Abstract
Obesity and osteoporosis are closely correlated genetically. FTO gene has been consistently identified to be associated with obesity phenotypes. A recent study reported that the mice lacking Fto could result in lower bone mineral density (BMD). Thus, we hypothesize that the FTO gene might be also important for osteoporosis phenotypes. To test for such a hypothesis, we performed an association analyses to investigate the relationship between SNPs in FTO and BMD at both hip and spine. A total of 141 SNPs were tested in two independent Chinese populations (818 and 809 unrelated Han subjects, respectively) and a Caucasian population (2,286 unrelated subjects). Combining the two Chinese samples, we identified 6 SNPs in FTO to be significantly associated with hip BMD after multiple testing adjustments, with the combined P values ranged from 4.99×10−4–1.47×10−4. These 6 SNPs are all located at the intron 8 of FTO and in high linkage disequilibrium. Each copy of the minor allele of each SNP was associated with increased hip BMD values with the effect size (beta) of ∼0.025 and ∼0.015 in the Chinese sample 1 and 2, respectively. However, none of these 6 SNPs showed significant association signal in the Caucasian sample, by presenting some extent of ethnic difference. Our findings, together with the prior biological evidence, suggest that the FTO gene might be a new candidate for BMD variation and osteoporosis in Chinese populations.
Introduction
Osteoporosis is a major public health problem with growing prevalence. Obesity, another common disease, has been demonstrated to be closely related with osteoporosis [1], [2], [3]. Adipocytes and osteoblasts arise from the same progenitor, bone marrow stromal cells, and can transdifferentiate into each other [4]. With aging, the composition of bone marrow shifts to favor the presence of adipocytes, osteoclast activity increases, and osteoblast function declines, leading to osteoporosis. Moreover, both osteoporosis and obesity have high genetic predisposition and the genetic correlation between them have been established across different ethnic groups [1], [3], [5]. Previous candidate gene and bivariate association studies have suggested some common genes both for obesity and osteoporosis, such as RANK [6], SP7 [7], and SOX6 [8].
Recently, several independent large-scale genome-wide association studies (GWAS) consistently identified a gene FTO (fat mass and obesity associated) to be associated with obesity-related traits and obesity risk [9], [10], [11]. The association has been further replicated in multiple studies in different populations [12], [13], [14], [15], resulting in much increasing attention on this gene. The FTO protein contains a double-stranded beta-helix fold homologous to those of Fe(II) and 2-oxoglutarate-dependent oxygenases, which might be involved in DNA demethylase [16]. Experimental animal studies provide direct functional evidence that FTO is a causal gene underlying obesity [17], [18]. Interestingly, a recent study found that the whole body Fto knockout mice displayed immediate postnatal growth retardation with shorter body length, lower body weight, and lower bone mineral density (BMD) than control mice [19]. This study reminded us that FTO might be a common genetic factor influencing not only obesity phenotypes, but also osteoporosis phenotypes, like BMD. To test for such hypothesis, we performed an association analyses to examine the relationship between the FTO gene and BMD. Our study was performed in three sample sets from two ethnicities, including two Chinese Han populations and a Caucasian population, in order to see whether the variants identified are common or ethnicity-specific.
Results
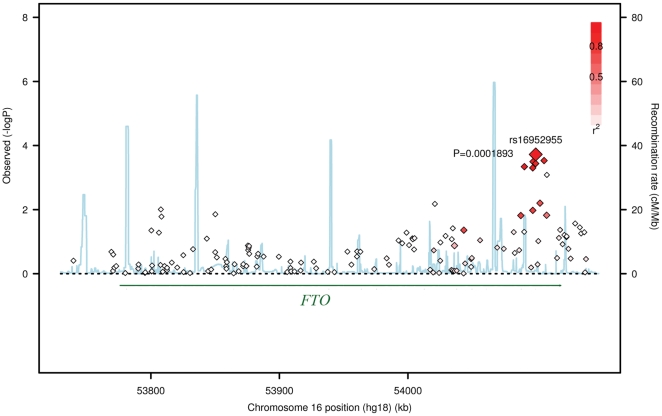
The basic characteristics of the study subjects are presented in Table 1. The major association results for SNPs in FTO with hip BMD are summarized in Table 2. Combining results from these two Chinese samples, 6 SNPs were identified to be significantly associated with hip BMD after multiple testing adjustments by FDR (Table 2). The most significant SNP was rs16952955, with the P values of 8.39×10−4, and 4.31×10−2 in the Chinese sample 1 and Chinese sample 2, respectively. After meta-analysis, the combined P value achieved a significant level of 1.47×10−4. Besides rs16952955, there were 5 additional SNPs (rs2540766, rs2540784, rs16952951, rs12447427, and rs2689247) showed significant association signals with hip BMD, with the combined P values ranged from 4.99×10−4–2.95×10−4 (Table 2). These 6 SNPs have a consistently protective effect on BMD, since each copy of the minor allele of each SNP was associated with increased hip BMD values with the effect size (beta) of ∼0.025 and ∼0.015 in the Chinese sample 1 and 2, respectively. All of these 6 SNPs are located at the intron 8 of FTO. We further characterized the LD for these SNPs using the regional association plot. As shown in Figure 1, these SNPs were in high LD with the top significant SNP rs16952955 (pairwise LD r 2>0.9). For the Caucasian sample, however, no significant results were found for these 6 SNPs and the MAF of each SNP was quite different between Caucasian sample and Chinese samples (P<0.01 by χ2 test).
