Abstract
Zinc accumulation is lost during prostate carcinogenesis. Recent studies reveal a strong association between prostate cancer progression and the downregulation of the zinc uptake transporters hZip1 and hZip3. The aim of this work was to assess the involvement of epigenetic processes in the disruption of zinc uptake homeostasis in prostate adenocarcinoma. In this report, we demonstrate an increase in hZip1 and hZip3 zinc transporters’ expression and zinc uptake by the prostate cancer cells DU-145 and LNCaP in response to 5-aza-2′-deoxycytidine. This effect is due to demethylation of the promoter region of the activator protein (AP)-2alpha protein, which is crucial for hZip1 and hZip3 genes expression. Loss of AP-2alpha expression in DU-145 and LNCaP prostate cancer cells is due to hypermethylation of its promoter region. Similarly, we found higher AP-2alpha promoter methylation levels in clinical samples of early-stage prostate adenocarcinoma when compared with adjacent non-malignant prostate tissue. Taken together, our findings provide a better understanding of the epigenetic mechanisms that are involved in the loss of AP-2alpha protein in prostate cancer cells which lead to decreased cellular zinc uptake—a sine qua non of prostate cancer development.
Introduction
The human prostate is unique in that it possesses the ability to accumulate high levels of intracellular zinc. Multiple studies have demonstrated that decreasing levels of intracellular zinc appear to be an important factor in the development and progression of prostate cancer (1,2). In fact, the inability to accumulate intracellular zinc by prostate cells often precedes the initial histopathological changes associated with prostate cancer. Cellular zinc handling becomes increasingly dysfunctional as prostate cancer progresses to castration-independent growth (3,4).
The zinc content of normal prostatic epithelium, benign prostatic hyperplastic tissue and cancerous prostate glands has been measured at 1018, 1142 and 146 μg/g of dry tissue, respectively (4). Recent mechanistic studies have revealed a strong association between the development of prostate cancer and downregulation of the zinc uptake transporters, hZIP1 and hZIP3. The expression of hZIP1 and hZIP3 genes was markedly downregulated in adenocarcinomatous glands and in prostatic intraepithelial neoplastic foci when compared with adjacent normal peripheral zone glandular epithelium and benign hyperplastic glands (5–7). Moreover, we recently reported that overexpression of hZip1 transporter has strong functional effect on the malignant potential of prostate cancer cells via inhibition of natural factor-kappaB-dependent pathways (8).
Although genetic alterations in prostate cancer have long been studied, the role of epigenetic changes during prostatic malignant transformation has recently garnered more attention. Epigenetic changes alter target gene expression without changing the cells DNA sequence. Inactivation of tumor suppressor genes by epigenetic changes is frequently observed in human cancers, particularly as a result of histone modification and/or DNA methylation.
Promoter methylation is one of the most common epigenetic events associated with altering gene expression. In a variety of tumors, CpG-rich regions, i.e. CpG islands, exhibit aberrant DNA hypermethylation resulting in abnormal transcriptional repression and gene inactivation (9). Specific to prostate cancer tumorogenesis, many of the inactivated genes in these CpG islands encode proteins that act as tumor suppressors, resulting in prostate cancer initiation, progression and, perhaps, an association with a more aggressive prostate cancer phenotype (10,11).
Recent studies have shown that the inhibition of DNA methyltransferase activity by 5-aza-2′-deoxycytidine (5-aza-CdR) prevented prostate cancer tumorigenesis in a mouse model (12). In the present report, we examine the effects of the demethylating agent 5-aza-CdR on the accumulation of intracellular zinc as well as the expression of zinc uptake transporters hZip1 and hZip3 in DU-145 and LNCaP prostate cancer cell lines. Recently, we reported that specificity protein 1 (SP1) and CAMP responsive element binding protein 1 are important transcription factors in the regulation of the hZip1 zinc transporter gene (13). In the current study, we also demonstrate the importance of SP1 and activator protein (AP)-2alpha proteins as transcription factors in the regulation of the hZip3 zinc transporter in RWPE-1 cells. Furthermore, we were able to document the critical role of AP-2alpha in regulating hZip1 gene transcription in the RWPE-1 normal prostatic epithelial cell line.
In addition, we show that the epigenetic mechanisms of gene silencing caused by promoter hypermethylation in prostate cancer cells are indirectly involved in transcriptional downregulation of the zinc transporters hZip1 and hZip3. Since the SP1 and AP-2alpha proteins play an important role in the transcriptional regulation of hZip1 and hZip3 genes, the loss of expression of any of these transcription factors would have a dramatic impact on the expression of the zinc uptake transporters. Specifically, we appear to demonstrate that hypermethylation of AP-2alpha’s promoter region results in decreased expression of AP-2alpha in DU-145 and LNCaP prostate cancer cells. This decreased expression results in a notable transcriptional down-regulation of the hZip1 and hZip3 zinc transporters and, thus, reduced ability to accumulate intracellular zinc.
Materials and methods
Cell lines and culture conditions
The hormone-dependent LNCaP, hormone-independent DU-145 prostate cancer cell lines and the RWPE-1 normal prostate epithelial cell line were obtained from ATCC (Rockville, MD). Cells were cultured in RPMI 1640 (Bio-Whittaker, Walkersville, MD) medium supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), gentamicin (50 mg/l), sodium pyruvate (1 mM) and non-essential amino acids (0.1 mM). RWPE-1 cells were maintained in Keratinocyte-Serum Free medium (Invitrogen, Carlsbad, CA) supplemented with 5 ng/ml of human recombinant epidermal growth factor and 0.05 mg/ml of bovine pituitary extract.
Antibodies and reagents
Antibodies to SP1 (sc17824X) and AP-2alpha (sc-184X) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to GAPDH (#2118) were obtained from Cell Signaling Technology (Danvers, MA). 5-aza-CdR was purchased from the Sigma–Aldrich Corporation (St Louis, MO).
Computer analysis of promoter regions
The analysis of 5′-flanking region of hZip3 gene was performed using Gene2Promoter online programs (www.genomatix.de). Prediction of transcriptional factor-binding sites was carried out using the AliBaba 2.0, TRANSFAC, Match (www.gene-regulation.com) and MatInspector (www.genomatix.de) online programs.
Plasmid construction
The 5′-flanking region of the hZip1 gene has been recently cloned into the luciferase reporter vector (13). Promoter region (−154/+100 relative to transcription start) of hZip3 gene site was amplified by polymerase chain reaction (PCR) with 5′-ATCTTGGGTACCGTTTCCCGCCGGGGCCGCTTGGTG-3′ forward and 5′-ATCTTGCTCGAGCGCTGCACGCCCAACGGCCG-3′ reverse primers containing restriction sites (underlined) using RWPE-1 genomic DNA as a template and cloned into Xho1/Kpn1 sites of the pGL3-basic reporter vector (Promega, Madison, WI). Site-directed mutagenesis of transcription factor-binding sites was performed by PCR with overlapped complementary primers spanning the targeted binding site (Supplementary Table 1 is available at Carcinogenesis Online). All mutations in reporter constructs were confirmed by sequencing using GL2 primer (Promega) in the Fox Chase Cancer Center Automated DNA Sequencing Facility.
