Abstract
Cell cycle progression contributes to the cellular response to DNA-damaging factors, such as chemotherapy and radiation. We hypothesized that the genetic variations in cell cycle pathway genes may modulate treatment responses and affect survival in patients with advanced non-small-cell lung cancer (NSCLC). We genotyped 374 single-nucleotide polymorphisms (SNPs) from 49 cell cycle-related genes in 598 patients with stages III–IV NSCLC treated with first-line platinum-based chemotherapy with/without radiation. We analyzed the individual and combined associations of these SNPs with survival and evaluated their gene–gene interactions using survival tree analysis. In the analysis of survival in all the patients, 39 SNPs reached nominal significance (P < 0.05) and 4 SNPs were significant at P <0.01. However, none of these SNPs remained significant after correction for multiple comparisons at a false discovery rate of 10%. In stratified analysis by treatment modality, after adjusting for multiple comparisons, nine SNPs in chemotherapy alone and one SNP in chemoradiation remained significant. The most significant SNP in chemotherapy group was CCNB2:rs1486878 [hazard ratio (HR) = 1.69, 95% confidence interval (CI), 1.25–2.30, P = 0.001]. TP73: rs3765701 was the only significant SNP in chemoradiation group (HR = 1.87; 95% CI = 1.35–2.59, P = 1.8 × 10−4). In cumulative analysis, we found a significant gene-dosage effect in patients receiving chemotherapy alone. Survival tree analysis demonstrated potential higher order gene–gene and gene–treatment interactions, which could be used to predict survival status based on distinct genetic signatures. These results suggest that genetic variations in cell cycle pathway genes may affect the survival of patients with stages III–IV NSCLC individually and jointly.
Introduction
Despite the development of many novel molecularly targeted agents and new chemotherapy regimens for cancer treatment in recent years, non-small-cell lung cancer (NSCLC), which accounts for ∼85% of all lung cancers (1), still remains the leading cause of cancer death around the world, with over 85% of patients surviving <5 years (2). In clinical practice, the standard therapy for patients with stages III–IV NSCLC is platinum-based chemotherapy, with or without radiotherapy. However, the major problem with these treatment options is that clinicians do not know which patient will benefit from which therapy (i.e. the sensitivity or resistance of the disease to chemotherapy and/or radiotherapy always remains unknown before the treatment is administered) (3). Thus, developing individualized treatments is critical to improving the clinical outcomes of stage III–IV NSCLC patients.
Most of the current chemotherapeutic agents (such as platinum and fluorouracil) and radiation target DNA and induce various types of DNA damage (4,5). In the presence of DNA damage, cell cycle checkpoints are activated and cell cycle progression is paused, allowing cells sufficient time to repair the damage before continuing cell division (6). Cell cycle checkpoints occur at the G1/S and G2/M transitions as well as at the intra-S-phase. One of the hallmarks of human cancers is the alteration of many signaling pathways, leading to the loss of basic cell cycle (7); this results in unrestrained cell proliferation, cell cycle deregulation and ultimately, cancer development (8).
In cells, the cyclins and cyclin-dependent kinases (CDKs) interact at specific stages of the cell cycle to drive the cell cycle from one phase to the next. For instance, the cell cycle is negatively regulated by several endogenous CDK inhibitors (e.g. CDKN1A, 1B and 1C and CDKN2A, 2B, 2C and 2D) that bind to and inactivate the cyclin–CDK complexes. CDK4 and CDK6 preferentially bind to D-type cyclins (cyclin D1, D2 or D3) (9,10) and drive progression through G1, whereas the CDK2/cyclin E complex is essential for the G1 to S transition. In the G1 phase, a second regulatory pathway, c-Myc, directly stimulates the expression of cyclin E and Cdc25a. In addition, Rb and its related proteins, Rbl1 and Rbl2, regulate the E2F family of transcription factors and affect the transition from G1 into S phase (11). DNA damage in the G1 phase always leads to the activation of the TP53 tumor suppressor, resulting in either G1 arrest or programmed cell death (12). A previous study showed that TP73 can partially substitute for TP53 as a tumor suppressor (13). The G2/M transition is monitored by the CDK1–cyclin B complex, and the G2/M checkpoint is initiated by the phosphorylation of checkpoint kinases (CHEK1 and CHEK2) and phosphatases (likely Cdc25c) (14). Besides the major cyclins, CDKs and CDK inhibitors mentioned above, many other regulatory proteins also participate in multiple phases of the cell cycle (15).
