized against corresponding actin band densities. The βc band density at the 0-h time point (after 1 h pre-treatment with cycloheximide) was designated as 100% and percent densities of subsequent time points were calculated based on this assumption. Curves were drawn with % βc band density (y axis) versus time following IL-5 stimulation (x axis).
Flow Cytometry
βc and IL-5Rα cell surface expression was measured by incubating WT βc and βc K16R-expressing HEK293 cells (∼100,000/tube) and primary eosinophils (50,000/tube) in PBS + 2% FBS with anti-βc (BD Biosciences) and anti-IL-5Rα antibodies (R&D Systems) for 20 min on ice according to standard protocols. Cells were then washed and incubated with PE-conjugated anti-mouse IgG1 and FITC-conjugated anti-goat IgG, respectively. Labeled proteins were analyzed immediately on an Accuri C6 (Accuri Cytometers) flow cytometer. The flow data were analyzed using CFlow Plus (Accuri Cytometers) and FCS Express (De Novo Software) software and graphed using Excel software. Mean fluorescence intensities (MFIs) and S.E. are described in the text and figure legends.
βc Endocytosis Assay
We established a flow cytometry-based βc internalization assay that measures the loss of cell surface βc immunoreactivity following IL-5 stimulation. Briefly, anti-βc antibodies (BD Biosciences), which do not inhibit IL-5 binding to the IL-5R,4 were added to pre-chilled HEK293 cells expressing WT βc, βc K16R, or βc K(1–3)R receptors for 30 min on ice. Cells were washed 3 times with ice-cold media to remove unbound, excess antibodies. IL-5 (10 ng/ml) was added to the cells, and aliquots of cells were transferred to 37 °C for 5, 10, and 15 min. Receptor internalization was terminated by adding ice-cold PBS to the cells at each time point. The remaining βc receptors on the cell surface were detected by incubating the anti-βc antibody-bound receptors with anti-mouse IgG1-PE and measured by flow cytometry. The MFI of immune reactive βc receptors in both cell lines at 0 min with IL-5 (unstimulated) was represented as 100% and the loss of immunoreactivity (MFI) was plotted for each time point.
Surface Biotinylation
HEK293 cells grown to 80% confluence in 100-mm plates were cell surface labeled with the nonpermeable sulfo-NHS-SS-biotin reagent (Thermo Fisher) following the manufacturer's instructions. Briefly, cells were washed 3 times on ice with cold PBS, pH 8.0, followed by addition of 5 ml of chilled PBS, pH 8.0, containing 200 μg of biotin reagent. Cell surface proteins were labeled with biotin for 30 min while rocking at 4 °C. Unbound biotin was removed by washing cells 3 times with cold PBS + 100 mm glycine. Chilled serum-containing DMEM was added back to the cells on ice, and either left unstimulated on ice (to inhibit endocytosis) or stimulated with 10 ng/ml of IL-5 for 1 h at 37 °C. After washing with cold PBS, intact cells were re-suspended in 500 μl of cold PBS and incubated with 0.5 μg of anti-βc mAb (BD Biosciences) for 2 h with rocking at 4 °C to bind biotinylated cell surface βc receptors. Cells were washed 3 times with PBS to remove unbound antibody then lysed with 500 μl of RIPA lysis buffer. Cell surface immune complexes were precipitated by addition of 10 μl of Protein G, separated by LDS-PAGE, and transferred to Immobilon-P PVDF membranes. Cell surface-labeled biotinylated proteins were detected by incubating membranes with Neutravidin-HRP reagent (Thermo Fisher).
JAK1 and JAK2 Combinatorial RNAi
HEK293 cells expressing WT IL-5Rs (40–50% confluent) were transfected with both SMART pool JAK2 (75 nm) and JAK1 siRNAs (50 nm), or a nontarget negative control siRNA (125 nm) with Lipofectamine 2000 (Invitrogen). Twenty-four hours post-transfection, cells were split into two plates and harvested 48 h post-transfection for analysis of gene silencing efficiency and functional assays.
Immunoprecipitation and Immunoblot Assays (IP/IB)
All IP/IB assays were done as previously described (4, 33) with the following antibodies: βc was IP with anti-βc 1C1 mAb (Santa Cruz Biotechnology); IBs were done with anti-βc polyclonal (H-300 (C-terminal), Santa Cruz Biotechnology); anti-βc polyclonal antibodies (N-terminal) and anti-IL-5Rα (both from R&D Systems); anti-ubiquitin mAb (P4D1) (all from Santa Cruz Biotechnology); anti-actin rabbit pAb (Sigma); anti-phosphotyrosine (mAb clone 4G10) and anti-pSTAT5 were purchased from Upstate Biotechnology. Anti-STAT5 was purchased from BD Biosciences. Proteins were visualized by incubation with enhanced chemiluminescence plus reagents (GE Healthcare) and images were captured with a FluorChem 8000 imaging system (Alpha Innotech).
RESULTS
Ubiquitination-deficient βc Receptors Accumulate on the Cell Surface
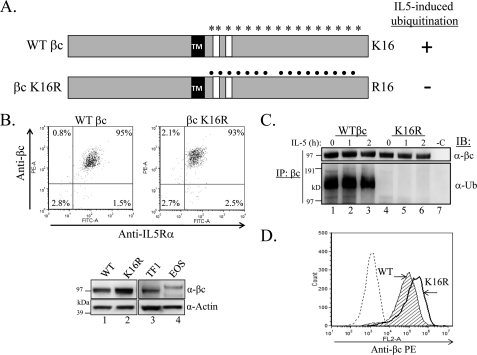
To investigate the role of βc ubiquitination in IL-5R endocytosis and signaling, we initially constructed a βc mutant in which all 16 cytoplasmic lysines were substituted with another positively charged amino acid, arginine (Fig. 1A, βc K16R). Lysine residues are canonical ubiquitination sites, whereas arginines are not. The HEK293 cell model system was chosen as a suitable background for our studies because: 1) they do not express endogenous IL-5Rs and thus phenotypes of mutant βc receptors can be examined; 2) we have shown previously that WT IL-5R signaling proceeds normally following IL-5 stimulation (31, 33); and 3) activated WT IL-5Rs traffic normally to lysosomes in this cell line (31, 33).4 WT βc and βc K16R constructs were individually transduced into HEK293 cells stably expressing IL-5Rα. Each cell line was sorted by flow cytometry for dual receptor-expressing cells (Fig. 1B), and lack of βc ubiquitination in K16R was confirmed by IP/IB (Fig. 1C).
FIGURE 1.
Generation of HEK293 cells stably expressing WT βc and βc K16R IL-5 receptors. A, schematic illustration of 16 intracellular lysines on WT βc (asterisks) and the same 16 lysines mutated to arginine on βc K16R (dots). TM, transmembrane domain. White boxes denote Box 1 and 2 motifs. B, flow cytometry analysis of sorted, dual receptor-expressing HEK293 cells stably transduced with WT βc (upper left panel) or βc K16R (upper right panel). Cells were labeled with anti-βc-PE and anti-IL-5Rα-FITC antibodies. IB analysis comparing the relative protein levels of WT βc and βc K16R (30 μg of whole cell lysates each) to endogenously expressed βc in TF1 cells (250,000 cells) and human primary eosinophils (1 × 106 eosinophils, lysed in LDS loading buffer) (lower panels). C, IL-5 time course of stably transduced cell lines shown in B, followed by IP of whole cell lysates with anti-βc antibodies (or Protein G only, -C) and immunoblotted as indicated. D, flow cytometry analysis of cells in B comparing cell surface expression of both receptor chains by labeling with anti-βc-PE antibodies.