AP-2alpha open-reading frame was amplified with specific 5′-ATCTTGAAGCTTGCCACCATGTTAGTTCACAGTTTTTCAGCCAT-3′ forward and 5′-ATCTTGGAATTCTCACTTTCTGTGCTTCTCCTCTTTGTC-3′ reverse primers containing restriction sites (underlined) and pCMV-SPORT6 vector containing full length AP-2alpha complementary DNA (Clone ID#4432023; Open Byosistems, Huntsville, AL) as a template and then was cloned into HindIII/EcoRI sites of pcDNA3.1 vector.
Transient transfection and luciferase reporter assays
Transfection of RWPE-1 cells was performed in 24-well plates using the TransIT Prostate transfection kit (Mirus, Madison, WI). Twenty-four hours after cell transfection, a luciferase assay was performed using DualGlo Luciferase Assay System (Promega).
DU-145 and LNCaP cells were transfected with pcDNA3.1 empty and pcDNA3.1-AP-2alpha vectors using the TransIT Prostate transfection kit. Additionally, the Label IT® Plasmid Delivery Control, Cy™3 (Mirus) was used for labeling cells for further sorting. Twenty-four hours after transfection, cells were subjected to sorting in the Fox Chase Cancer Center Flow Cytometry and Cell Sorting Facility to enrich for the population of transfected cells.
Electrophoretic mobility shift assay and Gel-Supershift assays
Electrophoretic mobility shift assay and Gel-Supershift assays were performed as described previously (13) using a set of double-stranded DNA oligos (Supplementary Table 2 is available at Carcinogenesis Online) and specific antibodies.
Assessment of cell growth
Cells were plated in 96-well plates (5 × 103 to 10 × 103 cells per well) and incubated for 48 h in complete cell growth medium. The metabolic capacity of cells was then evaluated using the CellTiter-Blue® Cell Viability Assay (Promega) according to the manufacturer instructions.
Reverse transcription and quantitative PCR
Total RNA was isolated from cells using the Mini RNA isolation-II Kit (Zymo Research Corporation, Orange, CA) followed by DNase-I (New England Biolabs, Ipswich, MA) treatment. RNA was then recovered and purified using an RNA Clean and Concentrator Kit (Zymo Research Corporation).
Reverse transcription (RT) of total RNA (1 μg) was then performed in a final volume of 20 μl with 100 U of Superscript III Reverse Transcriptase (Invitrogen, Gaithersburg, MD) and 50 ng of random hexamer primers. Then, complementary DNA samples were diluted and 5 μl of diluted complementary DNA were amplified by TaqMan quantitative polymerase chain reaction (qPCR) using hZip1- and hZip3-specific primers and probes (IDT DNA Technologies, Coralville, IA). GAPDH Mini qPCR assay (IDT DNA Technologies) was used as internal amplification control. Specific primers and probes are indicated in Supplementary Table 3 is available at Carcinogenesis Online.
Each sample was run in triplicate for hZip1, hZip3 and GAPDH in a 20 μl reaction using TaqMan Gene Expression Master Mix according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA). Reactions were carried out in an Applied Biosystems 7500 Real-Time PCR System. Relative hZip1 and hZip3 expression was analyzed using the2−ΔΔCt method (14).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed using the EZ-ChIP assay kit (Upstate, Charlottesville, VA). Briefly, 1 × 107 cells were cross-linked with 1% formaldehyde and washed twice with ice-cold phosphate-buffered saline containing a cocktail of protease inhibitors (Roche, Germany). Cells were resuspended in 500–1000 μl of sodium dodecyl sulfate-lysis buffer on ice and then sonicated with four sets of 10 s pulses by a Kontes Ultrasonic Cell Disrupter to an average DNA size of 200–400 bp. Protein concentrations of chromatin samples were measured using BCA™ Protein Assay (ThermoScientific, Rockford, IL) and 300 μg of chromatin protein were used in the immunoprecipitation step. The chromatin was precleared with protein A/G-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h followed by an overnight incubation at +4°C with 2 μg of specific antibodies. Normalized anti-mouse IgG (Santa Cruz Biotechnology) antibodies were used to control immunoprecipitations. Chromatin–antibody complexes were collected by reincubation with protein A/G PLUS-Agarose beads for 1 h. Chromatin was eluted from the beads and cross-links were reversed by incubation at 65°C for 4 h. ChIP DNA was recovered and purified using ChIP DNA Clean and Cncentrator Kit (Zymo Research Corporation).
About 2% of input DNA and 2–8% of eluted ChIP DNA were subjected to TaqMan qPCR with specific primers and probes (Supplementary Table 4 is available at Carcinogenesis Online). Samples were run in triplicates for both input DNA and ChIP-DNA in a 20 μl reaction using TaqMan Universal Master Mix according to the manufacturer’s instructions (Applied Biosystems). Reactions were carried out in an Applied Biosystems 7500 Real-Time PCR System.
Relative quantification of the ChIP-DNA was performed with the comparative Ct method. The ΔCt values were computed by subtracting the average of normalized against input Ct values of GAPDH ChIP-DNA samples from the average of normalized against input Ct values of hZip1 or hZip3 ChIP-DNA samples recovered with anti-AP-2alpha antibodies. In order to determine the fold change of target ChIP-DNA treated with 5-aza-CdR cells over target ChIP-DNA in untreated cells, the ΔΔCt was calculated by subtracting the ΔCt values of treated cells from the ΔCt values of untreated cells. The relative amount of target ChIP-DNA was finally calculated by the formula 2−(ΔΔCt).
Tissue samples
Tissue specimens were obtained from the Department of Urology, Cancer Research Center RAMS, Russia and analyzed at the Institute of Carcinogenesis, Cancer Research Center, Moscow, Russia. All of the prostate tumors had been histopathologically confirmed as stage pT2cN0M0 Gleason score 6 prostate cancer. All patients had not received any hormonal or radiation therapy prior to radical prostatectomy. Representative samples of confirmed prostate cancer as well as adjacent normal prostate tissue were taken from each prostate specimen and were freshly frozen at the time of surgery. Genomic DNA samples were isolated from the frozen tissues using ZR Genomic DNA™-Tissue MiniPrep (Zymo Research Corporation) according to manufacturer’s instructions.
