Abstract
14-3-3σ plays a regulatory role in epidermal epithelial differentiation and loss of 14-3-3σ leads to increased proliferation and impaired differentiation. A tumor suppressor function for 14-3-3σ has been proposed based on the fact that some epithelial-derived tumors lose 14-3-3σ expression. p63, a p53 family member, is a master regulator of epidermal epithelial proliferation and differentiation and is necessary for the epidermal development. The function of p63 in tumorigenesis is still controversial and poorly defined as multiple isoforms have been found to play either collaborative or opposing roles. By using ‘repeated epilation’ heterozygous (Er/+) mice containing a dominant-negative 14-3-3σ mutation, the functional relationship of p63 with 14-3-3σ in epidermal proliferation, differentiation and tumorigenesis was investigated. It was found that p63, particularly the ΔNp63α isoform, was strongly expressed in 14-3-3σ-deficient keratinocytes and knockdown of p63 remarkably inhibited proliferation in these cells. To study the functional roles of 14-3-3σ and p63 in epidermal tumorigenesis, we adopted a 7,12-dimethylbenzanthracene/12-O-tetradecanoyl-phorbol-13-acetate (DMBA/TPA) two-stage tumorigenesis procedure to induce formation of skin papillomas and squamous cell carcinomas in Er/+ mice and identified strong p63 expression in resultant tumors. The loss of one allele of p63 caused by the generation of Er/+/p63+/− double compound mice decreased the sensitivity to DMBA-/TPA-induced tumorigenesis as compared with Er/+ mice. This study shows that p63 and 14-3-3σ play opposing roles in the development of skin tumors and that the accumulation of p63 is essential for Ras/14-3-3σ mutation-induced papilloma formation and squamous cell carcinoma carcinogenesis.
Introduction
14-3-3σ belongs to a conserved family of regulatory proteins composed of seven isoforms. They are able to form soluble hetero- or homodimers that bind to functionally diversified multiple targets and mediate a wide range of cellular activities (1–3). Contrary to other family members, 14-3-3σ is limited to the stratified squamous epithelial cells and regulates their proliferation and differentiation. 14-3-3σ is functionally impaired in the repeated epilation mouse (Er/Er), which blocks epidermal differentiation during development (4,5). This protein is also known as a tumor suppressor, and loss of its expression due to gene promoter hypermethylation is frequently associated with epithelial-derived tumors (6,7). In response to DNA damage, 14-3-3σ is transcriptionally induced in a p53-dependent manner, and the increased 14-3-3σ protein subsequently causes cell cycle arrest at the G2/M checkpoint (8,9).
p63, a master regulator of epidermal cell proliferation, differentiation and maintenance, is expressed exclusively in the basal cells of the stratified epithelium. p63 null mice almost completely lose the epidermal epithelium (10,11). Despite the sequence similarity with the best-characterized tumor suppressor, p53, the function of p63 in tumorigenesis remains controversial. Two distinct promoters produce two types of p63 isoforms, e.g. TAp63 and ΔNp63, which either possesses or lacks the N-terminal transactivating domain, respectively. Alternative splicing at the C-terminus creates three distinct variants (α, β and γ) of TAp63 and ΔNp63. ΔNp63α is the predominant isoform expressed in epidermis and regarded as dominant negative since it counteracts the functions of TAp53 and TAp63 by competitively binding to the p53-response elements of their targeting genes. ΔNp63α can regulate cell proliferation by directly activating or repressing its target genes, such as 14-3-3σ, p21, p16 and p19 (12–14). Although p63 is rarely mutated, the ΔNp63α is frequently induced in cancer. Studies in tumorigenesis in p63+/− mice generated conflicting results and the discrepancies among these studies have not been solved so far (15,16). As such, the role of p63 in tumorigenesis needs further investigation.
We hypothesize that p63 and 14-3-3σ play opposite functions in epidermal epithelial proliferation, differentiation and tumorigenesis. The present study investigated the expression of p63 in the 14-3-3σ-mutated skin and keratinocytes and revealed that p63, specifically the ΔNp63α isoform were upregulated. We found that p63 was essential for maintenance of the keratinocyte proliferation. ΔNp63 isoform was also dramatically increased in the tumors induced with the 7,12-dimethylbenzanthracene (DMBA)/12-O-tetradecanoyl-phorbol-13-acetate (TPA) treatment, more significantly, the deletion of one p63 allele in Er/+ mice reduced tumor formation. These data suggest that p63 functions as an oncogene in the tumors induced by Ras/14-3-3σ mutation. Our data emphasize the importance of the 14-3-3σ and p63 interaction in keratinocyte homeostasis and tumorigenesis.
Materials and methods
Animals
Er/+ mutant mice in a mixed genetic background of C57BL/6J and CBA/CaGnLeJ (Stock #000515), the p63 heterozygous mutate mice in the C57BL/6J background (Stock # 003568) and the p53 heterozygous mutate mice in a C57BL/6J background (Stock # 002101) were purchased from the Jackson Laboratory, Bar Harbor, ME. The six strains used in the DMBA/TPA induction experiments are WT, Er/+, p53+/−, p63+/−, Er/+/p63+/− and Er/+/p53+/−. The double-mutant heterozygous mice were generated in our laboratory from the cross mating of the Er/+ mice with either p63+/− for Er/+/p63+/− mice or with p53+/− for Er/+/p53+/− mice. The Er/+ mice were genotyped based on the hairless phenotype and confirmed by western blot analysis of 14-3-3σ expression. Both p63+/− and p53+/− mice were genotyped by polymerase chain reaction based on the protocol provided by the Jackson Laboratory. Experimental animals were housed under pathogen-free conditions and handled in accordance with the guidelines approved by the Institutional Animal Care and Use Committee of the University of Louisville.