Western blot analysis comparing overall βc protein levels in our stably transduced cell lines to endogenous βc in TF1 and human eosinophils revealed similar expression between WT βc and TF1 cells. However, slightly higher βc protein expression was detected in our βc K16R-expressing cells as compared with TF1 cells, but especially as compared with eosinophils that are notorious for expressing low amounts of βc protein (Fig. 1B, bottom panel) (34). Surprisingly, flow cytometry analysis revealed higher cell surface expression of βc K16R receptors than WT βc (Fig. 1D), an observation that will be a huge focus of this study.
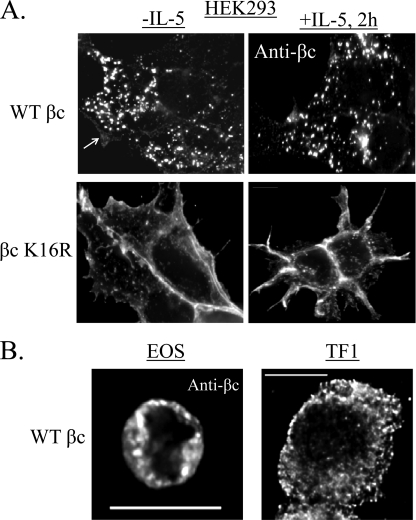
The first functional question we asked with our βc K16R construct was whether βc ubiquitination controlled its subcellular distribution. To this end, both cell lines were immunostained with anti-βc antibodies in the absence (−IL-5) or presence of IL-5 (+IL-5) and visualized by deconvolution fluorescence microscopy (Fig. 2A). Visualization of ubiquitination-competent WT βc receptors demonstrated a predominantly punctate intracellular pattern, both at steady-state and after IL-5 stimulation (Fig. 2A, upper panels). In contrast, only a faint outline of WT βc receptors was detected on the plasma membrane representing the cell surface pool (Fig. 2A, top panel, white arrow).
FIGURE 2.
Lack of βc ubiquitination results in cell surface accumulation of receptors. A, stably transduced HEK293 cells expressing either WT βc or βc K16R were grown on coverslips and either left unstimulated (−IL-5, left panels) or stimulated with 10 ng/ml of IL-5 for 2 h (+IL-5, right panels). Cells were permeabilized and stained with anti-βc antibodies followed by labeling with Alexa Fluor-488 secondary antibody. Deconvolution fluorescence microscopy was used to visualize βc subcellular distribution. Shown are single middle Z-stack images to represent the intracellular compartment. B, WT βc receptors localize to intracellular vesicles in both primary human eosinophils and TF1 cell lines. Cells were permeabilized and labeled with anti-βc antibodies, followed by staining with Alexa Fluor-594 secondary antibodies. Images were acquired and displayed as described in A.
Conversely, ubiquitination-deficient βc K16R revealed a remarkable difference. Rather than localizing predominantly to intracellular vesicles, the majority of βc K16R receptors were detected on the cell surface of HEK293 cells both before and after IL-5 stimulation (Fig. 2A, bottom panels). This result confirms the data obtained by flow cytometry analysis (Fig. 1D).
We next investigated whether the intracellular distribution pattern seen with WT βc receptors was unique to our stably transduced HEK293 cell line. Immunostaining of endogenously expressed WT βc receptors in freshly isolated human eosinophils and the human erythroleukemic cell line, TF-1-F11 clone with anti-βc antibodies, revealed the same punctate intracellular pattern seen in our stably transduced HEK293 cells, further supporting our HEK293 cell model system for these studies (Fig. 2B, middle optical Z-stack shown for both panels).
In summary, these data demonstrate that in the three cell types examined, the bulk of ubiquitinated WT βc receptors localize to punctate intracellular vesicles. Importantly, the data also show that the intracellular distribution of Ub-deficient βc K16R receptors is altered and leads to their accumulation on the cell surface, implicating this post-translational modification in controlling βc subcellular localization.
Reduced Intracellular Trafficking of βc K16R at Steady-state
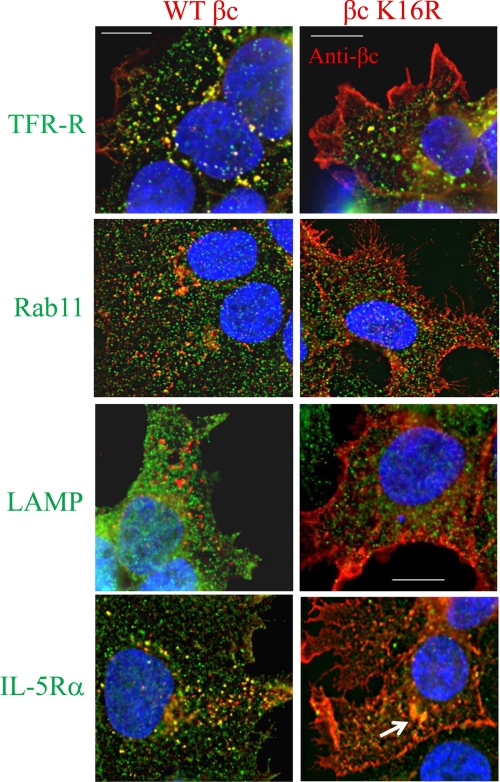
To gain further insight into the identity of the intracellular vesicles to which WT βc receptors localize constitutively, we performed βc co-localization assays with antibodies against various endocytic markers (Fig. 3). Cells expressing WT βc and βc K16R receptors were labeled with anti-βc antibodies (Fig. 3, all panels, red color) and co-stained with various antibodies: 1) anti-transferrin receptor (TFR-R), which is a marker for recycling endosomes (Fig. 3, upper panels, green); 2) anti-Rab11, a recycling endosomal marker (Fig. 3, middle panels, green); 3) LAMP-1, a lysosomal marker (third panels from top); and 4) IL-5Rα (Fig. 3, lower panels, green). In unstimulated cells, WT βc receptors co-localized with the TFR-R and Rab11 (yellow dots). Furthermore, as expected, WT βc-positive vesicles were negative for the lysosomal marker, LAMP-1, as the cells were not stimulated with IL-5. Importantly, however, staining with anti-IL-5Rα antibodies revealed that the majority of WT βc receptors co-localized with its ligand-binding partner in these intracellular vesicles (Fig. 3, bottom left panel, yellow dots).
FIGURE 3.
Reduced intracellular trafficking of βc K16R at steady state. Co-localization assays were performed in unstimulated WT βc and βc K16R-expressing HEK293 cell lines by staining all cells with anti-βc antibodies (red color) and anti-transferrin receptor (top panels, green), anti-Rab11 (second panels, green), anti-LAMP-1 (third panels, green), and anti-IL-5Rα antibodies (bottom panels, green). Localization of primary antibody binding was detected by labeling with Alexa Fluor-488 (all panels, green color) or Alexa Fluor-594 (all panels, red) secondary antibodies. Images were acquired and displayed as described in the legend to Fig. 2. Yellow vesicles indicate co-localization between the two antibodies used in the assay. Note how WT βc receptors (left panels) traffic through TFR-R and Rab11-positive vesicles (yellow dots), but not LAMP-1 (lysosomal marker). Also, note the strong co-localization between WT βc and IL-5Rα in intracellular vesicles under basal conditions (bottom left). Scale bar is 10 μm.