Somatic mutations have been observed in genes encoding many cell cycle checkpoint proteins, including cyclins, CDKs and CDK inhibitors, in lung and other human cancers (7,16,17). However, whether germ line genetic variations of cell cycle-related genes have an influence on the clinical outcomes of patients with NSCLC remains largely unknown. In this study, we used a pathway-based approach to systematically query genetic variations in major cell cycle pathway genes and evaluate their associations with the survival of stage III–IV patients receiving first-line platinum-based chemotherapy or chemoradiation.
Patients and methods
Patient population
The study population has been described previously (18). In brief, the subjects consisted of patients with Stage III–IV NSCLC from an epidemiological lung cancer study being conducted at The University of Texas M.D. Anderson Cancer Center. The patients were enrolled from 1995 to 2007. There were no recruitment restrictions on age, gender or ethnicity. All patients had newly diagnosed and histologically confirmed NSCLC and received platinum-based (carboplatinum or cisplatin) chemotherapy with/without radiotherapy at M.D. Anderson Cancer Center. None of the patients had been previously treated by surgery, chemotherapy and/or radiotherapy before enrollment into the study. This study was approved by the Institutional Review Board at M.D. Anderson and all patients signed an informed consent form before enrollment.
Collection of epidemiological and clinical data
The patients were interviewed by trained M.D. Anderson staff interviewers who used a structured questionnaire to collect information on demographic characteristics (age, gender, ethnicity, etc), work history, tobacco use history [including smoking status, as defined previously (19)], medical history and family history of cancer. Performance status was defined by the Eastern Cooperative Oncology Group (ECOG) scale at diagnosis (20). Peripheral blood samples were collected from patients at the end of each interview. Clinical and follow-up data were abstracted from patients’ medical records. Overall survival was used as the end point of this study.
Single-nucleotide polymorphism selection and genotyping
The selection of cell cycle pathway genes and single-nucleotide polymorphisms (SNPs) in each gene was done following a procedure that was described previously (21). Briefly, 49 cell cycle pathway genes were selected based on an extensive literature review of the PubMed database (5,7,10,13,22,23) and annotated by the Gene Ontology (GO) database (http://www.geneontology.org/). For SNPs, we utilized the International HapMap Project database (http://www.hapmap.org) (24) and dbSNP database (http://www.ncbi.nlm.nih.gov/SNP) (25) to select the tagging and potentially functional SNPs located within 10 kb upstream of the 5′ untranslated region and 10 kb downstream of the 3′ untranslated region of each gene. The tagging SNPs were chosen with a r2 ≥0.80 and a minor allele frequency ≥0.05 in Caucasian population. The potential functional SNPs had a minor allele frequency ≥0.01. Based on the information set of selected SNPs, a custom iSelect Infinium II Beadchip was designed by a proprietary program of Illumina. Genomic DNA was extracted from peripheral blood lymphocytes using the Human Blood Genomic DNA Extraction Kit (Qiagen, Valencia, CA). Genotyping was performed following the standard protocol for Illumina’s Infinium iSelect HD Custom Genotyping Beadchip (San Diego, CA). The BeadStudio software was used to autocall all genotypes. Each SNP had a call rate >95%.
Statistical analysis
In order to investigate the association between SNPs and treatment response, we stratified the studied population into two subgroups according to different therapeutic regimens: chemotherapy alone and chemotherapy with concurrent and/or subsequent radiotherapy. Pearson’s χ2-test or Student’s t-test were used to compare the distributions of the selected demographic and clinical variables by survival status. For each SNP, three different genetic models (dominant, recessive and additive) were analyzed, and the model with the highest statistical significance was considered to be the best-fitting model. We used a Cox proportional hazards model to evaluate the effects of the SNPs on overall survival. The survival time was defined as the time from the date of first treatment to the date of death or last follow-up. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated by the Cox model after adjustment for age, gender, ethnicity, smoking status, performance status and clinical stage. The Benjamini–Hochberg false discovery rate (FDR) method was used to obtain FDR-adjusted q-value to account for multiple comparisons (26). Kaplan–Meier curves and log-rank tests were also used to evaluate the effect of the SNPs on survival time.