The Er/Er homozygous mutants were generated from Er/+ heterozygous mice intercrossing. To label cell proliferation in embryos, the 5-bromo-2′-deoxyuridine (BrdU) was injected intraperitoneally into the pregnant mice at dose of 150 mg/kg body wt, 2h prior to collection of the embryos at embryonic day 18.5 (E18.5). The embryo tails were saved and processed for protein preparation for western analysis of the 14-3-3σ genotypes (5), and the rest of the embryos were fixed in 4% paraformaldehyde (PFA) at 4°C for overnight.
Two-stage DMBA/TPA induction of carcinogenesis
About 100 nmol of DMBA (cat # D3254; Sigma, St Louis, MO) in 0.2 ml acetone was topically applied to the shaved dorsal skins to induce DNA mutation. One week later, 17 nmol TPA (cat # P8139; Sigma) in 0.2 ml acetone was administered topically to the same area of the shaved dorsal skin with three administrations per week (Monday, Wednesday and Friday) for 19 weeks. Eight weeks after stop of TPA treatment, the tumors were dissected and processed for histopathological analysis. Mice were evaluated weekly for papilloma development. Only tumors that had attained a size of ≥1 mm were counted.
Histology and immunostaining
All tumors and control skins were dissected from euthanized mice and fixed immediately in 4% PFA at 4°C for overnight and then subjected to paraffin embedding and sectioning for histological studies. A small portion of tissue from each sample was snap frozen in liquid nitrogen for DNA isolation before PFA fix. Seven micrometers of paraffin-embedded sections was cut and deparaffinized prior to staining with hematoxylin and eosin. For most of the immunostaining, the tissue sections or cultured cell preparation were subjected to antigen-retrieving procedure by heating the slides at 95°C for 30 min in 10 mM Tris–ethylendiaminetetraacetic acid buffer (pH 9.0). The primary antibodies used in this study were mouse anti-p63 (1:200, cat #sc-8431; Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-ΔNp63 (1:200, cat #sc-8609; Santa Cruz Biotechnology), rabbit anti- p63α (1:200, cat #sc-8344; Santa Cruz Biotechnology), goat anti- TAp63 (1:100, cat #sc-8608, Santa Cruz Biotechnology), goat anti-C terminus of 14-3-3σ (1:200, cat # sc-7683; Santa Cruz Biotechnology), rat anti-BrdU (1:800, MAS 250c; Harlan-Sera Lab, Belton Loughborough, UK), rabbit anti-zonula occludens-1 (1:600, cat #61-7300; Zymed Laboratories, San Francisco, CA), mouse anti-proliferating cell nuclear antigen (1:200, cat #180110; Invitrogen, Carlsbad, CA), chicken anti-green fluorescent protein (GFP) (1:200, cat #GFP-1020; Aves Lab, Tigard, OR) and rabbit anti-filaggrin (1:500, PRB-417P; Covance Research Products, Denver, PA). The secondary antibodies conjugated with either carbocyanine 3 (Cy3) or fluorescein isothiocyanate were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). The horseradish peroxidase-conjugated secondary antibody was visualized with diaminobenzidene substrate following the manufacturer’s instructions (Cat # SK-4600; Vector Labs, Burlingame, CA).
Keratinocyte culture
Preparation and culture of the keratinocytes followed the published procedure (17). Briefly, the full thickness of skin was taken from the E18.5 embryos of Er/+ intercrosses and digested overnight at 4°C with 0.25% trypsin. The Er/Er keratinocytes were directly scraped from the outside surface of the Er/Er skin after overnight trypsin digestion. For preparation of the wild-type (WT) keratinocytes, the epidermal layer was first peeled from dermis and the keratinocytes were scrapped off from the inner surface of the WT epidermal sheet. Keratinocytes were cultured in Keratinocyte serum-free media (Invitrogen). For BrdU labeling, the cells were incubated in the medium containing 10 μM BrdU for 90 min and then fixed with 4% PFA for 15 min at room temperature before immunostaining with anti-BrdU antibody.
Calculation and statistical analysis of the BrdU- and p63-positive cell
The numbers of BrdU-positive, p63-positive and total 4′,6-diamidino-2-phenylindole (DAPI)-positive cells were counted from the digital photos taken at ×10 magnification from at least four sections in each group. The percentages of BrdU- or p63-positive cells per total cells (DAPI positive) were calculated. In p63 knockdown experiments, the percentages of the BrdU-positive cells in randomly selected 1000 of the total p63 small hairpin RNA (shRNA) lentivirus-transduced cells (labeled by coexpression of GFP) or in 1000 of uninfected cells (GFP-negative cells) were counted and calculated from three independent experiments. All data are shown as the average ± standard deviation (SD). Student's t-test was performed to determine the statistical difference. The differences were considered statistically significant if the P values were <0.05.
Lentiviral vector and viral production
The expression vectors encoding the p63 shRNA and scrambled control shRNA have been described previously (17). pPPTCMVΔNp63α is a lentiviral vector expressing ΔNp63α from a CMV promoter (kindly provided by Nobushige Tanaka). Lentiviruses were produced by a four plasmid (for third generation lentiviral vectors) transfection system as described by Tiscornia et al. (18).