Conversely, because the predominant localization of βc K16R receptors was on the plasma membrane, cells expressing mutant receptors had only a few co-localization events when stained with the three endocytic markers. However, compared with the endocytic markers, anti-IL-5Rα staining revealed a few more co-localization events between βc K16R and IL-5Rα in a perinuclear region (Fig. 3, bottom right panel, arrow). It is also worth noting that because βc K16R receptor expression is very high on the cell surface of these cells, the bright intensity of the antibody label might be obscuring any faintly stained intracellular vesicles.
In conclusion, these data suggest that the majority of ubiquitination-competent βc receptors traffic through early endosomes, some of which are recycling endosomes. Conversely, ubiquitination-deficient βc receptors show reduced intracellular trafficking and accumulate on the cell surface.
Absence of Receptor Ubiquitination Shows Partial Delay in IL-5-induced βc Degradation
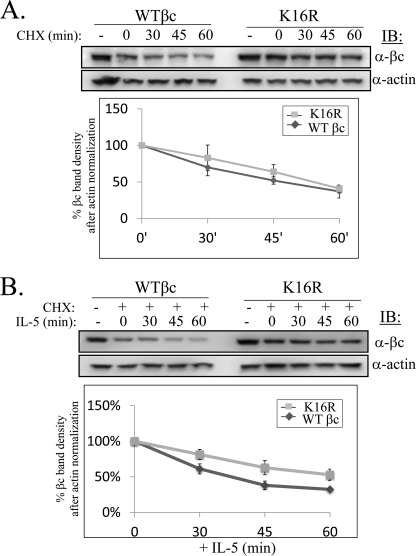
Because lack of βc ubiquitination resulted in increased cell surface expression of βc K16R, we wanted to determine the underlying molecular mechanism(s) for this observation. We reasoned that three possible explanations could account for this result: 1) delayed protein degradation of βc K16R; 2) reduced endocytosis of the mutant receptor; or 3) accelerated βc K16R recycling. We set out to test each possibility. Previously, we have shown that the ubiquitinated cytoplasmic tail of βc is a target of the proteasome wherein its partial degradation results in the generation of truncated βc products we termed βIP (4). As βc K16R lacks this molecular signature, we hypothesized that turnover of the ubiquitin-deficient βc mutant could be impaired and might explain the increased cell surface expression. To test this hypothesis, we compared degradation rates between the two receptors by pre-treating cells with cycloheximide to block new protein synthesis, and then “chased” βc protein levels at various time points by Western blot in either unstimulated (Fig. 4A) or IL-5-stimulated cells (Fig. 4B). At steady-state, the degradation curves between WT and βc K16R were very similar as shown in Fig. 4A, lower panel (the 60-min time point of βc K16R had 41% of starting protein levels remaining, whereas WT βc had 37%).
FIGURE 4.
Absence of receptor ubiquitination shows partial delay in βc degradation. A, WT βc and βc K16R-expressing HEK293 cell lines were either left untreated (−) or treated with 10 μg/ml of cycloheximide (CHX) (+) for 1 h followed by harvesting cells at the indicated time points. Whole cell lysates (50 μg/lane) were analyzed by IB with anti-βc and anti-actin antibodies. Degradation curves were generated by measuring βc band densities from IBs and normalized against corresponding actin band densities. The βc band density at the 0-h time point (after 1 h pretreatment with cycloheximide) was designated as 100% and percent densities of subsequent time points were calculated based on this assumption (n = 3). B, same as in A, except cells were stimulated with IL-5 (10 ng/ml) for the indicated times (n = 3).
In contrast, a more noticeable difference was detected after IL-5 stimulation between the two receptors. WT βc protein levels decreased rapidly following 30–60 min of IL-5 stimulation (only 32% of starting protein levels remained, plotted in Fig. 4B, lower panel), whereas βc K16R receptors degraded ∼20% slower, as 53% of the starting protein level remained after 60 min of IL-5 stimulation (Fig. 4B, plotted in the lower panel).
These data demonstrate that lack of βc ubiquitination results in a ∼20% slower turnover rate. However, this small decrease does not explain completely the huge increase in cell surface accumulation by βc Ub-deficient receptors, indicating that other regulatory mechanisms might be contributing to this effect.
Cell Surface Accumulation of βc K16R Receptors Is Not Due to Increased Recycling
Because WT βc receptors were detected in intracellular vesicles that stained positive for markers of recycling and early endosomes (TFR-R and Rab11, Fig. 3, upper left panels), it seemed reasonable to consider the possibility that an alteration in the recycling pathway of βc K16R receptors might explain its increased cell surface expression. The obvious alteration we considered was accelerated recycling back to the cell surface.
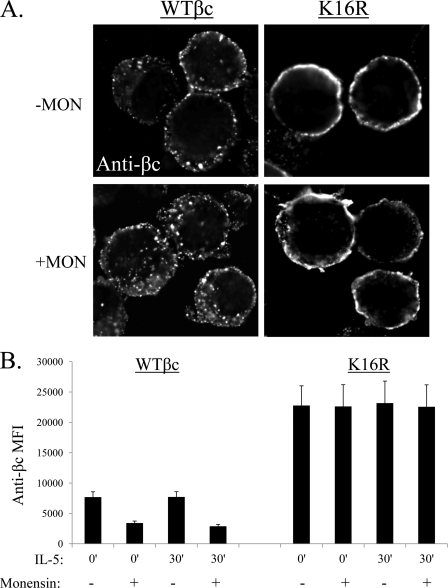
To explore this possibility, we used a recycling inhibitor, monensin (35, 36), which is a carboxylic ionophore that regulates proton movement across membranes, thereby possessing the capacity to increase the pH in acidic organelles, including endosomes. Treatment of cells with monensin leads to a block in transport of internalized cargo back to the plasma membrane resulting in the accumulation of endocytosed proteins in endosomes. We predicted that if βc K16R recycled back to the cell surface at a faster rate than WT βc, then treatment with the inhibitor would cause an increased intracellular accumulation of Ub-deficient receptors with a concomitant reduction in receptor cell surface expression as compared with WT βc receptors. To test our hypothesis, both cell lines were treated with or without monensin for 30 min in suspension, and then assayed for βc localization by either microscopy (Fig. 5A) or flow cytometry (Fig. 5B). Monensin-treated WT βc cells showed a significant increase in βc-positive intracellular vesicles as compared with untreated cells (Fig. 5A, left panels) with a simultaneous reduction in βc cell surface levels both before (55.6% reduction) and after IL-5 stimulation (62.5% reduction) (Fig. 5B, left bar graphs; MFIs in legend).
FIGURE 5.
