Abstract
E. coli 70S ribosomes tightly bind 8 equivalents of Zn(II), and EXAFS spectra indicate that Zn(II) may be protein-bound. Ribosomes were incubated with EDTA and Zn(II), and after dialysis, the resulting ribosomes bound 5 and 11 Zn(II) equivalents, respectively. EXAFS studies show that the additional Zn(II) in the zinc-supplemented ribosomes bind in part to the phosphate backbone of ribosome. Lastly, in vitro translation studies demonstrate that EDTA-treated ribosomes do not synthesize an active Zn(II)-metalloenzyme, while the as-isolated ribosomes do. These studies demonstrate that the majority of intracellular Zn(II) resides in the ribosome.
Keywords: E. coli, ribosomes, zinc
Zinc is an essential transition metal, required for life in all organisms2. It plays key catalytic roles in enzymes from all six major classes3, as well as a structural role in numerous transcriptional activators and regulators4, 5. While Zn(II) import and export is well understood2, 6–12, surprisingly little is known about the fate of intracellular zinc. The free Zn(II) concentration within a cell has been estimated to be in the femtomolar range, while the total cellular concentration has been established as approximately 200 μM13. However, only about 12% of the cellular Zn(II) has been accounted for as bound to Zn(II) metalloproteins13, leaving open the question as to where the remaining Zn(II) resides in the cell.
Previous studies have suggested that Zn(II) is associated with the ribosome. Atomic absorption spectroscopy of E. coli 70S ribosomes revealed 2 equivalents of bound Zn(II)14, while a PAR assay of ribosomes from B. subtilis indicated 2.5 eq of closely-associated Zn(II)15. Thus, while an association of zinc with the ribosome has been indicated previously, the amount of metal present in active ribosomes has not been accurately determined. Further, it has yet to be established whether ribosomal Zn(II) remains associated with the ribosome at all times or is labile. We report here biophysical studies that show strong and weak binding Zn(II) to the ribosome.
E. coli 70S ribosomes were isolated and quantified as described previously16. To verify that the isolated ribosomes were intact and functional, in vitro transcription/translation assays were performed using the PURESYSTEM Classic II mini alpha kit17 and plasmid pUB583018, which contains the gene for metallo-β-lactamase L1, which binds two Zn(II) ions18, from Stenotrophomonas maltophilia. After in vitro transcription/translation, the reaction mixture was assayed using nitrocefin as substrate, which indicated the production of 9.7 ± 1.9 μg of L1. Control reactions, conducted in the absence of ribosomes, did not generate metallo-β-lactamase activity, demonstrating the viability of some of the isolated ribosomes. In vitro transcription/translation experiments using the kit-provided, non-Zn(II) binding dihydrofolate reductase (DHFR) gene from E. coli were also conducted (see Supporting Information). These assays revealed that 0.072 ± 0.014 μg of DHFR were produced. Using the ICP-MS of the purified E. coli 70S ribosomes showed 8 equivalents of Zn(II) (Table 1). Only trace amounts of other transition metal ions, such as Co, Cu, Mn, Ni, and Fe, were present.
Table 1.
Metal content of E. coli 70S ribosomes.
| samples | Zn(II) content (eq/ribosome) | other metals (Co,Cu,Mn,Ni,Fe) |
|---|---|---|
| as-isolated | 7.9 ± 0.1 | 0.5 ± 0.3 |
| Zn(II)-supplemented | 11 ± 1 | 0.8 ± 0.3 |
| Zn(II)-depleted | 5.0 ± 0.1 | 0.8 ± 0.3 |
To examine the minimum and maximum Zn(II) content of the E. coli 70S ribosomes, ribosome samples were incubated with up to 100 eq of Zn(II) to fully populate weak binding sites, or up to 40 eq of EDTA to fully de-populate them, followed by exhaustive dialysis against lysis buffer (see Supporting Information). The concentration of Mg(II) was maintained at 10 mM throughout the incubation and dialysis steps and also during all in vitro transcription/translation assays since previous studies have suggested that ribosomes do not “fall apart” as long as the Mg(II) concentration does not fall under 5–7 mM19–21. The Zn(II)-supplemented ribosomes bound 11 eq of Zn(II), while the EDTA-treated ribosomes were found to contain only 5 eq of Zn(II) (Table 1), indicating 5 tight-binding Zn(II) sites and up to 6 weaker Zn(II) binding sites.