Analysis of the cumulative effects of unfavorable genotypes was also performed, and patients were divided into low-, medium- and high-risk groups based on the tertile distribution of the number of unfavorable genotypes. HRs and 95% CIs were calculated using the low-risk group as the reference group. A gene-based analysis was used to explore the associations between cell cycle genes and overall survival using the likelihood-ratio test as described previously (27). Survival tree analyses were used to identify higher order gene–gene interactions by using the modified STREE program (http://c2s2.yale.edu/software/stree/), which uses recursive partitioning to identify subgroups of individuals with different risks of death. The statistical analyses described above were completed using STATA software (version 10, College Station, TX) and SAS/Genetics, version 9.0 (SAS Institute, Cary, NC). P < 0.01 and q < 0.10 were considered statistically significant.
Results
Characteristics of patients
There were 598 stages III–IV NSCLC patients who had received platinum-based chemotherapy or chemoradiation enrolled in this study with a median follow-up time of 11.8 months. At the time of analysis, only 142 were still alive and 456 had died. The overall median survival time (MST) was 12.9 months. The mean age was 59.6 years (SD: 10.5, range: 28–81). The majority of patients (78.6%) were of European descent. There were no significant differences in terms of age (P = 0.884), ethnicity (P = 0.937), smoking status (P = 0.860), tumor grade (P = 0.482) or weight loss (P = 0.390) by vital status. There were significant differences in sex (P = 0.002), clinical stage (P = 0.0035) and performance status (P = 0.0015) between the two groups of patients (Supplementary Table 1 is available at Carcinogenesis Online). The chemoradiation group had a higher survival rate (27%) and then the group that received chemotherapy alone (19.62%) due to different stage distribution (31.8 and 87.7% stage IV patients for the former and the latter group, respectively, data not shown).
Associations between individual SNPs and survival among all patients
In the individual SNP analysis among all patients, there were 39 SNPs that reached nominal significance (P < 0.05) (Supplementary Table 2 is available at Carcinogenesis Online), among which four SNPs, TP73:rs3765703 (P = 0.0017); TP73:rs3765701 (P = 0.0040), CDC7:rs12125947 (P = 0.0054), and CDK8:rs4770974 (P = 0.0068), were significant at P < 0.01 (Supplementary Table 3 is available at Carcinogenesis Online). But after correction for multiple comparisons at an FDR of 10%, none of these SNPs remained significant.
Associations between SNPs and survival in patients stratified by treatment
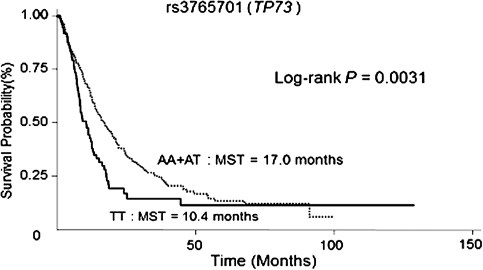
We then performed stratified analysis based on two different therapeutic regimens (chemotherapy alone and chemoradiation). There were 12 SNPs in chemotherapy alone group (Supplementary Table 4 is available at Carcinogenesis Online) and 11 SNPs in chemoradiation group (Supplementary Table 5 is available at Carcinogenesis Online) that were significant at P <0.01. After correcting for multiple comparisons at an FDR of 10%, nine SNPs in the chemotherapy group and one SNP in the chemoradiation group remained significant (Table I). The most significant SNP in the chemotherapy group was CCNB2:rs1486878: patients carrying at least one variant allele had a significantly increased risk of death (HR = 1.69; 95% CI = 1.25–2.30, P = 0.001). The seven other SNPs, CCNB2: rs10851643 (HR = 1.60; 95% CI = 1.19–2.14), CCND1:rs1683847 (HR = 1.35; 95% CI = 1.09–1.66), CCND2:rs12230555 (HR = 1.90; 95% CI = 1.26–2.87), CHEK2:rs743185 (HR = 1.58; 95% CI = 1.15–2.17), E2F3:rs942042 (HR = 1.63; 95% CI = 1.18–2.25), RB1:rs9568029 and rs7329938 (HRs = 1.64; 95% CI = 1.20–2.23 and 1.68; 95% CI = 1.17–2.41, respectively), were associated with an increased risk of death, whereas the remaining one,CDC2:rs2127355 (HR = 0.71; 95% CI = 0.57–0.88), was associated with a decreased risk of death. In the chemoradiation group, only one SNP, rs3765701 in TP73, remained significant: patients carrying the homozygous variant genotype had an increased risk of death (HR = 1.87; 95% CI = 1.35–2.59, P = 1.8 × 10−4) and a shorter MST compared with patients with at least one major allele (10.4 versus 17.0 months) (Figure 1).
Table I.