Absence of βc receptor ubiquitination does not lead to increased recycling. A, WT βc and βc K16R-expressing HEK293 cell lines were treated with or without 40 μm monensin in suspension for 30 min. After cytospinning, cells were fixed and immunostained with anti-βc antibodies. Note the increase in intracellular βc vesicles seen in monensin-treated cells (left panels), which were not observed in cells expressing βc K16R receptors (right panels). For this assay, we used HEK293 cell in suspension because βc cell surface labeling was difficult to interpret in adherent monensin-treated HEK293 cells. B, same as in A except βc cell surface levels were measured by flow cytometry and are expressed as mean MFI ± S.E. minus isotype control MFI in the absence or presence of inhibitors; n = 3. WT βc MFI, 0 min (−Mon) = 7,686 ± 875; 0 min (+Mon) = 3,413 ± 334; 30 min (−Mon) = 7,713 ± 869; 30 min(+Mon) = 2,887 ± 316. βc K16R MFIs: 0 min (−Mon) = 22,767 ± 3252; 0 min (+Mon) = 22,622 ± 3576; 30 min (−Mon) = 23,171 ± 3630; 30 min (+Mon) = 22,556 ± 3,623.
On the other hand, monensin treatment had a negligible effect on the intracellular accumulation of βc K16R-positive vesicles (Fig. 5, right panels), as well as βc K16R cell surface levels assayed by flow cytometry (Fig. 5B, bar graphs on right). Last, WT βc receptors expressed in freshly isolated eosinophils and TF1 cells demonstrated the same drop in cell surface expression when treated with monensin, confirming that ubiquitination-competent βc receptors traffic through the recycling pathway in other cells (supplemental Fig. S1). In conclusion, these data demonstrate that: 1) the majority of ubiquitination-competent βc receptors traffic through the recycling pathway in both unstimulated and IL-5-stimulated conditions; and 2) cell surface accumulation of βc K16R receptors is not due to accelerated recycling.
βc K16R Receptors Have Impaired IL-5-induced Endocytosis
The last possibility we considered to explain increased cell surface expression of βc K16R receptors was reduced endocytosis. To test this hypothesis, we employed three methods: 1) a flow cytometry-based internalization assay; 2) a cell surface IP biotinylation assay; and 3) a Cy3-IL-5 endocytosis assay.
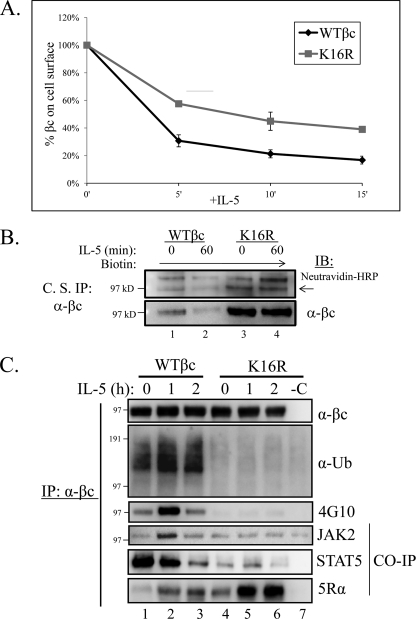
First, to compare the initial internalization rate between WT βc and βc K16R receptors, we established an IL-5-stimulated internalization assay that measures the loss of cell surface βc immunoreactivity by flow cytometry (see “Experimental Procedures”). As seen in Fig. 6A, IL-5-stimulated internalization of βc receptors for 15 min revealed a 22% slower internalization rate for βc K16R compared with WT βc (MFI WT βc at 15 min = 17 ± 3% versus βc K16R, 15 min = 39 ± 2%).
FIGURE 6.
Ubiquitination-deficient βc K16R receptors show impairment in IL-5-stimulated endocytosis and signaling. A, βc endocytosis assay. Briefly, pre-chilled HEK293 expressing WT βc and βc K16R receptors were incubated with anti-βc antibodies on ice. After washing, aliquots of cells were moved to 37 °C for the indicated time points. The remaining cell surface βc receptors were detected by incubating the anti-βc antibody-bound receptors with anti-mouse IgG1-PE and measured by flow cytometry. The MFI of immunoreactive βc receptors in both cell lines at the 0 min of IL-5 (unstimulated) was represented as 100% and the loss of immunoreactivity (MFI) was plotted for each time point (n = 3). MFIs, WT βc, 5′ = 31 ± 4%; βc K16R, 5 min = 58 ± 2%; WT βc, 10 min = 21 ± 3%; βc K16R, 10 min = 45 ± 7%; WT βc, 15 min = 17 ± 3%; βc K16R, 15 min = 39 ± 2%. B, WT βc and βc K16R-expressing HEK293 cell lines were cell surface labeled with the nonpermeable sulfo-NHS-SS-biotin reagent. Briefly, labeled cells were either left unstimulated or stimulated with 10 ng/ml of IL-5. Before cell lysis, anti-βc antibodies (BD Biosciences) were added to intact cells to bind labeled βc cell surface receptors for 2 h on ice. After cell lysis, βc immune complexes were analyzed by IB with neutravidin-HRP and anti-βc (H300) antibodies (n = 3). C, stably transduced HEK293 cells expressing WT or βc K16R receptors were stimulated with 10 ng/ml of IL-5 for the indicated times. Lysates were IP with anti-βc mAbs and analyzed by IB with the indicated antibodies. Note the correlation of lack of Ub with reduced JAK2 and STAT5 association, and Tyr phosphorylation in βc K16R receptors. n = 6.
We next confirmed this result by using a biochemical approach that labeled the bulk of cell surface proteins with biotin in both cell lines, followed by an IP protocol that specifically enriched labeled βc cell surface receptors with anti-βc antibodies (see “Experimental Procedures”) (Fig. 6B). IB with neutravidin-HRP conjugates revealed that biotin-labeled WT βc receptors decreased on the cell surface following IL-5 stimulation, whereas its βc K16R counterpart remained virtually unchanged (Fig. 6B, upper panel, arrow). IB with anti-βc antibodies confirmed the identity of the biotin-labeled βc bands, and also demonstrated that the cell surface levels of βc K16R did not change significantly after IL-5 stimulation (Fig. 6B, bottom panel).
Our standard Cy3-IL-5 endocytosis assay (31) was then used to visualize the uptake of fluorescently labeled IL-5 after initiating endocytosis at 37 °C in both cell lines (supplemental Fig. S2). In this assay, cells were not acid washed to ensure detection of the remaining Cy3-IL-5 on the cell surface after endocytosis. Microscopy analysis revealed that cells expressing WT βc receptors internalized Cy3-IL-5 very efficiently as evidenced by the pattern of punctate dots inside the cells (supplemental Fig. S2, left panel, white arrows). However, cells expressing βc K16R had fewer internalized Cy3-IL-5-labeled dots overall and concomitantly displayed more Cy3-IL-5 on their cell surface (supplemental Fig. S2, right panel, white arrow).
Together, these results demonstrate that ubiquitination-deficient βc receptors have a reduced internalization rate as compared with WT βc, indicating that βc ubiquitination is required for efficient ligand-induced IL-5R endocytosis. Given this result, it is highly likely that reduced βc K16R degradation (Fig. 4) could be a secondary effect resulting from decreased endocytosis.
βc K16R Receptors Have Blunted IL-5-induced Signaling
To investigate whether IL-5-induced signaling was affected in βc K16R-expressing cells, we used a biochemical approach to compare both cell lines (Fig. 6C). Surprisingly, IP/IB of both βc receptors revealed that following IL-5 stimulation Ub-deficient βc K16R receptors had attenuated signaling as evidenced by the absence of βc K16R tyrosine phosphorylation (Fig. 6C, third panel, IB: 4G10), and decreased association with both JAK2 and STAT5 (Fig. 6C, fourth and fifth panels). Moreover, lack of βc K16R phosphorylation was not due to its inability to associate with IL-5Rα as a strong interaction between the two proteins was detected by co-IP after IL-5 stimulation (Fig. 6C, bottom panel, lanes 5 and 6).