In vitro transcription/translation reactions were conducted on the Zn(II)-supplemented and EDTA-treated ribosomes, which were subjected to similar dialysis conditions, to determine if the treated ribosomes were active. Reactions with Zn(II)-supplemented ribosomes produced 5.4 ± 1.1 μg of L1 and 0.062 ± 0.012 μg of DHFR. EDTA-treated ribosomes produced no detectable L1 but produced 3.5 ± 0.6 μg of DHFR. The higher amounts of DHFR, as determined from activity assays, produced in the EDTA-treated reactions is probably due to inhibition of DHFR activity by mono- and divalent metal cations22. The production of no L1 in EDTA-treated reactions suggests a role of Zn(II) in the in vitro transcription/translation of this Zn(II)-metalloenzyme. Future studies will further address the role of Zn(II) in ribosome activity; nonetheless, these studies demonstrate that the EDTA/Zn(II) treatments and dialysis steps did not result in inactive ribosomes.
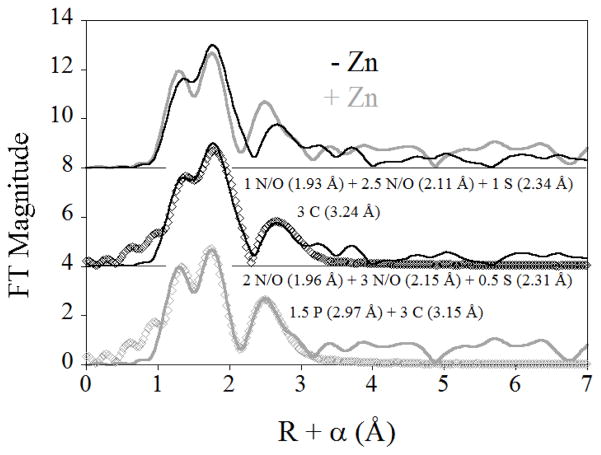
The nature of the Zn(II) binding sites were investigated by acquiring EXAFS spectra for Zn(II)-supplemented and EDTA-treated ribosomes (labeled as +Zn and −Zn in Figure 1, respectively). The two samples showed similar spectra (Figure 1, top), with the main peaks in their Fourier transforms nearly superimposable. Both samples show a single prominent outer shell feature, which moves to lower R in the Zn(II)-supplemented data. Fits to the data for EDTA-treated ribosomes, representing the average of the tight binding sites, indicate a primary coordination sphere of 1 N/O donor at 1.93 Å, 3 N/O donors at 2.11 Å, and 1 sulfur donor at 2.34 Å. The outer shell feature is best modeled as a shell of 3 carbon scatterers at 3.25 Å, presumably from carboxylate ligands (Figure 1, center, and Figure S1). Any attempt to model this feature with phosphorus, as might be anticipated if the ribosomal Zn(II) were interacting with the phosphate backbone of the ribosome’s constitutive RNA, was unsuccessful. In comparison, the EXAFS data for Zn(II)-supplemented ribosomes were best fit with a similar first shell of 2 N/O ligands at 1.96 Å, 3 N/O ligands at 2.14 Å and 0.5 sulfur donors at 2.29 Å. The outer shell feature, which shifts to R + α = 2.5 Å (from 2.9 Å in the Zn(II)-depleted data), is best modeled with a mixture of 3 carbon scatterers at 3.15 Å and 1.5 phosphorous at 2.97 Å (Figure 1, bottom, and Figure S2). Both contributions appear warranted, as inclusion of the carbon shell alone leads to a 3-fold improvement in fit residual, while inclusion of the phosphorus shell alone results in more than 4-fold improvement in the fit. Inclusion of both leads to a nearly 9-fold reduction in fit residual, and their refined distances are separated by more than the 0.16 Å resolution of the data. Thus, analysis of the XAS data indicates that the Zn(II) that is tightly associated with the ribosome (EDTA-treated) is most likely protein bound although we cannot rule out Zn(II) binding to non phosphate ligands of RNA that are buried or regions of interface between protein and RNA non phosphate ligands. However, there is at least some fraction of the more loosely associated Zn(II) (Zn(II)-supplemented), which interacts directly with the phosphate backbone of RNA.
Figure 1.
Comparison of the EXAFS Fourier transforms for Zn(II)-depleted (−Zn, solid black line) and Zn(II)-supplemented (+Zn, solid gray line) E. coli ribosomes (top), and the corresponding best fits (open symbols, see Table S1 for details).
The number of ribosomes present in an E. coli cell is, not surprisingly, dependent upon its growth stage. The number of ribosomes present has been estimated to fluctuate from 2,000 per cell at low growth to 70,000 at rapid growth23. Given the average volume of an E. coli cell, approximately 1.8 femtoliters,13 the present studies allow the total ribosomal Zn(II) content to be estimated. Using the as-isolated value of 8 equivalents of Zn(II) per ribosome, the concentration of Zn(II) contained within ribosomes is on average 20 μM at low growth, and 0.52 mM under rapid growth conditions. Estimates of total cellular Zn(II) range from 0.2 mM to 0.8 mM13, 24. Thus, the present studies suggest that a large portion of cellular Zn(II) is contained in the ribosome.