Associations between genetic variants in cell cycle pathway and survival in the patients with stages III–IV NSCLC stratified by therapeutic regimens
| Gene | SNP/genotype | Alive/dead, n | HR (95% CI)a | P |
| Chemotherapy alone | ||||
| CCNB2 | rs1486878 | |||
| CC | 26/92 | 1 (Reference) | ||
| CG | 20/93 | 1.69 (1.23–2.33) | 0.0011 | |
| GG | 5/24 | 1.69 (1.01–2.82) | 0.0457 | |
| Dominant | 1.69 (1.25–2.30) | 0.0007 | ||
| Recessive | 1.27 (0.78–2.06) | 0.3287 | ||
| Additive | 1.41 (1.13–1.76) | 0.0023 | ||
| rs10851643 | 1.60 (1.19–2.14) | 0.0017 | ||
| AA | 28/99 | 1 (Reference) | ||
| AC | 17/92 | 1.66 (1.22–2.25) | 0.0011 | |
| CC | 6/18 | 1.34 (0.79–2.28) | 0.2812 | |
| Dominant | 1.60 (1.19–2.14) | 0.0017 | ||
| Recessive | 1.06 (0.64–1.76) | 0.8196 | ||
| Additive | 1.31 (1.06–1.62) | 0.0119 | ||
| CCND1 | rs1683847 | |||
| GG | 23/57 | 1 (Reference) | ||
| GT | 23/99 | 1.38 (0.99–1.94) | 0.0598 | |
| TT | 5/53 | 1.81 (1.19–2.75) | 0.0056 | |
| Dominant | 1.48 (1.07–2.04) | 0.0168 | ||
| Recessive | 1.47 (1.03–2.10) | 0.0331 | ||
| Additive | 1.35 (1.09–1.66) | 0.0049 | ||
| CCND2 | rs12230555 | |||
| CC | 26177 | 1 (Reference) | ||
| CT | 33/92 | 0.76 (0.55–1.06) | 0.1069 | |
| TT | 9/46 | 1.60 (1.01–2.53) | 0.0473 | |
| Dominant | 0.86 (0.64–1.18) | 0.3728 | ||
| Recessive | 1.90 (1.26–2.87) | 0.0023 | ||
| Additive | 1.12 (0.88–1.42) | 0.3606 | ||
| CDC2 | rs2127355 | |||
| AA | 13/67 | 1 (Reference) | ||
| AG | 26/104 | 0.69 (0.50–0.96) | 0.0273 | |
| GG | 12/38 | 0.51 (0.33–0.79) | 0.0022 | |
| Dominant | 0.64 (0.47–0.87) | 0.0045 | ||
| Recessive | 0.65 (0.45–0.95) | 0.0267 | ||
| Additive | 0.71 (0.57–0.88) | 0.0017 | ||
| CHEK2 | rs743185 | |||
| CC | 38/142 | 1 (Reference) | ||
| CT | 13/59 | N/A | N/A | |
| TT | 0/8 | N/A | N/A | |
| Dominant | 1.58 (1.15–2.17) | 0.0050 | ||
| Recessive | N/A | N/A | ||
| Additive | N/A | N/A | ||
| E2F3 | rs942042 | |||
| AA | 45/147 | 1 (Reference) | ||
| AG | 6/58 | N/A | N/A | |
| GG | 0/4 | N/A | N/A | |
| Dominant | 1.63 (1.18–2.25) | 0.0031 | ||
| Recessive | N/A | N/A | ||
| Additive | N/A | N/A | ||
| RB1 | rs9568029 | |||
| CC | 30/82 | 1 (Reference) | ||
| CT | 13/84 | 1.78 (1.28–2.48) | 0.0007 | |
| TT | 8/43 | 1.36 (0.89–2.09) | 0.1540 | |
| Dominant | 1.64 (1.20–2.23) | 0.0018 | ||
| Recessive | 1.02 (0.69–1.50) | 0.9385 | ||
| Additive | 1.23 (1.02–1.50) | 0.0343 | ||
| rs7329938 | DOM | 1.68 (1.17–2.41) | 0.0046 | |
| CC | 43/165 | 1 (Reference) | ||
| CT | 7/42 | N/A | N/A | |
| TT | 1/2 | N/A | N/A | |
| Dominant | 1.68 (1.17–2.41) | 0.0046 | ||
| Recessive | N/A | N/A | ||
| Additive | N/A | N/A | ||
| Chemoradiation | ||||
| TP73 | rs3765701 | |||
| AA | 31/75 | 1 (Reference) | ||
| AT | 44/114 | 1.07 (0.77–1.49) | 0.6971 | |
| TT | 16/57 | 1.94 (1.32–2.86) | 0.0007 | |
| Dominant | 1.27 (0.93–1.73) | 0.1329 | ||
| Recessive | 1.87 (1.35–2.59) | 1.8 × 10−4 | ||
| Additive | 1.38 (1.12–1.69) | 0.0020 | ||
The significant results after multiple comparisons were shown in boldface (FDR-adjusted P-value, q < 0.1). N/A, the number of the rare homozygotes was <5%; therefore, only dominant model was applied.