Results
Treatment with 5-aza-CdR increases zinc uptake in prostate cancer cells
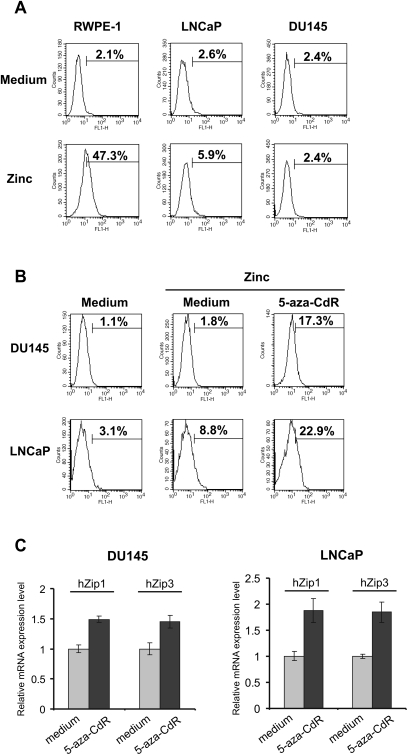
Since prostate cancer development and progression is characterized by a sequentially increasing dysregulation of zinc metabolism, we chose to use in our experiments the hormone-dependent LNCaP and the hormone refractory DU-145 prostate cancer cell lines as well as the normal prostate epithelial cell line RWPE-1. Firstly, we compared the ability of the normal prostate epithelial cell line RWPE-1 and the prostate cancer cell lines DU-145 and LNCaP to accumulate zinc by staining the cells with the fluorescent zinc indicator FluoZin-3. As demonstrated in Figure 1A, intracellular zinc levels were significantly decreased in the prostate cancer cell lines DU-145 and LNCaP when compared with RWPE-1 cells.
Fig. 1.
(A) Zinc accumulation in normal RWPE-1 and cancerous DU-145 and LNCaP prostate cells. Cells were cultured in complete cell media for 24 h followed by incubation with 2 μg/ml of zinc for 1 h in plain RPMI medium. Cells were washed twice in phosphate-buffered saline and incubated with the cell-permeable acetoxymethyl ester form of the fluorescent zinc indicator FluoZin-3 (5 μM) for additional 15 min. Fluorescence was examined by flow cytometry. The x-axis represents fluorescence intensity and the y- axis represents cell number. Figure represents data from one of three independent experiments. (B) The effect of treatment with 5-aza-CdR on cellular zinc uptake on the prostate cancer cell lines DU-145 and LNCaP. Cells were cultured with or without 5-aza-CdR (5 μM) for 48 h. Cell media was changed and incubation was continued for another 24 h in complete cell culture medium alone. Zinc accumulation was examined as described in panel A. (C) Upregulation of hZip1 and hZip3 expression was noted in DU-145 and LNCaP cells that were treated with 5-aza-CdR. Cells were treated with 5 μM of 5-aza-CdR for 48 h and then cell media was changed and incubation was continued for another 24 h. Cells were harvested and subjected to qRT–PCR reaction run in triplicate. Error bars represent mean (±SD) of triplicates.
To assess whether epigenetic factors participate in the regulation of intracellular zinc uptake in prostate cancer cells, DU-145 and LNCaP cells were treated with the inhibitor of DNA methyltransferases 5-aza-CdR. Treatment with 5-aza-CdR resulted in significantly increased intracellular zinc accumulation in DU-145 and LNCaP cells as shown in Figure 1B. The possible toxic effect of 5-aza-CdR on cell line survival was evaluated by propidium iodide staining following 48 h of incubation of a 5 μM concentration of 5-aza-CdR with LNCaP, DU-145 and RWPE-1 cells, respectively. PI staining did not reveal a significant increase in the induction of apoptosis between the cells treated with the demethylating and those not treated with the demethylating agent (data not shown).
Upregulation of hZip1 and hZip3 expression by 5-aza-CdR
Next, we sought to examine effects of 5-aza-CdR treatment on the transcriptional regulation of the hZip1 and hZip3 genes. Using RT–qPCR, we found that hZip1 and hZip3 messenger RNA levels were increased in both DU-145 and LNCaP cells treated with 5-aza-CdR (Figure 1C). These results imply that treatment with 5-aza-CdR engenders increased intracellular zinc uptake by upregulation of the main zinc transporters.
Given that treatment with 5-aza-CdR increases zinc uptake in prostate cancer cells and that this increase coincides with upregulated expression zinc transporters, we proposed that DNA methylation may be partially responsible for the decreased zinc accumulation in prostate cancer cells. Thus, we analyzed the methylation status of promoter regions of hZip1 and hZip3 genes. Using bisulfite sequencing, we found hZip1 and hZip3 promoter regions to be hypomethylated in both prostate cancer DU-145 and LNCaP cell lines (data not shown). Based on these findings, we hypothesized that 5-aza-CdR treatment could have a secondary effect on hZip1 and hZip3 genes by activation of silenced transcription factors involved in regulation of hZip1 and hZip3 expression.
Transcriptional regulation of hZip1 and hZip3 promoters: role of SP1 and AP-2alpha
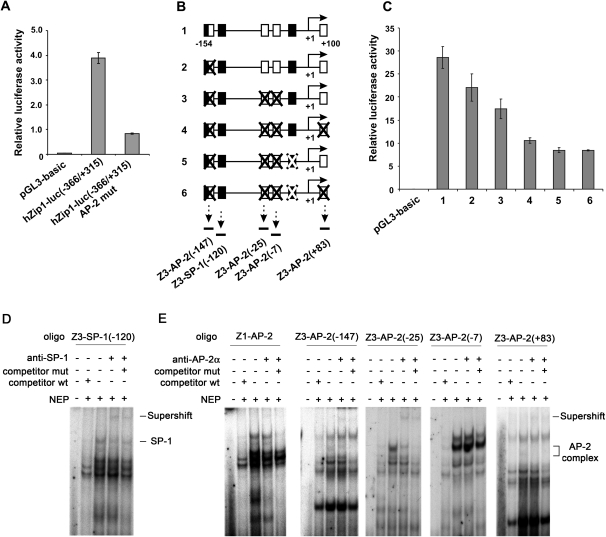
Since some of 5-aza-CdR’s effects on hZip1 and hZip3 expression appeared to involve the potential derepression of epigenetically silenced transcription factors, we investigated mechanisms of transcriptional regulation of hZip1 and hZip3 genes. Although the role of SP-1 and CAMP responsive element binding protein-1 proteins in the regulation of the hZip1 promoter was recently shown (13), the impact of the AP-2-binding site at −58/−67 position (relative to transcription start site) of the hZip1 promoter region still remains unclear. Since the AP-2 consensus sequence has been found, the pGL3-hZip1-luc (−366/+315) reporter vector was subjected to site directed mutagenesis. The Luciferase Reporter Assay revealed the high significance of the AP-2-binding site for transcriptional regulation of the hZip1 promoter. As demonstrated in Figure 2A, mutation of the AP-2alpha-binding site dramatically impaired transcriptional activity of the hZip1 (−366/+315) region.