RNA isolation and quantitative polymerase chain reaction
Primary cultured keratinocytes at 70–90% confluence were prepared at the indicated time points for RNA extraction using TRIzol reagent (Invitrogen). The A260/A280 ratio of all RNA samples was >2.0 as measured by Nanodrop. Double-stranded complementary DNA was reverse transcribed using random primers and SuperScript VILO cDNA synthesis kit (Invitrogen).
Real-time quantitative polymerase chain reaction (qPCR) was performed in an SYBR green-based polymerase chain reaction mixture on a MX3005p system (Agilent Technologies, Santa Clara, CA), with a program of a 10 min initial hot-start activation of Taq polymerase at 95°C, followed by 40 cycles of amplification (95°C for 25 s, 56°C for 30 s and 72°C for 30 s). The comparative Ct (threshold cycle) method normalized to β-actin was used to analyze relative changes in gene expression. The qPCR primer sequences for ΔNp63 and p63α have been published elsewhere (19,20).
Western blot analysis
Keratinocytes were lysed in cold radioimmunoprecipitation assay buffer (20 mM Tris–HCl, pH 7.4, 100 mM NaCl and 0.2% each of deoxycholate, Triton X-100 and Nonidet P-40) containing 1× complete protease-inhibitor mixture (Roche Diagnostics, Indianapolis, IN). Equal amounts of whole-cell lysates were separated on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. For immunoblotting, the mouse anti-p63 (1:200, cat #sc-8431; Santa Cruz Biotechnology), rabbit anti-filaggrin (1:500, PRB-417P; Covance Research Products, Denver, PA), goat anti-C terminus of 14-3-3σ (1:200, cat # sc-7683; Santa Cruz Biotechnology), and mouse anti-β-actin (1:1000, cat #A2228; Sigma) were used as primary antibodies. The horseradish peroxidase-conjugated goat anti-mouse secondary antibody and an enhanced chemiluminescence (ECL) system (Amersham Pharmacia, Piscataway, NJ) were used to visualize the signals.
Results
p63 expression is increased in the 14-3-3σ-deficient skin and keratinocytes
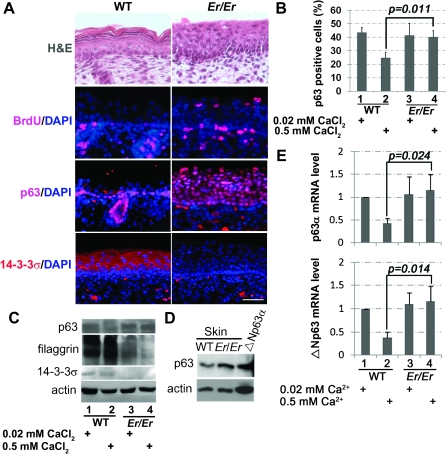
A single nucleotide insertion at the sfn allele (14-3-3σ gene) in the Er mouse produces a dominant negative, non-functional truncated protein (5,21). Er/Er skin lacking functional 14-3-3σ showed enhanced cell proliferation and defects in differentiation (5). BrdU incorporation assay showed that the keratinocyte proliferation was restricted to the basal layer of the epidermis in the normal E18.5 skin in vivo (Figure 1A). However, the BrdU-positive proliferating cells were expanded into the suprabasal layer in the 14-3-3σ-deficient epidermis (Figure 1A).
Fig. 1.
p63 expression is increased in 14-3-3σ-null keratinocytes. (A) Paraffin-embedded skin sections from WT and Er/Er E18.5 embryos were stained with hematoxylin and eosin or the specific antibodies against BrdU, p63 and 14-3-3σ as indicated. BrdU (red) labeled the proliferating cells in basal layer and hair follicles of both genotypes, but such BrdU-positive cells were also found in the suprabasal layer of the Er/Er skin. p63 immunostaining (red) was restricted to the epidermal basal progenitor cells in the WT control but extended to the suprabasal layer in Er/Er skin. 14-3-3σ immunostaining (red) was located in the suprabasal layer in the WT skin but lost in the Er/Er epidermis. DAPI (blue) was used for nuclear counterstaining. The scale bar represents 50 μm. (B) Quantification of p63-positive cells in the primary cultured keratinocytes isolated from E18.5 WT (1 and 2) or Er/Er (3 and 4) skins. The cells were cultured for 48 h in the medium containing 0.02 mM (maintenance medium, lanes 1 and 3) or 0.5 mM (differentiation medium, lanes 2 and 4) calcium. The p63-positive cells were immunostained with p63-specific antibody and calculated as percentage of total DAPI-labeled cells. The error bars represent the standard deviation from three independent experiments. Student's t-test was performed to determine the statistical difference. (C) Western blot analysis of protein lysates prepared from the primary WT and Er/Er keratinocytes using p63-specific, filaggrin-specific and 14-3-3σ-specific (C-18, recognizing only WT protein) antibodies. Actin was used as a loading control. Data were representative from three independent experiments. (D) Western blot analysis of protein lysates prepared from WT and Er/Er skin, 293T cells transfected with mouse Δp63α complementary DNA, using p63-specific antibody. Actin was used as a loading control. (E) Real-time qPCR analysis of ΔNp63 and p63α messenger RNAs in the WT and 14-3-3σ-null keratinocytes before (0.02 mM calcium) and after (0.5 mM calcium) differentiation induction. The error bars represent the standard deviation from three independent experiments.