Adjusted for age, gender, ethnicity, smoking status, performance status and clinical stage.
Fig. 1.
Kaplan–Meier curves for patients receiving chemotherapy with concurrent and/or subsequent radiotherapy by genetic variants of TP73: rs3765701.
Cumulative effects of multiple unfavorable genotypes on survival
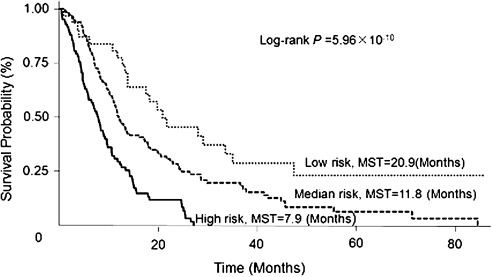
In the chemotherapy group, we performed a cumulative analysis of the nine SNPs that remained significant after multiple comparison correction and observed a significant gene-dosage effect. Compared with patients carrying no or only one unfavorable genotype (low risk), patients carrying two to four (median risk) and five to seven (high risk) unfavorable genotypes exhibited a progressively increased risk of death, with HRs of 2.47 (95% CI = 1.50–4.07) and 5.93 (95% CI = 3.44–10.30), respectively (P for trend = 2.91 × 10−6; Table II). We then plotted Kaplan–Meier curves for the different risk groups. The MSTs for patients in the median-risk and high-risk groups were 11.8 and 7.9 months, respectively, compared with 20.9 months for those in the low-risk group (Log-rank P = 5.96 × 10−10, Figure 2).
Table II.
Cumulative analysis of unfavorable genotypes in patients with stages III–IV NSCLC receiving chemotherapy alone
| No. of unfavorable genotypesa |
Alive/dead, n | HR (95% CI)b | P | |
| Low risk | 0–1 | 10/21 | 1 (Reference) | |
| Median risk | 2–4 | 34/112 | 2.47 (1.50–4.07) | 3.60 × 10−4 |
| High risk | 5–7 | 7/76 | 5.95 (3.44–10.30) | 1.87 × 10−10 |
| P for trend | 4.99 × 10−12 | |||
Unfavorable genotypes include CCNB2: rs1486878 CG + GG and rs10851643 AC + CC; CDC2: rs2127355 AA; RB1: rs9568029 CT + TT and rs7329938 CT + TT; CCND1: rs1683847 GT + TT; CCND2: rs12230555 TT; E2F3: rs942042 AG + GG and CHEK2: rs743185 CT + TT.
Adjusted for age, gender, ethnicity, smoking status, performance status and clinical stage.
Fig. 2.
Kaplan–Meier curves for patients receiving chemotherapy alone by different risk groups of unfavorable genotypes in cell cycle pathway genes.
Gene-based analysis for NSCLC survival stratified by treatment
We further explored the association of each gene in the cell cycle pathway with NSLC survival in each treatment group using the dominant and additive model (Supplementary Table 6 is available at Carcinogenesis Online). In the chemotherapy subgroup, the significant genes (global P < 0.05) for the gene-based analysis were CCNB2, CCND1, CDC2, E2F3, E2F8 and RB1. With the exception of E2F8, these genes are also related to the significant SNPs in the single SNP analysis. For patients treated with chemoradiation, the significant genes were CCND2, CDK8, E2F6, RB1 and TP73.