To rule out the possibility that impaired βc K16R signaling was not due to lack of IL-5 binding to these receptors, we used flow cytometry to measure the cell surface binding of Cy3-IL-5 in both cell lines (supplemental Fig. S3). Because we typically use 10 ng/ml of IL-5 for our stimulation assays, we decided to compare two different concentrations of Cy3-IL-5 for this assay, low and high (10 and 50 ng/ml). Statistical analysis of the data revealed no significant difference in Cy3-IL-5 cell surface binding between the two receptor-expressing cell lines (supplemental Fig. S3A, black and gray bars). Last, microscopy was used to visually confirm Cy3-IL-5 cell surface binding in these cell lines (supplemental Fig. S3B).
In all, our data indicate that ubiquitination-deficient βc receptors have blunted IL-5-induced signaling, which may be the result of reduced JAK2 association with the IL-5R. Moreover, based on this result, it is possible that impairment of IL-5-induced βc K16R endocytosis and degradation may be a result of decreased ligand-stimulated signaling.
Three βc Lysine Residues Are Critical for Receptor Ubiquitination, JAK2 Association, and βc Phosphorylation
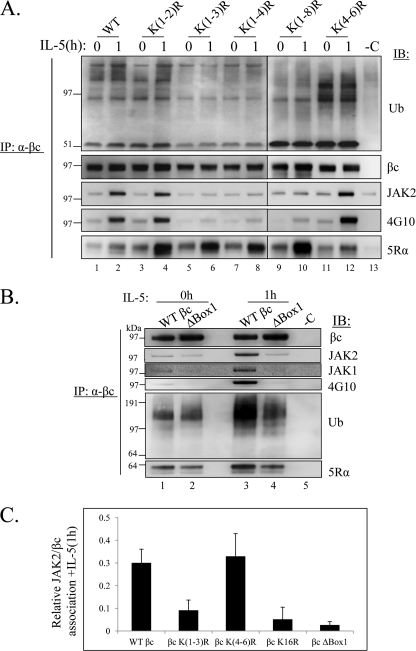
To identify the specific lysine residues necessary for βc ubiquitination, we returned to our panel of various βc Lys to Arg mutants that were generated during the stepwise mutagenesis process used to obtain βc K16R. In a preliminary screen, we investigated the contribution of the first 8 lysine residues in the βc cytoplasmic domain either alone or in various Lys combinations (Fig. 7A) by transient transfection of each construct into IL-5Rα-expressing HEK293 cells followed by IL-5-induced ubiquitination analysis with our standard IP/IB analysis (data summarized in Fig. 7). Most of the βc mutants examined were either ubiquitinated after IL-5 stimulation, or ubiquitinated prior to ligand stimulation (Fig. 7A, βc K(4–6)R). However, three mutants (βc K(1–3)R, βc K(1–4)R, and βc K(1–8)R) displayed a block in ubiquitination similar to that of βc K16R (summarized in Fig. 7A, shown in Fig. 8A, top panel). Interestingly, common to all three Ub-deficient βc receptors was the substitution of the first three lysines with arginines. Surprisingly, individual mutation of each Lys or even a combination of Lys(1–2) did not reproduce the Lys(1–3) Ub-deficient phenotype (summarized in Fig. 7).
FIGURE 8.
Three βc cytoplasmic lysines are necessary for βc ubiquitination, JAK2 association, and tyrosine phosphorylation. A, stably transfected HEK293 cells expressing the indicated βc KtoR mutants in pCMV-Script were left unstimulated (0 h) or stimulated with 10 ng/ml of IL-5 for 1 h. Lysates were prepared using RIPA buffer, IP with anti-βc mAbs and analyzed by IB with the indicated antibodies (n = 3). Note the correlation of Ub-deficient βc receptors with reduced JAK2 association and low βc Tyr phosphorylation. B, same as in A, except stably transfected HEK293 cells expressing βc ΔBox 1 mutant receptors were used for the IP/IB assays. Note how βc ΔBox 1 mutants have reduced, but not absent ubiquitination even though JAK2 or JAK1 are not associated with these receptors. C, quantification of the relative amount of JAK2 co-IP with βc following IL-5 stimulation was determined by measuring JAK2 band densities from IP/IBs in A and normalized against corresponding βc band densities (n = 3). Relative associations for each βc receptor are as follows: WT βc = 0.30 ± 0.06; βc (K1–3R) = 0.09 ± 0.04; βc K(4–6R) = 0.33 ± 0.1; βc K16R 0.05 ± 0.05; βc ΔBox 1 = 0.025 ± 0.016.
Furthermore, and consistent with the Ub-deficient receptor phenotype, microscopic and flow cytometry analyses revealed that βc K(1–3)R receptors accumulated on the cell surface (supplemental Fig. S4, A and B), and had impaired endocytosis (supplemental Fig. S4C). We, therefore, concluded that the presence of 3 βc lysines (Lys457, Lys461, and Lys467) were important for receptor Ub, and thus focused the rest of our studies on this βc mutant.
The next question we asked was whether any linear motif containing a cluster of 3 lysines was in fact the key mechanism for βc ubiquitination. To address this question, it seemed logical to examine the next cluster of 3 Lys to Arg mutants in the βc cytoplasmic domain (βc K(4–6)R). Surprisingly, βc K(4–6)R was ubiquitinated under basal conditions and remained ubiquitinated after IL-5 stimulation (Fig. 8, top panel, lanes 11 and 12), indicating that a linear sequence of just any clustered lysines is not the underlying mechanism mediating βc ubiquitination. The data thus suggested a strict requirement for the presence of the first 3 lysines in βc ubiquitination.
Closer examination of the location of βc Lys(1–3) revealed that the second Lys was located in the βc Box 1 motif, which is a JAK kinase binding site (reviewed in Ref. 37), whereas the first and third lysines lay just outside of this region (Fig. 7B). The connection between loss of βc ubiquitination with mutation of the βc JAK kinase binding site prompted us to question whether βc ubiquitination required: 1) the binding of JAK kinases to βc; 2) the presence of the βc Box 1 motif; or 3) the actual presence of JAK1/2 as both kinases are reportedly activated by IL-5 and other βc-engaging cytokines (33, 38–41).
Our first approach was to ask whether our panel of Ub-deficient βc Lys to Arg receptors were associated with the JAKs in our stably transfected cell lines (Fig. 8A, third panel). As compared with Ub-competent βc receptors (lanes 1–4 and 11–12), Ub-deficient βc receptors (βc K(1–3R), K(1–4)R, and K(1–8)R) demonstrated reduced interaction with JAK2, as well as decreased βc tyrosine phosphorylation (Fig. 8, A, fourth panel, lanes 5–10, and quantified in C). We confirmed that the defect in βc Tyr phosphorylation was not due to a block in IL-5Rα association as all receptors interacted with the α chain (Fig. 8A, bottom panel). Last, lack of signaling by βc K(1–3)R was not due to lack of IL-5 binding to these receptors as demonstrated under supplemental Fig. S3. In all, these data confirm a strong correlation between a βc Ub deficiency and reduced JAK2 association along with blunted βc tyrosine phosphorylation.