Table 1. Basic characteristics of the study subjects.
| Trait | Chinese Sample 1 | Chinese Sample 2 | Caucasian Sample 1 |
| Number | 818 | 809 | 2,286 |
| Female/Male | 412/406 | 413/396 | 1727/558 |
| Age (years) | 28.88 (5.18) | 40.18 (16.17) | 51.37 (13.76) |
| Weight (kg) | 57.37 (9.83) | 62.89 (10.39) | 75.27 (17.54) |
| Height (cm) | 163.51 (7.62) | 164.99 (8.62) | 166.35 (8.47) |
| BMI (kg/m2) | 21.38 (2.76) | 23.06 (3.05) | 27.14 (5.75) |
| Hip BMD (g/cm2) | 0.919 (0.129) | 0.921 (0.139) | 0.968 (0.175) |
| Spine BMD (g/cm2) | 0.960 (0.115) | 0.935 (0.138) | 1.025 (0.157) |
Note: Data are shown as mean (standard deviation, SD).
Table 2. Significant association results for SNPs in FTO with hip BMD.
| SNP | Position | Genic Position | A1/A2 | Chinese Sample 1 | Chinese Sample 2 | P combine | Caucasian sample | ||||||
| Freq | BETA | P-value | Freq | BETA | P-value | Freq | BETA | P-value | |||||
| rs16952955 | 54099469 | Intron8 | C/A | 0.146 | 0.0261 | 8.39×10−4 | 0.132 | 0.0150 | 4.31×10−2 | 1.47×10−4 | 0.034 | −0.0003 | 9.78×10−1 |
| rs2540766 | 54105971 | Intron8 | T/C | 0.144 | 0.0236 | 2.88×10−3 | 0.130 | 0.0165 | 3.25×10−2 | 2.95×10−4 | 0.042 | −0.0042 | 6.47×10−1 |
| rs2540784 | 54097334 | Intron8 | G/C | 0.146 | 0.0239 | 2.38×10−3 | 0.133 | 0.0159 | 4.02×10−2 | 3.18×10−4 | 0.035 | 0.0006 | 9.52×10−1 |
| rs16952951 | 54099427 | Intron8 | A/G | 0.144 | 0.0256 | 1.11×10−3 | 0.132 | 0.0138 | 7.56×10−2 | 3.65×10−4 | 0.034 | 0.0010 | 9.17×10−1 |
| rs12447427 | 54090589 | Intron8 | G/A | 0.145 | 0.0241 | 2.03×10−3 | 0.131 | 0.0145 | 6.15×10−2 | 4.57×10−4 | 0.034 | 0.0005 | 9.61×10−1 |
| rs2689247 | 54097159 | Intron8 | A/G | 0.147 | 0.0237 | 2.42×10−3 | 0.132 | 0.0145 | 5.91×10−2 | 4.99×10−4 | 0.035 | 0.0005 | 9.56×10−1 |
Freq, frequency is shown for allele A1. Meta-analysis was conducted for Chinese samples 1 and 2 using the METAL software taking into account sample size and direction of effect (P combine). We listed SNPs with P combine remaining significant after FDR adjustment.
Figure 1. Regional Association Plot for FTO on chromosome 16.
The color scheme of a white-to-black gradient reflects lower to higher linkage disequilibrium (LD, r2). The r2 is calculated between the top significant SNP rs16952955 and other SNPs. The scatter graph indicates the negative logarithm of P value for each SNP, which is based on the combined results in the two Chinese samples.
We further performed gender-specific association analyses for the above 6 SNPs with hip BMD in the Chinese samples. The association signals in each gender group were generally weaker than in the total sample, which might be largely due to the smaller sample sizes in each gender group. The associations were mainly driven by male subjects as reflected in the Chinese sample 1, with the P values ranged from 0.012–1.89×10−3 (Table 3).
Table 3. Gender-specific association results for the six SNPs identified for hip BMD in the Chinese samples.
| SNP | Chinese Sample 1 P value | Chinese Sample 2 P value | ||
| Male | Female | Male | Female | |
| rs16952955 | 1.89×10−3 | 0.164 | 0.114 | 0.252 |
| rs2540766 | 3.61×10−3 | 0.283 | 0.092 | 0.182 |
| rs2540784 | 0.012 | 0.086 | 0.123 | 0.176 |
| rs16952951 | 2.54×10−3 | 0.164 | 0.173 | 0.252 |
| rs12447427 | 6.02×10−3 | 0.142 | 0.153 | 0.226 |
| rs2689247 | 7.36×10−3 | 0.146 | 0.119 | 0.272 |
For the spine BMD, 5 of above 6 SNPs achieved significant level in the Chinese sample 1 (rs16952951: P = 8.78×10−5; rs16952955: P = 1.47×10−4; rs2540784: P = 1.97×10−4; rs2689247: P = 2.11×10−4; rs12447427: P = 4.79×10−4; and rs2540766: P = 1.99×10−3). However, when considering the Chinese samples 1 and 2 together, no SNPs remained significant after FDR adjustments. For the Caucasian sample, we found another 4 SNPs which are also located at the intron 8 of FTO to be nominally associated with spine BMD (rs11076017: P = 5.23×10−3; rs1420318: P = 6.14×10−3; rs1876942: P = 7.21×10−3; and rs17226942: P = 9.23×10−3).