Survival tree analysis
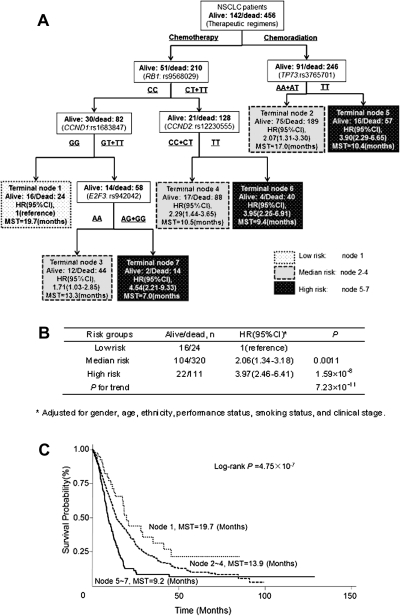
Survival tree analysis was then performed using the 10 significant SNPs (Table I) and the therapeutic modality (chemotherapy alone and chemoradiation). Treatment modality (chemotherapy versus chemoradiation) was chosen mostly based on clinical stage, and the tree model recognized the therapy as the initial split and the best predictor of survival (Figure 3A). RB1:rs9568029 and TP73:rs3765701 appeared at the second level of the tree structure, indicating that RB1 was the major factor determining the survival of patients receiving chemotherapy alone, whereas TP73 was the major factor determining the survival of patients receiving chemoradiation. The survival tree divided patients into seven terminal nodes with distinct survival patterns. The node 1 was the best survival group composed of patients receiving chemotherapy alone and carrying the RB1:rs9568029 CC genotype and the CCND1:rs1683847 GG genotype. Patients in this group had the longest MST of 19.7 months. The worst survival group was the node 7 consisting of patients of chemotherapy group with RB1:rs9568029 CT + TT, CCND1:rs1683847 GT + TT and E2F3:rs942042 AG + GG genotypes. Patients in this group had the shortest MST of 7 months. The terminal nodes were then grouped into three categories based on the tertile distribution of the estimated HRs for each node: low risk (node 1), medium risk (nodes 2–4) and high risk (nodes 5–7). Compared with the low-risk group, the HRs for the medium- and high-risk groups were 2.06 (95% CI = 1.34–3.18) and 3.97 (95% CI = 2.46–6.41), respectively (P for trend = 7.23 × 10−11), (Figure 3B). Figure 3C shows the Kaplan–Meier survival curves for these three risk groups. The estimated MST was 9.2 months for the high-risk group, 13.9 months for the medium-risk group and 19.7 months for the low-risk group, respectively (log-rank P = 4.75 × 10−7).
Fig. 3.
(A) Survival tree analysis showing the interactions among SNPs for the two therapeutic modalities; (B) Cox proportional model based on risk groups from the survival tree analysis; (C) Kaplan–Meier curves showing varied survival times in patients within different risk groups based on survival tree analysis.
Discussion
The cell cycle pathway is one of the most important cellular signal pathways that determine whether cells will survive or die when encountering any DNA-damaging factors (8). We have identified significant associations between several SNPs in cell cycle pathway and survival of advanced NSCLC patients receiving specific therapeutic modality. In individual SNP analysis of all patients combined, no SNPs remained significant after correction for multiple comparisons, suggesting different clinical variables and treatment modality exhibit profound effect on the survival of advanced NSCLC patients that overwhelms the effect of genetic variations and cannot be adjusted by statistical analyses. Therefore, for clinical outcome study of germ line genetic variations, it may be necessary to stratify by treatment modality.
In patients receiving chemotherapy alone, nine SNPs remained significant after adjusting for multiple comparisons and there was a significant gene-dosage effect, consistent with the critical role of cell cycle control in lung carcinogenesis and in DNA damage response of chemotherapeutic agents. The two most frequent mutational events in lung cancer occur to TP53 and Rb, both of which are critical players in cell cycle control (28). DNA damage induced by chemotherapeutic agents such as platinum activates cell cycle checkpoints and pauses cell cycle progression, which allows cells sufficient time to repair the damage before continuing cell division (6). Both DNA damage repair system and cell cycle control regulation are intimately involved in cellular response to chemotherapy. Many reports have shown that the expressions of DNA repair genes and genetic variations in DNA repair genes affect survival of NSCLC patients receiving platinum-based chemotherapy (29–31). However, there have only been scattered reports of genetic variations in cell cycle control pathway and clinical outcomes of NSCLC patients (32). To our knowledge, this current study is the most comprehensive largest study of cell cycle gene SNPs in lung cancer prognosis.
In the patients receiving chemoradiation, TP73:rs3765701 was the only SNP statistically associated with survival after adjusting for multiple comparisons. Previous studies have reported irradiation-induced TP73 expression in various tumor cells in vitro (33,34). In addition, TP73 may compensate for TP53 function by inducing apoptosis after irradiation (35). These results provide the biological plausibility for the involvement of TP73 in the therapeutic response of NSCLC patients to radiotherapy. There were more significant SNPs found in chemotherapy group alone than in the chemoradiation group, which may be explained by more homogeneous patient group and treatment for the former group, stronger effect of genetic variations on stage IV patients or potential gene–treatment interaction.