Fig. 2.
The normal prostate epithelial cell line RWPE-1 was transfected with the pGL3 luciferase reporter vectors. Twenty-four hours after transfection, luciferase activity was measured. Transcriptional activity of hZip1 and hZip3 promoter regions is represented by the histograms. The data are presented as the mean (±SD) of the three independent experiments performed in triplicates. (A) Transcriptional activity of the wild-type hZip1 (−331/+367) promoter region and its analog with a mutated AP-2-binding site. (B) Schematic illustration of native hZip3 (−193/+68) promoter region fragments and its mutated analogs cloned into the pGL3 luciferase reporter vector. Black squares represent SP1 binding sites, and white squares represent AP-2 binding sites. Mutated binding sites are crossed. Arrows with dotted lines under binding sites represent localization of the double-stranded DNA oligos used in Gel-Supershift assays displayed on panels D and E. (C) Transcriptional activity of hZip3 (−193/+68) promoter constructs indicated on panel B. (D and E) Gel-Supershift analysis of the ability of nuclear extract proteins (NEP) to bind double-stranded DNA oligos targeting the AP-2-binding site of the hZip1 promoter as well as the appropriate binding sites within the hZip3 promoter region indicated on panel B. NEP isolated from RWPE-1 cells were incubated with specific 32P-labelled double-stranded DNA oligos in the presence of 100-fold molar excess of their unlabeled wild-type (wt), mutant (mut) competitors or alone. Anti-AP-2alpha or anti-SP-1 antibodies were added to the NEP to demonstrate their binding specificity.
In order to ascertain which transcriptional factors play a significant role in the regulation of hZip3 promoter activity, we performed a computer analysis of this promoter region using the AliBaba 2.0, TRANSFAC, Match-1.0, and MatInspector™ computer programs. Since multiple SP-1 and AP-2-like consensus sequences were predicted in the hZip3 promoter region, a pGL3 luciferase reporter vector containing a hZip3 (−154/+100) promoter region was constructed. Transcriptional activity of the hZip3 promoter region was then measured after single and combined mutations of the transcription factor binding sites were created. The results of native and mutated hZip3 promoter activity demonstrated the important role of SP-1 and AP-2alpha in transcriptional regulation of the hZip3 promoter region as illustrated on Figure 2C.
Analysis of SP-1 and AP-2alpha binding to hZip1 and hZip3 promoters
Next, we performed Gel-Supershift assays to evaluate the ability of SP-1 and AP-2alpha proteins to bind hZip1 and hZip3 promoters. For the assays, several double-stranded DNA oligos and their mutated analogs spanning the SP-1 or AP-2alpha-binding sites within hZip1 and hZip3 promoters were generated. The SP-1 protein was able to bind the hZip3 promoter region in the −120/−114 (relative to transcription start site) position (Figure 2D). As demonstrated on Figure 2E, using the following set of probes spanning the appropriate AP-2-binding sites within hZip1 and hZip3 promoters, we were able to demonstrate the ability of the AP-2alpha transcription factor to bind hZip1 (−67/−58), hZip3 (−147/−138), hZip3 (−25/−15) and hZip3 (+83/+92) promoter regions.
AP-2alpha expression is downregulated in DU-145 and LNCaP cells due to promoter hypermethylation
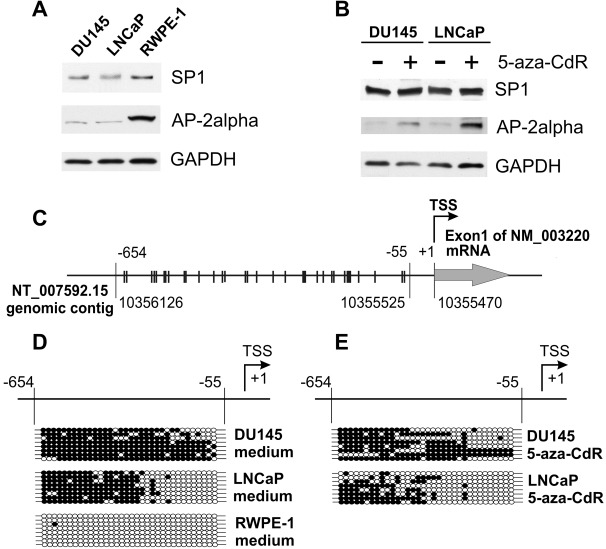
Since SP-1 and AP-2alpha have key roles in hZip1 and hZip3 promoter activities, we compared the levels of expression of these proteins in DU-145 and LNCaP prostate cancer cell lines as well as in the normal prostate epithelial cell line RWPE-1. SP1 is a ubiquitously expressed protein and western blot assay revealed no difference in the level of SP1 protein expression in prostate cancer and normal cells. In contrast, the level of the AP-2alpha protein was found to be decreased in both the DU-145 and LNCaP cell lines as compared with the RWPE-1 cell line (Figure 3A).
Fig. 3.
Hypermethylation of the AP-2alpha promoter region is associated with decreased expression of AP-2alpha in DU-145 and LNCaP prostate cancer cell lines. AP-2alpha expression can be restored by treatment of these cell lines with 5-aza-CdR. Western blot analysis of the levels of AP-2alpha and SP1 transcription factors in DU-145 and LNCaP prostate cancer cell lines: (A) the comparison with normal prostate epithelial cells RWPE-1; (B) DU-145 and LNCaP cells had been treated with 5 μM of 5-aza-CdR for 48 h followed by an additional 24 h of incubation in cell medium alone. (C) Schematic illustration of the AP-2alpha promoter region analyzed by bisulfite sequencing. The analyzed region is −654/−55 relative to the transcription start site of the indicated AP-2alpha transcript and corresponds to 10356126–10355525 nucleotide positions of the human genomic contig (NCBI accession # NT_007592.15). The NCBI genomic contig is represented schematically in the opposite direction for ease of depiction due to the fact that the AP-2alpha gene is encoded by the ‘minus’ chain. Each line crossing −654/−55 promoter region represents an individual CpG dinucleotide. (D) The bisulfite sequencing analysis of methylation of the AP-2alpha promoter region in DU-145, LNCaP and RWPE-1 cells. Each line represents an allele; each circle represents a CpG dinucleotide. Filled circles represent methylated CpG dinucleotides and clear circles represent unmethylated CpG dinucleotides. The 5′-flanking region of AP-2alpha gene is hypermethylated in DU-145 and LNCaP prostate cancer cell lines, whereas in normal prostate epithelial RWPE-1 cells, it remains unmethylated. (E) The bisulfite sequencing analysis was performed after DU-145 and LNCaP cells had been treated with 5 μM of 5-aza-CdR.