Previous studies have suggested as many as eight ribosomal proteins that are capable of binding Zn(II) (20, 22). The B. subtilis ribosomal protein L31 has clearly been shown to bind Zn(II)25. The solution structure of ribosomal protein L36 from T. thermophilus revealed a Zn(II)-ribbon-like fold26, suggesting it, too, may bind Zn(II). However, none of the available crystal structures show Zn(II) bound to L31 (Hensley, Crowder, unpublished results). Further proteomic studies have suggested that E. coli ribosomal proteins L2, L13, S2, S15, S16, and S17 could bind Zn(II) (Figure 2)27. None of these proteins are in close proximity to the peptide exit site and none, except L3628, have been shown to bind Zn(II) in current crystal structures of ribosomes (Hensley, Crowder, unpublished results). This result may be due to the procedure used to purify ribosomes16, which contains EDTA, or to misidentification of Zn(II) as a Mg(II) in some of the early crystal structures. Our finding that 8 Zn(II) are bound to the ribosome under normal conditions indicates that perhaps all of the proteomics-identified proteins bind Zn(II).
Figure 2.
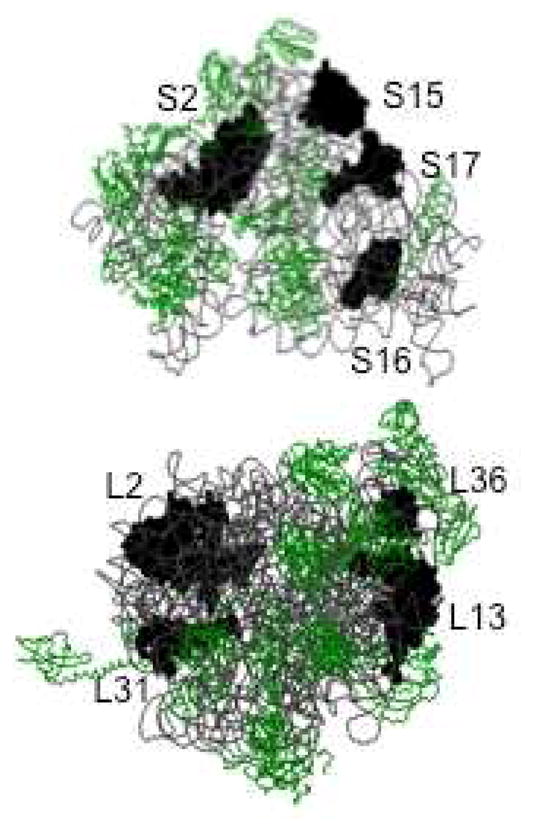
Structure of E. coli 30S (top) and 50S (bottom) ribosome with potential Zn(II) binding proteins labeled. This figure was rendered with Raswin using Protein Databank coordinates 2AVY and 2AW41.
The current report supports the possible presence of four Zn(II) binding proteins in the large ribosomal subunit of the 70S E. coli ribosome (L36, L31, L13, and L2) and four Zn(II) binding proteins in the small subunit (S17, S16, S15, and S2) (Figure 2). Ribosome-associated Zn(II) identified in this study could possibly serve in a structural (non-catalytic) role for the ribosomal proteins. Further study on these potential Zn(II) binding proteins is required to validate that these proteins bind Zn(II). Currently, investigations by our group into the metal binding capabilities of recombinant E. coli L36, L31, and L13 have indicated all these proteins are able to bind Zn(II) (M. P. Hensley and M. W. Crowder, unpublished). Future studies will focus identifying the remaining Zn(II)-binding, ribosomal proteins and probing their physiological role(s).
Supplementary Material
Acknowledgments
The authors wish to thank Dr. Gary Janssen, Jacqueline Giliberti, and Racheal Desmone, for their assistance in ribosome isolation and handling. This work was supported by the National Institutes of Health (GM079411 (MWC) and GM093987 (MWC/DLT)). The XAS measurements were made possible by the Case Center for Synchrotron Biosciences, grant P30-EB-009998, from the National Institute of Biomedical Imaging and Bioengineering (NIBIB).
Footnotes
Supporting Information Available: Detailed experimental procedures and a description of the XAS data analysis, including one Figure and one Table, are provided as Supplementary Material. This material is available free of charge at http://pubs.acs.org.