Survival tree analysis allowed us to explore how gene–gene and gene–treatment interactions modulate clinical outcomes. As expected, the therapeutic modality (chemotherapy versus chemoradiation) was the initial split in the tree structure because treatment modality was decided mostly based on clinical stage, which is the major determining factor for survival. SNPs under the initial split revealed the potential interactions among those genes. It is well known that there are complex genetic and molecular networks that determine the cellular function and response to therapy. In our study, the tree structure might provide some clues for these complex interactions. As shown in Figure 3A, the CCND2:rs12230555 and RB1:rs9568029 interacts, which is supported by the previous report that CCND2 could inactivate RB1 by phosphorylation (36). The CCND1:rs1683847 also interacts with RB1:rs9568029, in accordance with previous studies reporting that overexpression of CCND1 was associated with RB1 dysfunction (37) and cisplatin resistance (38). The appearance of an E2F3 SNP in the branch of RB1 and CCND1 SNPs is consistent with the fact that E2F proteins are key proteins for RB1 to control the G1/S transition of cell cycle (39). Nevertheless, it should be noted that this survival tree analysis is a post hoc exploratory analysis and the observed interactions are statistical interactions. Several nodes have very few patients in the survivor group and the analysis may be less stable. Caution should be taken when interpreting these results. The results need to be validated in independent studies.
Gene-based analysis in patients stratified by treatment showed that for the most part, the top genes from the individual SNP analysis were also found to be significant in gene-based analysis. Interestingly, in the chemoradiation subgroup, gene-based result revealed more than one gene significantly associated with survival, unlike the individual SNP analysis, suggesting that gene-based analysis may reveal additional information compared with individual SNP analysis.
In conclusion, our study suggests that genetic variations in the cell cycle pathway have significant effects on the survival of patients with stages III–IV NSCLC treated with chemotherapy and chemoradiation. Based on different therapeutic modality, we found distinctive genotypes for predicting a better or worse survival. Although we reported the SNPs after correction for multiple comparisons, these findings need to be validated in independent studies. The validated genomic makers could provide a clinically applicable genetic signature with a minimal number of genes and then integrate with other clinical and epidemiologic profiles to improve prediction efficiency of clinical outcome.
Supplementary material
Supplementary Tables 1–6 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute (R01CA111646, P50 CA070907, R01CA055769 and R03 CA128079).
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- CDK
cyclin-dependent kinase
- FDR
false discovery rate
- HR
hazard ratio
- MST
median survival time
- NSCLC
non-small-cell lung cancer
- SNP
single-nucleotide polymorphism
References
- 1.Aggarwal C, et al. Biomarkers with predictive and prognostic function in non-small cell lung cancer: ready for prime time? J. Natl Compr. Canc. Netw. 2010;8:822–832. doi: 10.6004/jnccn.2010.0059. [DOI] [PubMed] [Google Scholar]
- 2.Coate LE, et al. Molecular predictive and prognostic markers in non-small-cell lung cancer. Lancet Oncol. 2009;10:1001–1010. doi: 10.1016/S1470-2045(09)70155-X. [DOI] [PubMed] [Google Scholar]
- 3.Petrelli F, et al. Is there a role for maintenance therapy in non-small-cell lung cancer? An emerging issue. Expert Rev. Anticancer Ther. 2009;9:1455–1465. doi: 10.1586/era.09.112. [DOI] [PubMed] [Google Scholar]
- 4.Poehlmann A, et al. Importance of DNA damage checkpoints in the pathogenesis of human cancers. Pathol. Res. Pract. 2010;206:591–601. doi: 10.1016/j.prp.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Vermeulen K, et al. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwell LH, et al. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 7.Malumbres M, et al. To cycle or not to cycle: a critical decision in cancer. Nat. Rev. Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- 8.Park MT, et al. Cell cycle and cancer. J. Biochem. Mol. Biol. 2003;36:60–65. doi: 10.5483/bmbrep.2003.36.1.060. [DOI] [PubMed] [Google Scholar]
- 9.De Luca A, et al. Cyclin T: three forms for different roles in physiological and pathological functions. J. Cell. Physiol. 2003;194:101–107. doi: 10.1002/jcp.10196. [DOI] [PubMed] [Google Scholar]
- 10.Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 11.Neganova I, et al. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 2008;213:30–44. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, et al. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 13.Tozluoglu M, et al. Cataloging and organizing p73 interactions in cell cycle arrest and apoptosis. Nucleic Acids Res. 2008;36:5033–5049. doi: 10.1093/nar/gkn481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lobrich M, et al. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 15.Malumbres M. Cyclins and related kinases in cancer cells. J. BUON. 2007;12(suppl 1):S45–S52. [PubMed] [Google Scholar]