It has previously been shown that downregulation of the AP-2alpha transcription factor is caused by hypermethylation of its promoter region in breast cancer cells (15). Additionally, it was reported that the AP-2alpha protein expression is lost in prostate cancer cells (16). However, the methylation status of the AP-2alpha promoter region in the prostate cancer cell lines DU-145 and LNCaP to date had not been characterized. Thus, we analyzed the methylation of −654/−55 promoter region relative to the +1 position of AP-2alpha transcript (accession # NM_003220) by bisulfite sequencing. As demonstrated in Figure 3D, the AP-2alpha promoter region was found to be unmethylated in the normal prostate epithelial cell line RWPE-1. In contrast, we observed high methylation levels of this region in both the DU-145 and LNCaP prostate cancer cell lines.
Treatment with 5-aza-CdR results in AP-2alpha promoter demethylation, upregulation of its expression and binding to hZip1 and hZip3 promoters
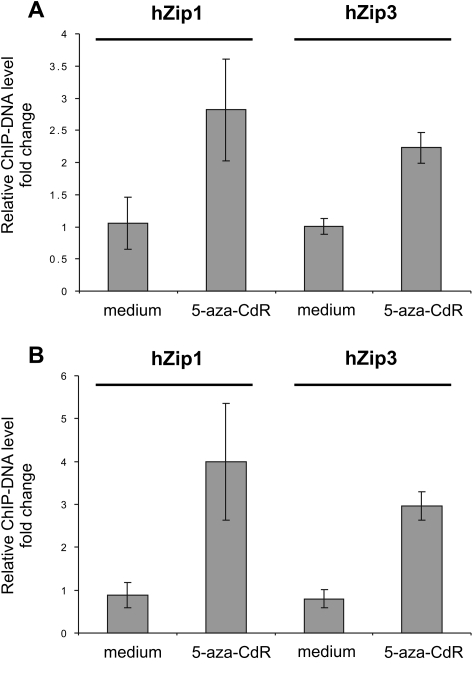
Based on the data from our preceding experiments, we examined the effect of 5-aza-CdR treatment on the methylation level of the AP-2alpha promoter region in DU-145 and LNCaP cells. As demonstrated in Figure 3E, 5-aza-CdR treatment resulted in reduced methylation of the AP-2alpha promoter regions in both DU-145 and LNCaP cells. Furthermore, demethylation of the AP-2alpha promoter coincided with an increased expression of AP-2alpha protein levels in those cell lines (Figure 3B), whereas SP-1 protein levels remained without any changes. Next, ChIP–qPCR analysis was performed to examine the effect of 5-aza-CdR treatment on binding of AP-2alpha with hZip1 and hZip3 promoters in vivo in DU-145 and LNCaP cells. As we theorized, increased binding of the AP-2alpha transcription factor with hZip1 and hZip3 promoter regions occurred in both DU-145 and LNCaP cells after 5-aza-CdR treatment (Figure 4).
Fig. 4.
ChIP–qPCR assay representing association of AP-2alpha at hZip1 and hZip3 promoter regions in vivo. Target DU-145 (A) or LNCaP (B) ChIP-DNA increased after cells were treated with 5-aza-CdR. Bars represent fold change of 2−(ΔΔCt) of target ChIP-DNA in treated cells over 2−(ΔΔCt) of target ChIP-DNA in untreated cells. Normalized against input Ct values of hZip1 and hZip3 promoters ChIP-DNA were normalized to the GAPDH promoter ChIP-DNA samples recovered with anti-AP-2alpha antibodies. DU-145 and LNCaP cells were preincubated with 5 μM of 5′-aza-CdR for 48 h with an additional 24 h incubation in cell medium alone. After incubation, cells were harvested and ChIP–qPCR was performed. The qPCR reactions were ran in triplicates. The representative data of one of the three ChIP–qPCR experiments are presented. Error bars represent ±SD of triplicates.
The impact of AP-2alpha transcription factor in regulation of hZip1 and hZip3 genes expression, cell growth and zinc accumulation in prostate cancer cells
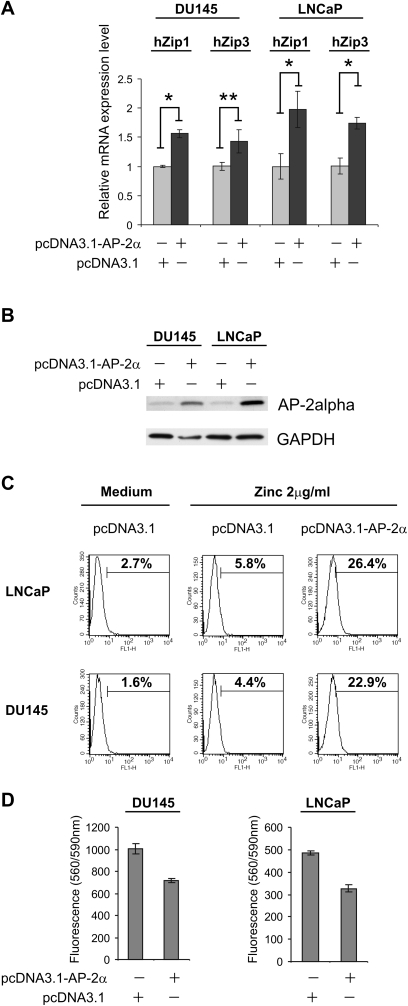
To investigate if restored expression of AP-2alpha would result in upregulated zinc transporters expression and increased zinc accumulation, we overexpressed this transcription factor in DU-145 and LNCAP prostate cancer cell lines. We performed the transient transfection of DU-145 and LNCaP cells with a pcDNA3.1 vector containing AP-2alpha open-reading frame. The relative messenger RNA levels of hZip1 and hZip3 zinc transporters were measured by qRT–PCR in DU-145 and LNCaP cells transfected with pcDNA3.1-AP-2alpha vector and control pcDNA3.1 vector. Twenty-four hours after transfection, the population of transfected cells was separated from non-transfected cells using fluorescence-activated cell sorting and incubated for additional 24 h. Cells were then harvested and subjected to RT–qPCR analysis and western blotting. As illustrated in Figure 5A, we observed increased hZip1 and hZip3 messenger RNA levels in DU-145 and LNCaP cells transfected with AP-2alpha expression construct. We then sought to examine the functional significance of increased zinc transporters expression. Consistent with the ability of hZip1 and hZip3 to enhance intracellular zinc uptake (1,17), the accumulation of labile zinc was notably increased in cells transfected with the AP-2alpha expression vector (Figure 5C) cultured in the presence of physiologically relevant concentrations of zinc. Also, we evaluated the effect of AP-2alpha expression in DU-145 and LNCaP prostate cancer cells on cellular growth using the CellTiter-Blue Assay. As displayed in Figure 5D, we observed a 25 and 33.1% inhibition in prostate cancer cell growth for DU-145 cells and LNCaP calls, respectively.