Given the importance of p63 in keratinocyte proliferation and differentiation, we then asked whether p63 expression pattern was altered in the 14-3-3σ mutant epidermis in vivo. Antibody immunostaining revealed that both the basal and suprabasal layers in the 14-3-3σ-deficient epidermis were stained p63 positive, in contrast to the restricted basal layer staining in the WT control (Figure 1A). The primary WT mouse keratinocytes can be maintained at an undifferentiated stage when cultured in 0.02 mM of low calcium-containing medium and undergo differentiation when the calcium concentration in the culture medium is switched to 0.5 mM. This high calcium-induced keratinocyte differentiation was characterized by reduced p63-positive progenitor population (Figure 1B). However, keratinocytes isolated from the 14-3-3σ-deficient embryos still maintained a high percentage of the p63-positive population at the high-(differentiating)-calcium culture conditions (Figure 1B). Such increased p63 expression in the 14-3-3σ-deficient skin or in the keratinocytes at the high-calcium medium was further confirmed by western blot analysis (Figure 1C and D). Consistent with lacking of differentiation, 14-3-3σ-deficient keratinocytes failed to express differentiation marker, filaggrin (Figure 1C). In addition, the p63 protein detected by western blotting showed a molecular weight similar to ΔNp63α, suggesting that ΔNp63α was the major p63 isoform induced (Figure 1D). Although specific detection of ΔNp63α messenger RNA by qPCR is impossible due to its sequence similarity with other isoforms, the messenger RNA levels of both ΔNp63 and p63α isoforms in the mutant keratinocytes were not reduced by calcium switch, in contrast to the downregulation of their transcriptions in the WT keratinocytes upon the high calcium-induced differentiation (Figure 1E). A similar increased expression of both ΔNp63 and p63α isoforms were also found in the mutant skin in vivo, as compared with the WT controls (data not shown). On the other hand, the TAp63 isoform was not detectable in those RNA samples by real-time qPCR method (data not shown). Consistently, immunostaining with antibodies specific for ΔNp63, p63α or TAp63 detected the expressions of ΔNp63 and p63α but not TAp63 in Er/Er suprabasal layers (data not shown). These data suggest that ΔNp63α expression level is inversely correlated to the level of 14-3-3σ in epidermis and that the ΔNp63α overexpression may count for the increased proliferation and impaired differentiation observed in the 14-3-3σ-deficient epidermis.
p63 is critical for maintenance of the keratinocyte proliferation but not differentiation
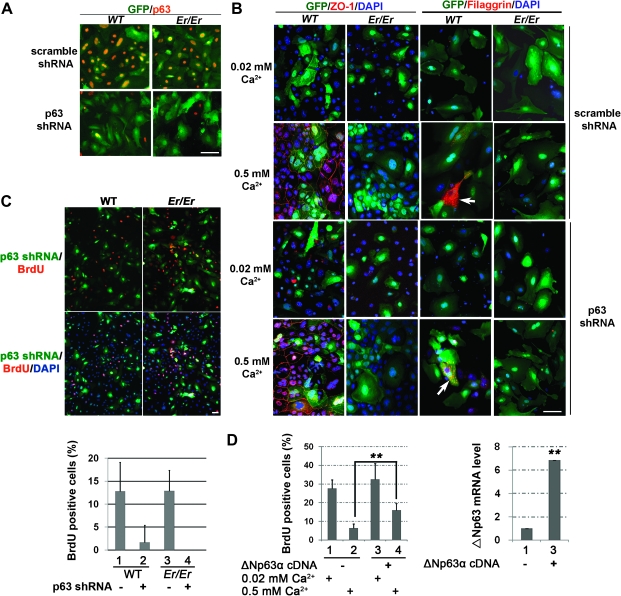
To further test whether the increased ΔNp63α expression in 14-3-3σ-deficient keratinocytes is responsible for the undifferentiated phenotype, we knocked down the p63 expression in those mutant cells using lentivirus-carried shRNA. Immunostaining with p63 antibody was subsequently used to confirm the efficient knockdown of p63 expression in keratinocytes (Figure 2A). The differentiation stage induced by high-calcium culture was evaluated by immunostaining of the tight junction marker, zonula occludens-1, which is localized to the junction formed around the cell borders once the keratinocytes are differentiated (Figure 2B). The keratinocytes without a functional 14-3-3σ were unable to differentiate and did not form the tight junctions, as evidenced by lacking a continuous zonula occludens-1 staining pattern around cell borders in a calcium switch assay (Figure 2B). In addition, the filaggrin granules could be detected only in the differentiated WT but not Er/Er keratinocytes, even when p63 expression was knocked down (Figure 2B). Thus, the p63 knockdown did not rescue the defective differentiation phenotype in the 14-3-3σ-deficient keratinocytes. This result suggests that p63 is not a downstream factor of 14-3-3σ to promote keratinocyte differentiation.
Fig. 2.