- 16.Danesi R, et al. Pharmacogenomics in non-small-cell lung cancer chemotherapy. Adv. Drug Deliv. Rev. 2009;61:408–417. doi: 10.1016/j.addr.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Kastan MB, et al. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 18.Dai J, et al. Genetic variations in the regulator of G-protein signaling genes are associated with survival in late-stage non-small cell lung cancer. PLoS One. 2011;6:e21120. doi: 10.1371/journal.pone.0021120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pu X, et al. PI3K/PTEN/AKT/mTOR pathway genetic variation predicts toxicity and distant progression in lung cancer patients receiving platinum-based chemotherapy. Lung Cancer. 2010;71:82–88. doi: 10.1016/j.lungcan.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oken MM, et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am. J. Clin. Oncol. 1982;5:649–655. [PubMed] [Google Scholar]
- 21.Wu X, et al. Novel susceptibility loci for second primary tumors/recurrence in head and neck cancer patients: large-scale evaluation of genetic variants. Cancer Prev. Res. (Phila.) 2009;2:617–624. doi: 10.1158/1940-6207.CAPR-09-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norbury C, et al. Animal cell cycles and their control. Annu. Rev. Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 23.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 24.International HapMap Consortium. The International HapMap Project. (2003) Nature. 426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 25.Sherry ST, et al. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. [PubMed] [Google Scholar]
- 26.Benjamini Y, et al. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B. Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 27.Garcia-Closas M, et al. Large-scale evaluation of candidate genes identifies associations between VEGF polymorphisms and bladder cancer risk. PLoS Genet. 2007;3:e29. doi: 10.1371/journal.pgen.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mountzios G, et al. Histopathologic and genetic alterations as predictors of response to treatment and survival in lung cancer: a review of published data. Crit. Rev. Oncol. Hematol. 2010;75:94–109. doi: 10.1016/j.critrevonc.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Olaussen KA, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N. Engl. J. Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 30.Zhou W, et al. Excision repair cross-complementation group 1 polymorphism predicts overall survival in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin. Cancer Res. 2004;10:4939–4943. doi: 10.1158/1078-0432.CCR-04-0247. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, et al. Germline genetic variations in drug action pathways predict clinical outcomes in advanced lung cancer treated with platinum-based chemotherapy. Pharmacogenet. Genomics. 2008;18:955–965. doi: 10.1097/FPC.0b013e32830efdd4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma H, et al. Potentially functional polymorphisms in cell cycle genes and the survival of non-small cell lung cancer in a Chinese population. Lung Cancer. 2010;73:32–37. doi: 10.1016/j.lungcan.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 33.Pucci B, et al. pRb2/p130 promotes radiation-induced cell death in the glioblastoma cell line HJC12 by p73 upregulation and Bcl-2 downregulation. Oncogene. 2002;21:5897–5905. doi: 10.1038/sj.onc.1205750. [DOI] [PubMed] [Google Scholar]
- 34.Lin KW, et al. Multiple stress signals induce p73beta accumulation. Neoplasia. 2004;6:546–557. doi: 10.1593/neo.04205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakatsuki M, et al. p73 protein expression correlates with radiation-induced apoptosis in the lack of p53 response to radiation therapy for cervical cancer. Int. J. Radiat. Oncol. Biol. Phys. 2008;70:1189–1194. doi: 10.1016/j.ijrobp.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 36.Baker GL, et al. Multiple functions of D-type cyclins can antagonize pRb-mediated suppression of proliferation. Cell Cycle. 2005;4:330–338. [PubMed] [Google Scholar]
- 37.Knudsen ES, et al. Tailoring to RB: tumour suppressor status and therapeutic response. Nat. Rev. Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noel EE, et al. The association of CCND1 overexpression and cisplatin resistance in testicular germ cell tumors and other cancers. Am. J. Pathol. 2010;176:2607–2615. doi: 10.2353/ajpath.2010.090780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ewen ME. The cell cycle and the retinoblastoma protein family. Cancer Metastasis Rev. 1994;13:45–66. doi: 10.1007/BF00690418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.