Fig. 5.
The DU-145 and LNCaP cell lines were transiently transfected with the pcDNA3.1-AP-2alpha expression vector and pcDNA3.1 empty vector. Twenty-four hours after transfection, cells were subjected to fluorescence-activated cell sorting. After another 24 h of incubation in cell medium, cells were harvested and RT–qPCR assay (A), western blot assay (B) were performed. (C) In order to asses whether transfection of DU-145 and LNCaP cells with the pcDNA-AP-2alpha expression vector has an effect on the regulation of intracellular zinc uptake, transfected DU-145 and LNCaP cells were incubated with 2 μg/ml of zinc for 1 h, washed twice in phosphate-buffered saline and incubated with the cell-permeable fluorescent zinc indicator FluoZin-3 (5 μM) for additional 15 min. Fluorescence was examined by flow cytometry. Data is representative of three independent experiments. *P < 0.01; **P < 0.05 compared with pcDNA3.1-AP-2alpha transfected cells. (D) The effect of AP-2alpha expression on cell growth was examined using the CellTiter-Blue Assay. DU-145 and LNCaP prostate cancer cells were transfected as described above, plated in a 96-well plate (5 × 103 to 10 × 103 cells per well) in triplicates and incubated for 48 h in complete cell growth medium. After the 48 h incubation period ended, the CellTiter-Blue Assay was performed. Error bars represent ±SD of triplicate fluorescence means.
Analysis of AP-2alpha promoter methylation in clinical specimens
Results of several previous studies have shown that the loss of AP-2alpha expression occurs early in the development of prostate adenocarcinoma (16) as well as during breast cancer progression (15). Our results demonstrate that AP-2alpha promoter region hypermethylation in DU-145 and LNCaP prostate cancer cell lines coincides with decreased AP-2alpha expression. Based on these results, we wanted to examine whether AP-2alpha promoter hypermethylation occurs in tissue samples from prostate glands surgically removed for localized prostate cancer. Samples of genomic DNA were isolated from frozen prostate tumors and adjacent normal tissues of five patients. After bisulfite conversion, DNA samples were amplified with specific primers and then cloned into pGEM-T Easy vector. Eight individual clones of each PCR product representing individual alleles of the AP-2alpha 5′-flanking region were sequenced. Figure 6 demonstrates that methylation of the AP-2alpha 5′-flanking region in all prostate cancer tissues was found to be markedly increased in comparison with the adjacent normal prostatic tissue except for tumor #2 where the methylation level of the AP-2alpha 5′-flanking region remained hypomethylated compared with the adjacent normal tissue.
Fig. 6.
Schematic illustration of the AP-2alpha (−654/−55) promoter region (for more detailed information, see Figure 3C) and bisulfite sequencing analysis of its methylation in prostate tumors (T) and normal adjacent tissues (N) taken from five different patients who underwent surgical removal of their prostate for localized prostate cancer. Analysis of tissue samples demonstrated distinct patterns of methylation. Individual CpG dinucleotides in the AP-2alpha promoter region are significantly hypermethylated in prostate tumor tissue in comparison with adjacent normal prostatic tissue. Each line represents an allele; each circle represents a CpG dinucleotide. Filled circles represent methylated CpG dinucleotides and clear circles represent unmethylated CpG dinucleotides.
Discussion
The process whereby a normal prostatic epithelial cell transforms into a cancerous cell involves the loss of the ability to accumulate intracellular zinc. This transformation remains one of the sine qua non cellular changes during prostatic carcinogenesis. Recent studies reveal a strong association between the development of prostate cancer and the downregulation of zinc uptake transporters hZip1 and hZip3 (6,18). Therefore, in the present study, we examined the epigenetic mechanisms that might be responsible for the downregulation of hZip1 and hZip3 genes in DU-145 and LNCaP prostate cancer cell lines.
DNA hypermethylation is one of the best-characterized epigenetic changes noted in neoplastic development and can be associated with gene silencing (10,11,19). Recent studies have shown that 5-aza-CdR appears to interrupt tumorigenesis by inhibition of DNA methyltransferase. In the present study, the expression of hZip1 and hZip3 zinc uptake transporters as well as the ability to accumulate intracellular zinc was notably increased after 5-aza-CdR treatment. Interestingly, an analysis of the promoter regions of the hZip1 and hZip3 genes did not exhibit hypermethylation in either cell line. Thus, we do not regard hypermethylation of the hZip1 and hZip3 promoters as the primary mechanism responsible for downregulation of these genes in DU-145 and LNCaP prostate cancer cells. It is probably that the epigenetic regulation of hZip1 and hZip3 gene expression is a secondary event. Indeed, it is known mechanism while downregulation of transcriptional regulators caused by their promoters hypermethylation has affected expression of their target genes (20,21).
Recently, we were able to characterize the transcriptional regulation of the hZip1 promoter (13) in contradistinction to the hZip3 promoter, whose regulatory control remained unclear. In our present work, we determined the essential role of SP1 and AP-2alpha proteins in transcriptional regulation of hZip3 expression. Additionally, we characterized the AP-2-like binding site in the hZip1 promoter, which has been shown to have an important regulatory role on hZip1 expression. Since SP1 and AP-2alpha proteins were found to be critical for transcriptional regulation of both hZip1 and hZip3 genes, we compared the expression of these transcriptional factors in DU-145 and LNCaP prostate cancer cell lines as well as in the normal prostatic epithelial cell line RWPE-1. The level of SP1 expression remained unchanged; therefore, we focused our investigations on the AP-2alpha transcription factor, which has been shown to be downregulated in both DU-145 and LNCaP cells in comparison with RWPE-1 cells.
AP-2alpha is a well-characterized transcription factor that is required for growth, normal morphogenesis and apoptosis in mammalian cells (22–24). It binds to the GC-rich promoter regions of various genes (25) where it has been shown to be involved in both transcriptional activation of some genes (26–28) and repression of other genes (29–31). Furthermore, the AP-2alpha has been associated with cellular transformation and anchorage-independent growth (32) as well as being under the stimulatory control of the ERBB2 oncogene (33).
Conversely, different investigators have demonstrated an antitumor activity of the AP-2alpha protein (29,34). For example, the loss of AP-2alpha has been strongly associated with the aggressiveness and progression of numerous types of cancer, such as gliomas, melanoma, breast, prostate and colorectal cancers (15,35–37). In particular, the AP-2alpha protein has been associated with luminal differentiation of prostatic epithelium, and its expression is lost in high-grade prostatic intraepithelial neoplastic and prostate cancer (16,38). It was also reported that the loss of AP-2alpha affects the normal expression profiles of many genes involved in carcinogenic pathways (29,39).