Next, the requirement of the βc Box 1 motif in receptor ubiquitination was evaluated by constructing a mutant that was deleted in this region (βc ΔBox 1, deleted in 458–465 aa, Fig. 7, A and B). This deleted sequence included a deletion of βc K2 (Lys461), but still contained K1 and K3 (Lys457 and Lys467). After stable transfection into IL-5Rα-expressing cells, mutant βc receptors were analyzed by our standard IP/IB analysis (Fig. 8B). As expected, βc ΔBox 1 receptors displayed almost undetectable JAK2 and JAK1 association (Fig. 8,B, second and third panels, and quantified in C) and decreased βc Tyr phosphorylation (Fig. 8B, fourth panel). Interestingly, βc ΔBox 1 receptors were ubiquitinated to the same extent as WT receptors in unstimulated cells (Fig. 8A, fifth panel, lanes 1 and 2), but ubiquitination levels were visibly reduced following IL-5 stimulation (Fig. 8B, compare lanes 3 and 4). However, the reduction in βc ubiquitination was not to the same extent as seen with βc K(1–3R) and K16R (Figs. 6C and 8A), probably because K1 and K3 were still present in this mutant. This result demonstrates that although JAK1/2 were not associated with βc, the receptor was still visibly ubiquitinated, indicating that JAK kinase binding to βc contributes to optimal receptor ubiquitination, but might not be the only mechanism involved. Last, IB with anti-IL-5Rα confirmed the association between βc ΔBox 1 and the α chain, albeit at a slightly reduced level (Fig. 8B, bottom panel, lanes 3 and 4).
In conclusion, our data demonstrate that the Box 1 motif is only partially required for optimal βc ubiquitination, but fully required for JAK1/2 binding and βc tyrosine phosphorylation, as has been shown for other Type I receptors (Reviewed in 42, 43–46). Furthermore, these findings support our earlier hypothesis that the presence of βc K1–3 (Lys457, Lys461, and Lys467) is required for efficient βc ubiquitination and suggest that these residues possibly act cooperatively to promote this activity. Last, loss of these 3 βc Lys residues does not appear to inhibit IL-5Rα binding to a significant degree.
The Presence of JAK1 and JAK2 Is Partially Required for IL-5-induced βc Ubiquitination
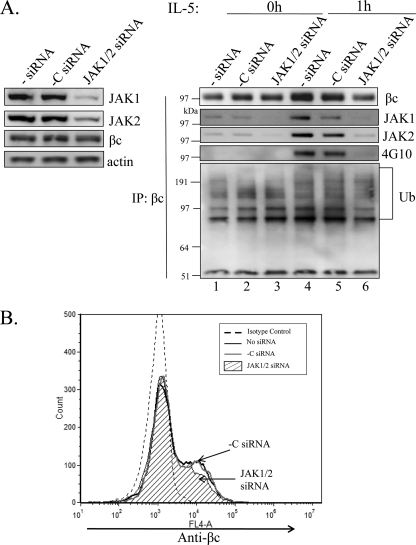
Because many reports have shown that IL-5R signaling requires both JAK1 and JAK2 for βc tyrosine phosphorylation and signal transduction (33, 38–41), we sought to investigate whether βc ubiquitination had the same requirements. We used combinatorial RNAi to simultaneously silence both JAKs in HEK293 cells stably expressing the WT IL-5R (Fig. 9). After confirming significant knock-down of both JAKs (Fig. 9A, left panels), we examined the ubiquitination and phosphorylation status of βc by our standard IP/IB analysis before and after IL-5 stimulation (Fig. 9A, right panel). As expected, depletion of JAK2 and JAK1 expression completely abolished IL-5-induced βc tyrosine phosphorylation (Fig. 9A, right panel, lanes 4–6) due to lack of JAK kinase co-IP with βc immune complexes (Fig. 9A, right panels, lanes 4–6).
FIGURE 9.
βc ubiquitination is partially dependent on the presence of JAK2 and JAK1. A, HEK293 cells expressing WT IL-5Rs were transfected with both JAK1 and JAK2 siRNAs, or scrambled -C siRNAs for 48 h as described under “Experimental Procedures.” Whole cell lysates from unstimulated cells were analyzed by IB with the indicated Abs to determine knockdown efficiencies (left panel). Whole cell lysates from A and IL-5 stimulated cells (10 ng/ml, 1 h) were IP with anti-βc antibodies and IB with indicated Abs (right panel). Note how the absence of JAK1/2 partially reduces βc-Ub. B, cells from A were labeled with anti-βc-activated protein C antibodies and analyzed by flow cytometry. Note how JAK1/2 gene silencing slightly reduced βc cell surface expression (hatched histogram).
IB with anti-Ub antibodies revealed partially reduced IL-5-induced βc ubiquitination in the absence of JAK1/2 as compared with the scrambled siRNA control (Fig. 9A, right bottom panel, lanes 4–6). Conversely, the absence of JAK1/2 did not have much of an effect on basal βc ubiquitination (Fig. 9A, right bottom panel, lanes 1–3). We did, however, observe a slight decrease in βc cell surface expression by flow cytometry in the absence of JAK1/2 (Fig. 9B, hatched histogram), as well as in the βc IP (Fig. 9A, right panel, lane 6). It is possible that reduced βc protein levels might be contributing to the reduced appearance of βc ubiquitination.
In summary, JAK1 and JAK2 gene silencing partially prevented βc ubiquitination and its cell surface stability, consistent with other reports demonstrating that JAK expression influences plasma membrane stability (reviewed in Refs. 42, 47, and 48). Overall, these data support a model wherein JAK kinases are important players in optimal βc ubiquitination. However, the observation that the βc Box 1 deletion mutant as well as JAK gene silencing techniques still resulted in residual βc ubiquitination suggests that other ubiquitination mechanisms must be involved. For example, future studies investigating the role of βc serine phosphorylation in receptor ubiquitination should provide a more complete understanding of this process.
DISCUSSION
We previously showed that the cytoplasmic domain of the hematopoietic receptor, βc, which is shared by IL-5, IL-3, and GM-CSF, is ubiquitinated and degraded by the proteasomes in response to ligand stimulation by each of these three cytokines (4). Here, we provide new molecular details demonstrating that ubiquitination of the βc cytoplasmic tail is predominantly regulated by the presence of three critical lysine residues, Lys457, Lys461, and Lys467. Absence of these lysine residues significantly impaired JAK kinase binding to βc, which lead to defective receptor ubiquitination and signaling. Importantly, our study also demonstrated that βc ubiquitination regulates IL-5-stimulated βc endocytosis and turnover.
In general, it is well accepted that a canonical P-X-P motif in the Box 1 region of Type I and Type II cytokine receptors is necessary for JAK kinase binding because mutation of these proline residues leads to reduced binding and activation of the JAKs (37, 42–46). It was therefore quite unexpected to find that substitution of 3 lysine residues to arginine in the βc intracellular domain consistently resulted in severely impaired JAK kinase binding to the receptor even though the βc P-X-P motif was still present. Although our findings do not refute the necessity of the Box 1 P-X-P sequence for JAK kinase binding in Type I cytokine receptors, they do demonstrate that, for βc, the presence of a cluster of 3 lysine residues in the vicinity of the Box 1 region is critically important for optimal JAK binding.