p63 shRNA silencing inhibits keratinocyte proliferation and Δp63α complementary DNA overexpression increases keratinocyte proliferation upon differentiation. (A) p63 immunostaining showed that p63 expression was efficiently and specifically knocked down in the primary WT and Er/Er keratinocytes after 3 days of transduction with a lentivirus expressing p63 shRNA, as shown by lacking of p63 immunoreactivity. The primary WT (left) and Er/Er (right) keratinocytes were transduced with either p63 specific (bottom) or scrambled shRNA (top) lentiviruses (both vectors coexpress GFP). (B) The defective differentiation phenotype of Er/Er keratinocytes cannot be rescued by p63 knockdown upon high calcium-induced differentiation, as evidenced by lacking zonula occludens-1-positive tight junction formation and filaggrin expression. Primary cultured WT and Er/Er keratinocytes were transduced with scramble- (top two rows) and p63- (bottom two rows) specific shRNA lentiviruses (visualized by green fluorescence) for 3 days and then replaced with fresh medium containing 0.5 mM calcium for additional 48 h. The calcium switch-induced tight junction formation and filaggrin granules formation were visualized by zonula occludens-1 and filaggrin immunostaining, respectively, and the nuclei were counterstained by DAPI. The arrows indicate the filaggrin-positive cells. The representative images are showed. (C) Decreased cell proliferation in both WT and Er/Er keratinocytes transduced with p63-specific shRNA lentiviruses. Primary WT (left) and Er/Er (right) keratinocytes were transduced with p63-specific shRNA lentiviruses for 3 days and then the proliferating cells were labeled by BrdU incorporation. The percentages of BrdU-positive cells in either non-tranduced GFP-negative cells or tranduced GFP-positive cells were summarized in the table on the bottom. (D) Quantification of BrdU-positive cells in WT keratinocytes transduced with lentiviral vector only (1 and 2) or the virus expressing Δp63α (3 and 4) for 2 days before culture in 0.02 mM (1 and 3) and 0.5 mM (2 and 4) calcium-containing medium for additional 2 days. Overexpression of Δp63α was confirmed by qPCR assay and shown on right. The error bars represent the standard deviation from three independent experiments. Student's t-test was performed to determine the statistical difference. **P < 0.005. The scale bars represent 50 μm in (A–C).
We next examined the effect of p63 shRNA knockdown on keratinocyte proliferation by BrdU incorporation assay. The proliferation rate was calculated based on the percentage of BrdU-positive cells in the total transduced cells marked by GFP coexpressed from the shRNA lentivirus vector. p63 knockdown in the WT cells reduced the BrdU-positive cells from 12% in the GFP-negative cells (non-transduced) to 2% of the GFP-positive cells (transduced) (Figure 2C), further confirming the functional role of p63 in keratinocyte proliferation. Similarly, p63 knockdown also prevented Er/Er keratinocyte proliferation (Figure 2C). The reduction in BrdU incorporation was specific for p63 shRNA since the scramble shRNA control showed no such effect (Supplementary Figure S1 is available at Carcinogenesis Online). On the other hand, we also examined effect of ΔNp63α overexpression on the WT keratinocyte proliferation. The overexpression of ΔNp63α increased BrdU incorporation significantly in WT cells under the differentiation (high calcium) culture conditions (Figure 2D). These results suggest that constant ΔNp63α expression in the 14-3-3σ-deficient keratinocytes under the differentiation (high calcium) culture conditions is responsible for maintenance of rapid cell growth.
DMBA/TPA-induced tumors in Er/+ mice express ΔNp63
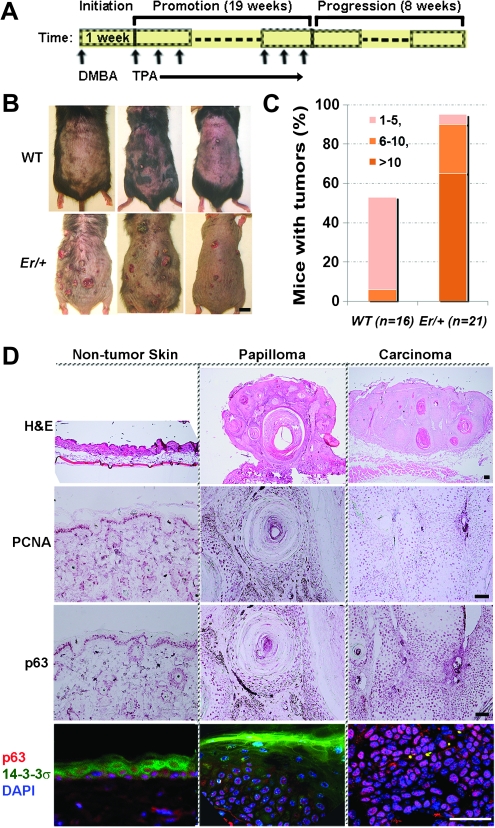
Given that p63 controls keratinocyte proliferation, we then asked whether p63 expression was relevant to tumorigenesis in Er/+ heterozygous mice. The DMBA/TPA two stages of tumor induction procedure was performed and diagramed in Figure 3A. In agreement with the tumor suppressing function of 14-3-3σ (22), loss of one allele of 14-3-3σ rendered the Er/+ mice more sensitive to the chemical-induced tumorigenesis than the WT controls (Figure 3B). Although approximate half (53%) of the WT mice developed papillomas by 28 weeks post-DMBA administration, there were >95% of Er/+ mice developing papillomas (Figure 3C). Er/+ mice that developed papillomas had significantly more and larger papillomas than the WT mice (Figure 3C). For example, 65% of individual Er/+ mice but none of WT mice had >10 papillomas (Figure 3C). Most cells in those DMBA-/TPA-induced papillomas and tumors expressed the cell proliferation markers, proliferating cell nuclear antigen and p63 (Figure 3D). There were very few small papillomas and no carcinoma formed in the WT animals treated with DMBA/TPA in our condition. p63 immunostaining in those papilloma sections showing p63 expression was limited or immediately adjacent to basal layer of papillomas (Supplementary Figure S2 is available at Carcinogenesis Online). Coimmunofluorescence staining further showed non-overlapping expression of 14-3-3σ and p63; 14-3-3σ expression was low in undifferentiated p63-positive papilloma and tumor cells but high in p63-negative differentiated papilloma cells (Figure 3D).