The downregulation of AP-2alpha protein expression caused by hypermethylation of its promoter region has been reported in hematological as well as in breast cancer and other solid cancers (15,40). Little is presently known regarding the methylation patterns of AP-2alpha promoter in prostate cancer cell lines. Thus, based on our finding of AP-2alpha downregulation in prostatic cancer cell lines, we hypothesized that the mechanism of AP-2alpha downregulation in prostate cancer cells involves hypermethylation of its promoter region. Using bisulfite sequencing, we were able to confirm this hypothesis by finding high methylation levels of the AP-2alpha (−755/−150) promoter region in DU-145 and LNCaP prostate cancer cells. In contrast, the same promoter region in RWPE-1 normal prostate epithelial cells was unmethylated. Next, we examined the effects of the demethylating agent 5-aza-CdR on methylation of AP-2alpha promoter regions in the DU-145 and LNCaP cell lines. Bisulfite sequencing revealed a reduction in methylation levels in both DU-145 and LNCaP cells, which coincided with an elevation of AP-2alpha protein levels in those cells.
Our data coupled with previous published reports suggest that the loss of the AP-2alpha protein is caused by its promoter hypermethylation during breast cancer progression (15) as well as being an early event in prostate cancer development (16). To test this supposition, we examined methylation patterns of the AP-2alpha promoter region from patients’ tissues who had Gleason score 6 prostate cancer. Gleason score 6 prostate cancer is considered to be typical for early well-differentiated disease. Patients with Gleason score 6 disease generally have a very favorable prognosis and a low risk of recurrence after therapy (41). Bisulfite sequencing of the AP-2alpha promoter region in five human tissue samples with prostate cancer revealed that 80% of these patients had a high methylation level at the AP-2alpha promoter region. Despite the fact that the level of AP-2alpha promoter methylation was lower in human prostate cancer tissue than compared with the prostate cancer cell lines DU-145 and LNCaP, the degree of methylation in human prostate cancer tissues was still significantly higher than compared with adjacent normal prostate tissue as well as in normal prostate epithelial RWPE-1 cells. These data suggest that the methylation of AP-2alpha promoter region in prostate cancer cells is an early event in neoplastic development.
In order to test our theory that the AP-2alpha protein plays a significant role in the transcriptional regulation of both hZip1 and hZip3 genes, we re-established an altered expression of AP-2alpha in the DU-145 and LNCaP prostate cancer cell lines. We transfected DU-145 and LNCaP prostate cancer cells with an AP-2alpha expression vector that resulted in the upregulation of the hZip1 and hZip3 genes. As a result of this upregulation, these transfected cells demonstrated a significantly higher zinc uptake.
The re-establishment of normal intracellular zinc levels is a particularly attractive target for therapeutic manipulation given the importance of dysregulation of zinc metabolism in the development and progression of prostate cancer. Recent studies have demonstrated that the activation of Ras responsive element binding protein-1 resulted in hZip1 zinc transporter downregulation in prostate cancer cells (42). In addition to this work, the present study, to our knowledge, represents the first evidence that decreased zinc uptake and expression of zinc transporters in prostate cancer cells have been particularly the result of epigenetic repression of AP-2alpha which plays an important role in zinc transporters transcriptional regulation. These findings certainly represent future therapeutic targets for vigorous investigation in the treatment of prostate cancer.
Supplementary material
Supplementary Tables 1–4 can be found at http://carcin.oxfordjournals.org/
Funding
This work was supported in part by National Institutes of Health (Grant RO1 CA108890, RO1 CA134463 to V.M.K.); CCSG Pilot Project Award (V.M.K.); American Institute for Cancer Research Grant (R.G.U.); Department of Defense, Physician Research Training Award (A.K.).
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AP
activator protein
- 5-aza-CdR
5-aza-2′-deoxycytidine
- SP1
Specificity Protein 1
References
- 1.Gaither LA, et al. The human ZIP1 transporter mediates zinc uptake in human K562 erythroleukemia cells. J. Biol. Chem. 2001;276:22258–22264. doi: 10.1074/jbc.M101772200. [DOI] [PubMed] [Google Scholar]
- 2.Costello LC, et al. The intermediary metabolism of the prostate: a key to understanding the pathogenesis and progression of prostate malignancy. Oncology. 2000;59:269–282. doi: 10.1159/000012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogunlewe JO, et al. Zinc and cadmium concentrations in indigenous blacks with normal, hypertrophic, and malignant prostate. Cancer. 1989;63:1388–1392. doi: 10.1002/1097-0142(19890401)63:7<1388::aid-cncr2820630725>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 4.Zaichick V, et al. Zinc in the human prostate gland: normal, hyperplastic and cancerous. Int. Urol. Nephrol. 1997;29:565–574. doi: 10.1007/BF02552202. [DOI] [PubMed] [Google Scholar]
- 5.Huang L, et al. Decreased intracellular zinc in human tumorigenic prostate epithelial cells: a possible role in prostate cancer progression. Cancer Cell Int. 2006;6:10. doi: 10.1186/1475-2867-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin RB, et al. hZIP1 zinc uptake transporter down regulation and zinc depletion in prostate cancer. Mol. Cancer. 2005;4:32. doi: 10.1186/1476-4598-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desouki MM, et al. hZip2 and hZip3 zinc transporters are down regulated in human prostate adenocarcinomatous glands. Mol. Cancer. 2007;6:37. doi: 10.1186/1476-4598-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golovine K, et al. Overexpression of the zinc uptake transporter hZIP1 inhibits nuclear factor-kappaB and reduces the malignant potential of prostate cancer cells in vitro and in vivo. Clin. Cancer Res. 2008;14:5376–5384. doi: 10.1158/1078-0432.CCR-08-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ushijima T, et al. Aberrant DNA methylation in contrast with mutations. Cancer Sci. 2010;101:300–305. doi: 10.1111/j.1349-7006.2009.01434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pierconti F, et al. Epigenetic silencing of SOCS3 identifies a subset of prostate cancer with an aggressive behavior. Prostate. 2011;71:318–325. doi: 10.1002/pros.21245. [DOI] [PubMed] [Google Scholar]