Indeed, similar observations have been made with the IFNα receptor (IFNAR1) (49, 50). In one report, investigators described the role of a cluster of 2 lysine residues important for Tyk2 binding to IFNAR1 (49), whereas the second identified a different cluster of three lysines in the same receptor, which were critical for receptor ubiquitination, endocytosis, and turnover (50). Interestingly, the latter study demonstrated that ubiquitination of the three lysines resulted in the exposure of a linear endocytic motif that promoted IFNAR1 endocytosis (50).
Furthermore, examination of the IL-5Rα cytoplasmic domain revealed a similar cluster of three lysine residues (Lys370, Lys379, and Lys383) in the membrane proximal domain whose spatial distribution was very similar to βc. It will be interesting to see if mutation of some of these residues in the context of an intact proline-rich sequence will decrease JAK binding to IL-5Rα.
Currently, we do not have a clear understanding of how a cluster of 3 closely spaced intracellular lysines regulate JAK kinase binding, which leads to enhanced βc ubiquitination, but based on some molecular clues provided in our study, we can offer a few possibilities. First, and the most obvious possibility, is that because the critical lysines reside in and around the Box 1 motif, their substitution to arginine could significantly alter the structural configuration of this region and thus preclude JAK1 and JAK2 binding. Indeed, investigators have previously demonstrated for the EPO and gp130 receptors that their transmembrane domains consist of a rigid α-helical structure that extends into the intracellular (as well as extracellular) regions, and this structure provides the correct orientation for JAK2 association with the receptors (42, 51, 52). Based on this information, it is quite conceivable that substitution of the 3 lysines with arginines in this βc membrane proximal domain alters the orientation of the JAK/receptor interaction interface and thus inhibits JAK binding.
A second possibility could be that actual ubiquitination of these 3 lysine residues is required for the recruitment of JAK kinases or another unidentified protein, which facilitates JAK kinase recruitment. In this scenario, the βc ubiquitination machinery would bind normally at a distant site; however, substrate lysine residues would not be available for ubiquitination. A third possibility could be that these 3 lysines are part of a binding site for the βc ubiquitination machinery, and without their presence, the machinery cannot bind. Bioinformatics analysis of the βc proximal membrane region (around the Box 1 motif) did not identify any consensus ubiquitin ligase binding sites in this region. However, it is possible that the proper folding of this region with the presence of these 3 lysines is absolutely required for the ubiquitination machinery to bind at a distant site.
We are currently examining the role of candidate βc ubiquitin ligases whose binding sites have been identified in the βc cytoplasmic domain. However, it is well documented that JAK1 and JAK2 associate with suppressors of cytokine signaling 3 (SOCS3) and SOCS1 (53, 54), respectively, and it is well established that SOCS proteins are negative feedback inhibitors of cytokine receptor signaling (55). SOCS1/3 are reported to interact with JAK1/2 via a central Src homology 2 domain and then act as adaptors via their SOCS box for the cellular ubiquitination machinery involving the Cullin·Elongin B/C complex (53, 54). In this regard, it is tempting to speculate that in addition to ubiquitinating JAK1/2, the SOCS1/3·Elongin B/C ubiquitin ligase complex might also contribute to βc ubiquitination. We have not tested this possibility, but will certainly not rule it out.
Another significant finding from this study was the requirement of βc ubiquitination for its endocytosis, which then controls receptor subcellular distribution (Figs. 3 and 5). Indeed, studies with the glycine (GLYT 1b), glutamate, and dopamine transporters have reported similar findings (35, 56–58). Importantly, we also detected the presence of IL-5Rα and WT βc in recycling endosomes under basal conditions by a co-localization assay, whereas βc K16R-expressing cells lacked this pattern completely (Fig. 3, bottom panels). Together, these findings indicate that even before ligand binding, both receptor chains are in close proximity as they traffic in recycling endosomes. This spatially favorable environment might possibly promote higher order receptor aggregation as was shown for the GM-CSFR (59).
Because IL-5 plays a critical role in regulating eosinophil biology, identifying mechanisms that control IL-5 signaling are important for discovering new therapies to block disease progression. A recent study has identified βc as a relevant target for novel therapies to treat allergic inflammatory disorders (60). In this report, investigators demonstrated in a mouse model of allergic asthma that βc-deficient mice had a significant reduction in the allergen-induced expansion and accumulation of eosinophils in the lung resulting in decreased tissue damage. Moreover, our data show that optimal IL-5-mediated signaling requires JAK kinase binding to βc via the presence of 3 critical lysine residues. Because the GM-CSF and IL-3 receptors share βc for signaling (61), and both require JAK1 and JAK2 activity, it will be interesting to see if these receptor systems require the presence of the first 3 βc intracellular lysine residues. If they do, then designing small molecular inhibitors or peptides with the ability to mask these residues for JAK kinase binding could be a feasible approach to block proinflammatory signals elicited by βc-engaging cytokines.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI063178 from the NIAID (to M. M.-M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
J. T. Lei, T. Mazumdar, and M. Martinez-Moczygemba, unpublished observations.
- IL-5R
- IL-5 receptor
- βIP
- βc intracytoplasmic proteolysis
- βc
- β chain
- Ub
- ubiquitin
- MFI
- mean fluorescence intensity
- TFR-R
- transferrin receptor
- IB
- immunoblot
- IP
- immunoprecipitation
- SOCS
- suppressors of cytokine signaling
- PE
- phosphatidylethanolamine
- βc
- common beta chain.