Fig. 3.
p63-positive cells are associated with DMBA-/TPA-induced tumors derived from Er/+ mice. (A) Schematic diagram shows a DMBA/TPA two-stage tumor induction procedure. About 100 nmol of DMBA in 0.2 ml acetone was applied topically to the shaved dorsal skin. One week after the single DMBA treatment, TPA was (17 nmol in 0.2 ml acetone) applied topically to the shaved dorsal skin for three times per week for 19 weeks. Eight weeks after stopping TPA treatment, the tumors were collected and processed for histopathological examination. The vertical arrows indicate the drug administration times. (B) Representative images showing induced tumors on the WT (top) and Er/+ (bottom) mice by the end of the two-stage DMBA/TPA tumorigenesis protocol. Scale bar: 1 cm. (C) The percentage of mice carrying the indicated numbers of papillomas in total DMBA-/TPA-treated mice is shown (WT, n = 16; Er/+, n = 21). The numbers in the table legend represent the number of tumors per tumor-carrying mouse. (D) Representative photographs of hematoxylin and eosin, immunohistochemistry (cell proliferating nuclear marker or p63), and double immunofluorescence of 14-3-3σ (green) and p63 (red) counterstained by DAPI in blue on the skin tissues without tumor (left), papillomas (middle) and carcinomas (right) collected from the tumor-induced Er/+ mice. Scale bars: 50 μm.
The DMBA-/TPA-induced tumorigenesis in the Er/+ mice depends on the presence of p63
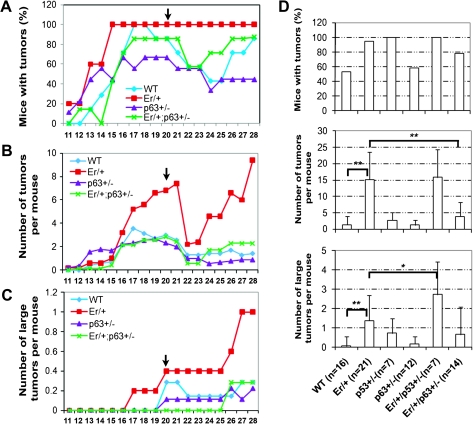
To determine functional interaction of 14-3-3σ and p63 in skin carcinogenesis, we treated Er/+, p63+/−, Er/+/p63+/− and WT control adult mice with DMBA/TPA two-stage tumor induction procedure. It has been well known that the carcinogen DMBA induces activating ras mutations and the subsequent multiple TPA treatments ensure tumor promotion (18,19). The numbers and the sizes of developing papillomas were monitored during entire drug treatment. Papillomas began to appear in all groups at ∼11 to 13 weeks after the initial DMBA administration. Remarkably, numerous papillomas in Er/+ mice grew progressively fast during the treatment and almost all the treated Er/+ mice developed papillomas at multiple spots within the treated skin area (Figure 4A–C). In contrast, Er/+/p63+/− mice exhibited notably resistant to papillomagenesis, as featured with fewer tumors that were smaller in size (Figure 4A–C). By the endpoint of the experiments, the average number of papillomas in each Er/+/p63+/− mouse was ∼3-fold less than this found in the Er/+ counterpart (Figure 4D). Such a significant difference in the number of the induced papillomas and tumors between the Er/+ and the Er/+/p63+/− mice (Figure 4D) suggests that p63 mediates papilloma development in the Er/+ mice. Consistent with previous studies, mice heterozygous for p63 gene alone developed the similar number and size of papillomas as WT mice (15,16) (Figure 4D). In contrast, consistent with the fact that p53 functions as tumor suppressor and its mutation induces tumorigenesis, the Er/+/p53+/− compound mutant mice developed tumors larger in size, as compared with the single heterozygous mice in either 14-3-3σ or p53 genes (Figure 4D). These results further demonstrate that p63 and p53 play different roles in epidermal tumorigenesis.
Fig. 4.
p63+/−/Er/+ double heterozygous mice develop fewer DMBA-/TPA-induced papillomas than Er/+ mice. (A) Papilloma incidence (percentage of mice with papillomas) in WT (n = 6), Er/+ (n = 6), p63−/+ (n = 9) and Er/+/p63+/− (n = 7) mice during the time course (weeks) after the initial DMBA treatment. Mice were monitored weekly for papilloma development. The arrows indicate the endpoint for TPA treatment. (B) Papilloma multiplicity (average number of papillomas per mouse) during the time course (weeks) after the initial DMBA treatment. (C) Average number of large (diameter ≥ 4 mm) tumors per mouse during the time course (weeks) after the initial DMBA treatment. (D) Papilloma incidence (percentage of mice with papillomas) (the top panel), papilloma multiplicity (average number of papillomas per mouse (the middle panel) and average number of large (diameter ≥ 4 mm) tumors per mouse (the low panel) at 28 weeks after DMBA application for each groups of WT, Er/+, p53+/−, p63+/−, Er/+/p53+/− and Er/+/p63+/− mice. Student's t-test was performed to determine the statistical difference. **P < 0.005 and *P < 0.05.
Discussion
p63 is structurally related to p53 and p73 proteins and a member of the p53 family of transcriptional factors. p63 is mainly expressed by epidermal basal layer cells in the stratified squamous epithelium and its expression is sharply reduced when the basal cells move to the suprabasal layers and commit to differentiation. p63−/− mice showed major defects in limb and craniofacial development due to lacking stratified epidermis and consequently impaired epithelial–mesenchymal signaling (10,11).