- 11.Collard RL, et al. Methylation of the ASC gene promoter is associated with aggressive prostate cancer. Prostate. 2006;66:687–695. doi: 10.1002/pros.20371. [DOI] [PubMed] [Google Scholar]
- 12.McCabe MT, et al. Inhibition of DNA methyltransferase activity prevents tumorigenesis in a mouse model of prostate cancer. Cancer Res. 2006;66:385–392. doi: 10.1158/0008-5472.CAN-05-2020. [DOI] [PubMed] [Google Scholar]
- 13.Makhov P, et al. Transcriptional regulation of the major zinc uptake protein hZip1 in prostate cancer cells. Gene. 2009;431:39–46. doi: 10.1016/j.gene.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, et al. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Douglas DB, et al. Hypermethylation of a small CpGuanine-rich region correlates with loss of activator protein-2alpha expression during progression of breast cancer. Cancer Res. 2004;64:1611–1620. doi: 10.1158/0008-5472.can-0318-2. [DOI] [PubMed] [Google Scholar]
- 16.Ruiz M, et al. Activator protein 2alpha transcription factor expression is associated with luminal differentiation and is lost in prostate cancer. Clin. Cancer Res. 2001;7:4086–4095. [PubMed] [Google Scholar]
- 17.Kelleher SL, et al. Zip3 (Slc39a3) functions in zinc reuptake from the alveolar lumen in lactating mammary gland. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;297:R194–R201. doi: 10.1152/ajpregu.00162.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rishi I, et al. Prostate cancer in African American men is associated with downregulation of zinc transporters. Appl. Immunohistochem. Mol. Morphol. 2003;11:253–260. doi: 10.1097/00129039-200309000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Ribarska T, et al. Epigenetic inactivation of the placentally imprinted tumor suppressor gene TFPI2 in prostate carcinoma. Cancer Genomics Proteomics. 2010;7:51–60. [PubMed] [Google Scholar]
- 20.Guo X, et al. Homeobox gene IRX1 is a tumor suppressor gene in gastric carcinoma. Oncogene. 2010;29:3908–3920. doi: 10.1038/onc.2010.143. [DOI] [PubMed] [Google Scholar]
- 21.Schayek H, et al. Progression to metastatic stage in a cellular model of prostate cancer is associated with methylation of the androgen receptor gene and transcriptional suppression of the insulin-like growth factor-I receptor gene. Exp. Cell. Res. 2010;316:1479–1488. doi: 10.1016/j.yexcr.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schorle H, et al. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell PJ, et al. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 24.Orso F, et al. The AP-2alpha transcription factor regulates tumor cell migration and apoptosis. Adv. Exp. Med. Biol. 2007;604:87–95. doi: 10.1007/978-0-387-69116-9_6. [DOI] [PubMed] [Google Scholar]
- 25.Eckert D, et al. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena P, et al. Activator protein-2 mediates transcriptional activation of the CYP11A1 gene by interaction with Sp1 rather than binding to DNA. Mol. Endocrinol. 1999;13:1402–1416. doi: 10.1210/mend.13.8.0335. [DOI] [PubMed] [Google Scholar]
- 27.Tan CC, et al. Transcription factor Ap2delta associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription. Proc. Natl Acad. Sci. USA. 2008;105:7472–7477. doi: 10.1073/pnas.0711896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S, et al. Extracellular matrix fibronectin increases prostaglandin E2 receptor subtype EP4 in lung carcinoma cells through multiple signaling pathways: the role of AP-2. J. Biol. Chem. 2007;282:7961–7972. doi: 10.1074/jbc.M610308200. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz M, et al. Activator protein 2alpha inhibits tumorigenicity and represses vascular endothelial growth factor transcription in prostate cancer cells. Cancer Res. 2004;64:631–638. doi: 10.1158/0008-5472.can-03-2751. [DOI] [PubMed] [Google Scholar]
- 30.Zhu CH, et al. Dominant negative interference of transcription factor AP-2 causes inhibition of ErbB-3 expression and suppresses malignant cell growth. Breast Cancer Res. Treat. 2002;71:47–57. doi: 10.1023/a:1013378113916. [DOI] [PubMed] [Google Scholar]
- 31.Ren Y, et al. Transcription factor AP-2 functions as a repressor that contributes to the liver-specific expression of serum amyloid A1 gene. J. Biol. Chem. 2001;276:17770–17778. doi: 10.1074/jbc.M010307200. [DOI] [PubMed] [Google Scholar]
- 32.Kannan P, et al. N-ras oncogene causes AP-2 transcriptional self-interference, which leads to transformation. Genes Dev. 1994;8:1258–1269. doi: 10.1101/gad.8.11.1258. [DOI] [PubMed] [Google Scholar]
- 33.Begon DY, et al. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J. Biol. Chem. 2005;280:24428–24434. doi: 10.1074/jbc.M503790200. [DOI] [PubMed] [Google Scholar]
- 34.Sumigama S, et al. Suppression of invasion and peritoneal carcinomatosis of ovarian cancer cells by overexpression of AP-2alpha. Oncogene. 2004;23:5496–5504. doi: 10.1038/sj.onc.1207723. [DOI] [PubMed] [Google Scholar]
- 35.Huang S, et al. Loss of AP-2 results in downregulation of c-KIT and enhancement of melanoma tumorigenicity and metastasis. EMBO J. 1998;17:4358–4369. doi: 10.1093/emboj/17.15.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jean D, et al. Loss of AP-2 results in up-regulation of MCAM/MUC18 and an increase in tumor growth and metastasis of human melanoma cells. J. Biol. Chem. 1998;273:16501–16508. doi: 10.1074/jbc.273.26.16501. [DOI] [PubMed] [Google Scholar]
- 37.Heimberger AB, et al. Loss of the AP-2alpha transcription factor is associated with the grade of human gliomas. Clin. Cancer Res. 2005;11:267–272. [PubMed] [Google Scholar]
- 38.Lipponen P, et al. Expression of activator protein 2 in prostate cancer is related to tumor differentiation and cell proliferation. Eur. Urol. 2000;37:573–578. doi: 10.1159/000020195. [DOI] [PubMed] [Google Scholar]
- 39.Marreiros A, et al. KAI1 promoter activity is dependent on p53, junB and AP2: evidence for a possible mechanism underlying loss of KAI1 expression in cancer cells. Oncogene. 2005;24:637–649. doi: 10.1038/sj.onc.1208216. [DOI] [PubMed] [Google Scholar]
- 40.Dunwell T, et al. A genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Mol. Cancer. 2010;9:44. doi: 10.1186/1476-4598-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blute ML, et al. Use of Gleason score, prostate specific antigen, seminal vesicle and margin status to predict biochemical failure after radical prostatectomy. J. Urol. 2001;165:119–125. doi: 10.1097/00005392-200101000-00030. [DOI] [PubMed] [Google Scholar]
- 42.Milon BC, et al. Ras responsive element binding protein-1 (RREB-1) down-regulates hZIP1 expression in prostate cancer cells. Prostate. 2010;70:288–296. doi: 10.1002/pros.21063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.