REFERENCES
- 1. Hogan S. P., Rosenberg H. F., Moqbel R., Phipps S., Foster P. S., Lacy P., Kay A. B., Rothenberg M. E. (2008) Clin. Exp. Allergy 38, 709–750 [DOI] [PubMed] [Google Scholar]
- 2. Blanchard C., Rothenberg M. E. (2009) Adv. Immunol. 101, 81–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tavernier J., Devos R., Cornelis S., Tuypens T., Van der Heyden J., Fiers W., Plaetinck G. (1991) Cell 66, 1175–1184 [DOI] [PubMed] [Google Scholar]
- 4. Martinez-Moczygemba M., Huston D. P. (2001) J. Clin. Invest. 108, 1797–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 6. Hicke L. (2001) Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 7. Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 8. Pickart C. M. (2004) Cell 116, 181–190 [DOI] [PubMed] [Google Scholar]
- 9. Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. (2003) Nat. Cell Biol. 5, 461–466 [DOI] [PubMed] [Google Scholar]
- 10. Haglund K., Di Fiore P. P., Dikic I. (2003) Trends Biochem. Sci. 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 11. Pickart C. M., Fushman D. (2004) Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 12. Bonifacino J. S., Weissman A. M. (1998) Annu. Rev. Cell Dev. Biol. 14, 19–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hicke L., Dunn R. (2003) Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 14. Miranda M., Sorkin A. (2007) Mol. Interv. 7, 157–167 [DOI] [PubMed] [Google Scholar]
- 15. Acconcia F., Sigismund S., Polo S. (2009) Exp. Cell Res. 315, 1610–1618 [DOI] [PubMed] [Google Scholar]
- 16. Govers R., ten Broeke T., van Kerkhof P., Schwartz A. L., Strous G. J. (1999) EMBO J. 18, 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Kerhof P., Sachse M., Klumperman J., Strous G. J. (2001) J. Biol. Chem. 276, 3778–3784 [DOI] [PubMed] [Google Scholar]
- 18. Geetha T., Jiang J., Wooten M. W. (2005) Mol. Cell 20, 301–312 [DOI] [PubMed] [Google Scholar]
- 19. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 20. Varghese B., Barriere H., Carbone C. J., Banerjee A., Swaminathan G., Plotnikov A., Xu P., Peng J., Goffin V., Lukacs G. L., Fuchs S. Y. (2008) Mol. Cell Biol. 28, 5275–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ceresa B. P., Schmid S. L. (2000) Curr. Opin. Cell Biol. 12, 204–210 [DOI] [PubMed] [Google Scholar]
- 22. González-Gaitán M. (2003) Nat. Rev. Mol. Cell Biol. 4, 213–224 [DOI] [PubMed] [Google Scholar]
- 23. Polo S., Di Fiore P. P. (2006) Cell 124, 897–900 [DOI] [PubMed] [Google Scholar]
- 24. Sorkin A., Goh L. K. (2008) Exp. Cell Res. 314, 3093–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sorkin A., von Zastrow M. (2009) Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scita G., Di Fiore P. P. (2010) Nature 463, 464–473 [DOI] [PubMed] [Google Scholar]
- 27. Finger E. C., Lee N. Y., You H. J., Blobe G. C. (2008) J. Biol. Chem. 283, 34808–34818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krantz D. E., Waites C., Oorschot V., Liu Y., Wilson R. I., Tan P. K., Klumperman J., Edwards R. H. (2000) J. Cell Biol. 149, 379–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paing M. M., Temple B. R., Trejo J. (2004) J. Biol. Chem. 279, 21938–21947 [DOI] [PubMed] [Google Scholar]
- 30. de Pablo Y., Pérez-García M. J., Georgieva M. V., Sanchis D., Lindqvist N., Soler R. M., Comella J. X., Llovera M. (2008) J. Neurochem. 104, 124–139 [DOI] [PubMed] [Google Scholar]
- 31. Lei J. T., Martinez-Moczygemba M. (2008) J. Leukocyte Biol. 84, 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ragimbeau J., Dondi E., Alcover A., Eid P., Uzé G., Pellegrini S. (2003) EMBO J. 22, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martinez-Moczygemba M., Huston D. P., Lei J. T. (2007) J. Leukocyte Biol. 81, 1137–1148 [DOI] [PubMed] [Google Scholar]
- 34. Liu L. Y., Sedgwick J. B., Bates M.E., Vrtis R. F., Gern J. E., Kita H., Jarjour N. N., Busse W. W., Kelly E. A. (2002) J. Immunol. 169, 6459–6466 [DOI] [PubMed] [Google Scholar]
- 35. Haugsten E. M., Malecki J., Bjørklund S. M., Olsnes S., Wesche J. (2008) Mol. Biol. Cell 19, 3390–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fernández-Sánchez E., Martínez-Villarreal J., Giménez C., Zafra F. (2009) J. Biol. Chem. 284, 19482–19492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miyajima A., Kitamura T., Harada N., Yokota T., Arai K. (1992) Annu. Rev. Immunol. 10, 295–331 [DOI] [PubMed] [Google Scholar]
- 38. Ogata N., Kouro T., Yamada A., Koike M., Hanai N., Ishikawa T., Takatsu K. (1998) Blood 91, 2264–2271 [PubMed] [Google Scholar]
- 39. Okuda K., Foster R., Griffin J. D. (1999) Ann. N.Y. Acad. Sci. 872, 305–312; discussion 312–313 [DOI] [PubMed] [Google Scholar]
- 40. Kouro T., Takatsu K. (2009) Int. Immunol. 21, 1303–1309 [DOI] [PubMed] [Google Scholar]
- 41. Hansen G., Hercus T. R., McClure B. J., Stomski F. C., Dottore M., Powell J., Ramshaw H., Woodcock J. M., Xu Y., Guthridge M., McKinstry W. J., Lopez A. F., Parker M. W. (2008) Cell 134, 496–507 [DOI] [PubMed] [Google Scholar]
- 42. Haan C., Kreis S., Margue C., Behrmann I. (2006) Biochem. Pharmacol. 72, 1538–1546 [DOI] [PubMed] [Google Scholar]
- 43. Kouro T., Kikuchi Y., Kanazawa H., Hirokawa K., Harada N., Shiiba M., Wakao H., Takaki S., Takatsu K. (1996) Int. Immunol. 8, 237–245 [DOI] [PubMed] [Google Scholar]
- 44. Alves dos Santos C. M., van Kerkhof P., Strous G. J. (2001) J. Biol. Chem. 276, 10839–10846 [DOI] [PubMed] [Google Scholar]
- 45. Pelletier S., Gingras S., Funakoshi-Tago M., Howell S., Ihle J. N. (2006) Mol. Cell Biol. 26, 8527–8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haan C., Heinrich P. C., Behrmann I. (2002) Biochem. J. 361, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He K., Loesch K., Cowan J. W., Li X., Deng L., Wang X., Jiang J., Frank S. J. (2005) Endocrinology 146, 4755–4765 [DOI] [PubMed] [Google Scholar]
- 48. Meenhuis A., Irandoust M., Wölfler A., Roovers O., Valkhof M., Touw I. P. (2009) Biochem. J. 417, 737–746 [DOI] [PubMed] [Google Scholar]
- 49. Yan H., Krishnan K., Lim J. T., Contillo L. G., Krolewski J. J. (1996) Mol. Cell. Biol. 16, 2074–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kumar K. G., Barriere H., Carbone C. J., Liu J., Swaminathan G., Xu P., Li Y., Baker D. P., Peng J., Lukacs G. L., Fuchs S. Y. (2007) J. Cell Biol. 179, 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Constantinescu S. N., Huang L. J., Nam H., Lodish H. F. (2001) Mol. Cell 7, 377–385 [DOI] [PubMed] [Google Scholar]
- 52. Greiser J. S., Stross C., Heinrich P. C., Behrmann I., Hermanns H. M. (2002) J. Biol. Chem. 277, 26959–26965 [DOI] [PubMed] [Google Scholar]
- 53. Ungureanu D., Saharinen P., Junttila I., Hilton D. J., Silvennoinen O. (2002) Mol. Cell. Biol. 22, 3316–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boyle K., Zhang J. G., Nicholson S. E., Trounson E., Babon J. J., McManus E. J., Nicola N. A., Robb L. (2009) Cell Signal. 21, 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Piessevaux J., Lavens D., Peelman F., Tavernier J. (2008) Cytokine Growth Factor Rev. 19, 371–381 [DOI] [PubMed] [Google Scholar]
- 56. Hanyaloglu A. C., von Zastrow M. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 537–568 [DOI] [PubMed] [Google Scholar]
- 57. Sorkina T., Hoover B. R., Zahniser N. R., Sorkin A. (2005) Traffic 6, 157–170 [DOI] [PubMed] [Google Scholar]
- 58. González-González I. M., García-Tardón N., Giménez C., Zafra F. (2008) Glia 56, 963–974 [DOI] [PubMed] [Google Scholar]
- 59. Hercus T. R., Thomas D., Guthridge M. A., Ekert P. G., King-Scott J., Parker M. W., Lopez A. F. (2009) Blood 114, 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Asquith K. L., Ramshaw H. S., Hansbro P. M., Beagley K. W., Lopez A. F., Foster P. S. (2008) J. Immunol. 180, 1199–1206 [DOI] [PubMed] [Google Scholar]
- 61. Martinez-Moczygemba M., Huston D. P. (2003) J. Allergy Clin. Immunol. 112, 653–665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.