14-3-3σ belongs to a seven members of 14-3-3 scaffold protein family, which binds to >100 of ligand proteins involving oncogenic signaling and cell cycle regulation (1). 14-3-3σ is uniquely expressed by the stratified squamous epithelium, strongly upregulated in the suprabasal cells, immediately after their differentiation from the basal progenitor cells (23). 14-3-3σ mutation generating a truncated protein lacking 40 amino acids at C-terminus causes the death of new born Er/Er pups due to lacking of functioning epidermis resulting from unrestricted expanding of the epidermal progenitor cells and failure of differentiation (5). It is probably that the increasing expression of 14-3-3σ in the differentiating cells blocks the transcription of p63 and its downstream target genes, which are necessary for maintenance of the undifferentiating stage. Without 14-3-3σ, those transcriptional factors continue to function and keep the cells in high proliferative undifferentiating stage; this was indeed what we observed on the 14-3-3σ homozygous mutants in vivo and the isolated mutant keratinocytes in vitro in the present study. We also showed that the papilloma cells induced from Er/+ mice not only expressed high level of p63 but also displayed high proliferation phenotype as shown by increased immunoreactivity to cell proliferating nuclear marker. Such high proliferation phenotype was also observed in 14-3-3σ-mutant keratinocytes upon differentiation condition with the increased level of ΔNp63α as revealed by qPCR and western blot. Since Er/Er mutant cells resist to high calcium-induced differentiation in vitro, we knocked down the p63 expression in those cells by shRNA to assess whether p63 silencing would rescue the differentiation phenotype. Our data showed that this was not the case since the mutant cells still failed to differentiate in the calcium switch experiment when p63 was silenced, suggesting that high level of p63 in the Er/Er keratinocytes under the differentiating culture condition was not responsible for the impaired differentiation. However, p63 shRNA knockdown reduced BrdU incorporation significantly, suggesting that the cell proliferation requires the presence of p63. Furthermore, overexpression of ΔNp63α alone sufficiently increased the keratinocyte proliferation upon the differentiating condition.
The function roles of p63 in tumorigenesis are still controversial and poorly defined, as multiple isoforms have been found to play either collaborative or opposing roles. In contrast to p53, which has been well established as a tumor suppressor and found mutated in many human cancers, p63 is rarely mutated in human. However, ΔNp63α overexpression is often associated with the tumors derived from bladder, lung, oral, nasopharyngeal and skin epithelium (24–30) and with an aggressive clinical course and poor prognosis (31). There were some contrary reports, for instance, the loss of ΔNp63 was associated with an aggressive phenotype (32). Such controversy has been further widen by two reports from the studies of the p63+/− and p53+/− double heterozygous mice (15,16), with one study suggesting that loss of one allele of p63 in p53+/− or p73+/− mice predisposes the mice to more tumor development (15), whereas the other study showed that the p63 heterozygosity was not prone to tumor formation (16). Some of those contradictive results may rise from the opposite functions of TAp63 and ΔNp63, while the TAp63 acts as tumor suppressor and the ΔNp63 is considered oncogenic. The outcome effects of p63 in tumor depend on the cancer cell origin, the unique molecular signaling signature and the relevant levels of TAp63 and ΔNp63.
We show in this study that ΔNp63α is a predominant isoform of p63 in the 14-3-3σ-null keratinocytes and its increased expression in the Er/+ epithelium contributes to tumorigenesis initiated by 14-3-3σ/Ras mutation. Reducing p63 expression by deleting one allele of the p63 gene in p63+/−/Er/+ compound heterozygous mice showed resistant to the DMBA/TPA tumor inductions, which suggests that p63 plays an oncogenic role in the Ras/14-3-3σ mutation-induced skin tumorigenesis, presumably by promoting cancer stem cell survival and proliferation. 14-3-3σ has been shown to inhibit the ΔNp63α function by causing its nuclear export and retaining its cytoplasmic localization (33). Our data also indicated that loss of 14-3-3σ increased ΔNp63α transcription in keratinocytes, although the mechanism is not clear, this might be caused by indirect regulation of p63 transcription.
The underlying molecular mechanism by which ΔNp63α promotes tumor formation is still ambiguous. ΔNp63α could play a dominant-negative effect by competing with tumor suppressor p53 for transcription regulation, a good example of this is that the ΔNp63 influences cell cycle checkpoint by modulating p53-dependent gene p21 expression (14). On the other hand, ΔNp63α could also directly activate transcription of the cell cycle control genes (34). ΔNp63α has recently been identified as oncogene that promotes skin stem cell proliferation and tumorigenesis by targeting to the chromatin remodeler lsh (35). In line of this study, our results showed that reducing p63 expression in vivo reduced susceptibility of the Er/+ mice to the DMBA-/TPA-induced skin tumorigenesis. Our experiments provide a mouse model for investigation of p63 roles in cancer. Further exploration of molecular networks that both 14-3-3σ and p63 involved may lead to discoveries of novel therapeutic methodologies to prevent skin tumor development.
Supplementary material
Supplementary Figures S1 and S2 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (RR018733 and EY019891 to Q.Li) and (EY018830 to Q.Lu); Research to Prevent Blindness of NYC for general support and the Histocore funded by National Institutes of Health (EY015636).
Supplementary Material
Acknowledgments
Conflict of Interest Statement. None declared.