References
- 1.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the Bacterial Ribosome at 3.5 Å Resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 2.Hantke K. Bacterial zinc uptake and regulators. Curr Opin Microbiol. 2005;8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Vallee BL, Auld DS. Zinc Metallochemistry in Biochemistry. In: Joiles P, Jornvall H, editors. Interface Between Chemistry and Biochemistry. Birkhauser Verlag; Basel, Switzerland: 1995. pp. 259–277. [Google Scholar]
- 4.Andreini C, Banci L, Bertini I, Rosato A. Zinc throughout the three domains of life. J Proteome Res. 2006;5:3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- 5.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 6.Grass G, Fan B, Rosen BP, Franke S, Nies DH, Rensing C. ZitB (YbgR), a Member of the Cation Diffusion Facilitator Family, is an Additional Zinc Transporter in Escherichia coli. J Bact. 2001;183:4664–4667. doi: 10.1128/JB.183.15.4664-4667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C, Wong MD, Rosen BP, Smith RL. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum ZupT is a Zn(II) uptake system in Escherichia coli. J Bacteriol. 2005;187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu M, Fu D. Structure of the zinc transporter YiiP. Science. 2007;317:1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- 9.Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol. 1999;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 10.Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. ZupT is a Zn(II) Uptake System in Escherichia coli. J Bact. 2002;184:864–866. doi: 10.1128/JB.184.3.864-866.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patzer SI, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 12.Patzer SI, Hantke K. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem. 2000;275:24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- 13.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 14.Chan Y, Suzuki K, Olvera J, Wool IG. Zinc finger-like motifs in rat ribosomal proteins S27 and S29. Nucleic Acid Res. 1993;21:649–655. doi: 10.1093/nar/21.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabriel SE, Helmann JD. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bacteriol. 2009;191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spedding G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes, and organelles. In: Spedding G, editor. Ribosomes and protein synthesis: a practical approach. Oxford University Press; New York, NY: 1990. pp. 1–29. [Google Scholar]
- 17.Shimizu Y, Kuruma Y, Ying BW, Umekage S, Ueda T. Cell-free translation systems for protein engineering. FEBS J. 2006;273:4133–4140. doi: 10.1111/j.1742-4658.2006.05431.x. [DOI] [PubMed] [Google Scholar]
- 18.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. Overexpression, Purification, and Characterization of the Cloned Metallo-b-Lactamase L1 from Stenotrophomonas maltophilia. Antimicrobial Agents and Chemotherapy. 1998;42:921–926. doi: 10.1128/aac.42.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King TC, Rucinsky T, Schlessinger D, Milanovich F. Escherichia coli ribosome unfolding in low Mg2+ solutions observed by laser Raman spectroscopy and electron microscopy. Nucl Acids Res. 1980;9:647–661. doi: 10.1093/nar/9.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazawa F, Dick VC, Tamaoki T. Reversible dissociation of Escherichia coli ribosomes by nitrogen mustard. Biochim Biophys Acta. 1968;155:193–201. doi: 10.1016/0005-2787(68)90349-3. [DOI] [PubMed] [Google Scholar]
- 21.Weller DL, Shechter Y, Musgrave D, Rougvie M, Horowitz J. Conformational changes in Escherichia coli ribosomes at low magnesium ion concentrations. Biochemistry. 1968;7:3668–3675. doi: 10.1021/bi00850a046. [DOI] [PubMed] [Google Scholar]
- 22.Baccanari D, Phillips A, Smith SO, Sinski D, Burchall J. Purification and properties of Escherichia coli dihydrofolate reductase. Biochemistry. 1975;14:5267–5273. doi: 10.1021/bi00695a006. [DOI] [PubMed] [Google Scholar]
- 23.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eide DJ. Zinc transporters and the cellular trafficking of zinc. Biochim Biophys Acta. 2006;1763:711–722. doi: 10.1016/j.bbamcr.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, Kudo T, Kobayashi K, Ogasawara N, Park SM, Ochi K, Kawamura F. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol Microbiol. 2004;52:273–283. doi: 10.1111/j.1365-2958.2003.03972.x. [DOI] [PubMed] [Google Scholar]
- 26.Hard T, Rak A, Allard P, Kloo L, Garber M. The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc ribbon like fold. J Mol Biol. 2000;296:169–180. doi: 10.1006/jmbi.1999.3433. [DOI] [PubMed] [Google Scholar]
- 27.Katayama A, Tsujii A, Wada A, Nishino T, Ishihama A. Systematic search for zinc-binding proteins in Escherichia coli. Eur J Biochem. 2002;269:2403–2413. doi: 10.1046/j.1432-1033.2002.02900.x. [DOI] [PubMed] [Google Scholar]
- 28.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.