Glossary
Abbreviations
- BrdU
5-bromo-2′-deoxyuridine
- DAPI
4′,6-diamidino-2-phenylindole
- DMBA
7,12-dimethylbenzanthracene
- GFP
green fluorescent protein
- PFA
paraformaldehyde
- qPCR
quantitative polymerase chain reaction
- shRNA
small hairpin RNA
- TPA
12-O-tetradecanoyl-phorbol-13-acetate
- WT
wild-type
References
- 1.Benzinger A, et al. Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer. Mol. Cell. Proteomics. 2005;4:785–795. doi: 10.1074/mcp.M500021-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Bridges D, et al. 14-3-3 proteins: a number of functions for a numbered protein. Sci. STKE. 2005;296:re10. doi: 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- 3.Hermeking H, et al. 14-3-3 proteins in cell cycle regulation. Semin. Cancer. Biol. 2006;16:183–192. doi: 10.1016/j.semcancer.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Herron BJ, et al. A mutation in stratifin is responsible for the repeated epilation (Er) phenotype in mice. Nat. Genet. 2005;37:1210–1212. doi: 10.1038/ng1652. [DOI] [PubMed] [Google Scholar]
- 5.Li Q, et al. Identification of 14-3-3sigma mutation causing cutaneous abnormality in repeated-epilation mutant mouse. Proc. Natl Acad. Sci. USA. 2005;102:15977–15982. doi: 10.1073/pnas.0508310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson AT, et al. High frequency of hypermethylation at the 14-3-3 sigma locus leads to gene silencing in breast cancer. Proc. Natl Acad. Sci. USA. 2000;97:6049–6054. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lodygin D, et al. Analysis of 14-3-3 sigma expression in hyperproliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma. Oncogene. 2003;22:5519–5524. doi: 10.1038/sj.onc.1206854. [DOI] [PubMed] [Google Scholar]
- 8.Chan TA, et al. 14-3-3 Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 9.Hermeking H, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 10.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 11.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 12.Su X, et al. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 2009;28:1904–1915. doi: 10.1038/emboj.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truong AB, et al. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westfall MD, et al. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell. Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Keyes WM, et al. p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc. Natl Acad. Sci. USA. 2006;103:8435–8440. doi: 10.1073/pnas.0602477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin Y, et al. IKK1 control of epidermal differentiation is modulated by notch signaling. Am. J. Pathol. 2011;178:1568–1577. doi: 10.1016/j.ajpath.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiscornia G, et al. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 19.Lo Iacono N, et al. Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development. 2008;135:1377–1388. doi: 10.1242/dev.011759. [DOI] [PubMed] [Google Scholar]
- 20.King KE, et al. Unique domain functions of p63 isotypes that differentially regulate distinct aspects of epidermal homeostasis. Carcinogenesis. 2006;27:53–63. doi: 10.1093/carcin/bgi200. [DOI] [PubMed] [Google Scholar]
- 21.Xin Y, et al. 14-3-3sigma controls corneal epithelial cell proliferation and differentiation through the Notch signaling pathway. Biochem. Biophys. Res. Commun. 2010;392:593–598. doi: 10.1016/j.bbrc.2010.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy AL, et al. Increased sensitivity to two-stage skin carcinogenesis of mice heterozygous for the repeated epilation mutation (Er) Int. J. Cancer. 1990;46:928–930. doi: 10.1002/ijc.2910460529. [DOI] [PubMed] [Google Scholar]
- 23.Dellambra E, et al. Downregulation of 14-3-3sigma prevents clonal evolution and leads to immortalization of primary human keratinocytes. J. Cell Biol. 2000;149:1117–1130. doi: 10.1083/jcb.149.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibi K, et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl Acad. Sci. USA. 2000;97:5462–5467. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patturajan M, et al. DeltaNp63 induces beta-catenin nuclear accumulation and signaling. Cancer Cell. 2002;1:369–379. doi: 10.1016/s1535-6108(02)00057-0. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi, et al. Frequent gain of the p40/p51/p63 gene locus in primary head and neck squamous cell carcinoma. Int. J. Cancer. 2000;86:684–689. doi: 10.1002/(sici)1097-0215(20000601)86:5<684::aid-ijc13>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 27.Parsa R, et al. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J. Invest. Dermatol. 1999;113:1099–1105. doi: 10.1046/j.1523-1747.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 28.Nylander K, et al. Characterization of the expression pattern of p63 alpha and delta Np63 alpha in benign and malignant oral epithelial lesions. Int. J. Cancer. 2000;87:368–372. [PubMed] [Google Scholar]
- 29.Park BJ, et al. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res. 2000;60:3370–3374. [PubMed] [Google Scholar]
- 30.Crook T, et al. High level expression of deltaN-p63: a mechanism for the inactivation of p53 in undifferentiated nasopharyngeal carcinoma (NPC)? Oncogene. 2000;19:3439–3444. doi: 10.1038/sj.onc.1203656. [DOI] [PubMed] [Google Scholar]
- 31.Karni-Schmidt O, et al. Distinct expression profiles of p63 variants during urothelial development and bladder cancer progression. Am. J. Pathol. 2011;178:1350–1360. doi: 10.1016/j.ajpath.2010.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koga F, et al. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin. Cancer. Res. 2003;9:5501–5507. [PubMed] [Google Scholar]
- 33.Fomenkov A, et al. RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle. 2004;3:1285–1295. doi: 10.4161/cc.3.10.1155. [DOI] [PubMed] [Google Scholar]
- 34.Lefkimmiatis K, et al. p73 and p63 sustain cellular growth by transcriptional activation of cell cycle progression genes. Cancer Res. 2009;69:8563–8571. doi: 10.1158/0008-5472.CAN-09-0259. [DOI] [PubMed] [Google Scholar]
- 35.Keyes WM, et al. DeltaNp63alpha is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell. 2011;8:164–176. doi: 10.1016/j.stem.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.