For the previously reported SNP rs9939609 identified by GWAS for obesity phenotypes [9], [10], [11], we only detected nominally significant association with spine BMD in the Chinese sample 1 (P = 0.037).
Discussion
The FTO gene has become the hotspot for researchers since it was reported as a novel obesity-susceptibility gene by a number of GWAS and follow-up replication studies. Given the evidence that obesity and osteoporosis share some common genetic determinations, we first performed an association study examining the SNPs in FTO for association with BMD. We identified a cluster of SNPs in FTO to be significantly associated with hip BMD in Chinese populations.
The human FTO gene is expressed in many tissues including mesenteric fat, pancreas, liver and adipose tissue, with the highest concentrations found in the hypothalamus [10], [20]. Functional studies have demonstrated the direct effect of FTO on obesity. For example, Fischer et al. have reported that the loss of Fto in mice leads to postnatal growth retardation and a significant reduction in adipose tissue and lean body mass [18]. Church et al. have shown that a mutation in the mouse Fto gene results in reduced fat mass, increased energy expenditure, and unchanged physical activity [17]. In respect to FTO's relevance to osteoporosis, a recent study by Gao et al. has found that Fto plays an essential role in postnatal growth. The mice lacking Fto completely displayed postnatal growth retardation manifested as reduced body weight and length, lower BMD [19]. Taking into account of this biological evidence and our statistical findings, we suggest that FTO might have potential role on BMD or osteoporosis.
It is necessary to examine the associations in different populations from different ethnicities, since the genomic variation is greater when compared across ethnicities. Our study identified consistent association for 6 SNPs in intron 8 of FTO with hip BMD in two Chinese populations. Unfortunately, such results were not replicated in the Caucasian population, which implied some extent of ethnic difference. Such ethnic difference could be explained from two aspects. First, the allele frequencies are quite different between the Chinese and Caucasian populations. The minor alleles of these 6 significant SNPs were much common in the Chinese than in the Caucasians, which may contribute to the overall effect. Second, the difference might be age-specific. The two Chinese populations were relatively younger than the Caucasians. Although we have included age as a covariate to adjust BMD, it could not eliminate the potential confounding effect of age on BMD variations thoroughly. Since animal studies have reported that Fto plays an important role in postnatal growth [19], this might suggest that the FTO gene might be a candidate genetic marker for peak bone mass acquisition. However, it is still too early to get the conclusion. We found several other SNPs which are also in intron 8 of FTO for nominally significant association with spine BMD in the Caucasian population, but not in the Chinese populations. Additional studies are needed to investigate the true effect of FTO on BMD in Caucasian populations.
The association of FTO with spine BMD was less significant than with hip BMD in our study, since we only observed significant associations between FTO and spine BMD in the Chinese sample 1. When considering the two Chinese samples together, no SNPs remained significant after multiple testing adjustments. Our results suggest that the effect of FTO on hip BMD might be stronger than spine BMD. However, it is still too soon to get such conclusion. The sample size of the Chinese samples was relatively small, which might decrease the statistical power to detect genetic associations. Increasing the sample size in further studies is needed to validate our results, and we are hopeful that publication of our findings will facilitate replication analyses by other groups.
In summary, our data provide novel evidence that a cluster of SNPs in FTO are associated BMD variations in Chinese populations. Considering the prior biological findings, we suggest that FTO might be a new candidate for osteoporosis. Further studies are warranted to explore the generality of our findings and elucidate the true functional variant.
Materials and Methods
Subjects
The study was approved by the Institutional Review Board or Research Administration of Xi'an Jiaotong University, Hunan Normal University, Creighton University and University of Missouri-Kansas City. Signed informed consent documents were obtained from all study participants before entering the study.
Chinese samples
The Chinese sample 1 comprised 818 unrelated subjects, which were recruited from Southwest Chinese Han adults living in Changsha city and its neighboring areas. The Chinese sample 2 comprised 809 unrelated subjects drawing from Northwest Chinese Han adults in Xi'an City and its neighboring areas. For all the subjects, the exclusion criteria have been detailed in our earlier publication [21]. Briefly, subjects with chronic diseases and conditions that might potentially affect bone mass, structure, or metabolism were excluded from the study to minimize the influence of known environmental and therapeutic factors on bone variation. BMD at hip and spine was measured using Hologic 4500 W machines (Hologic Inc., Bedford, MA, USA) under the same strict protocols. The coefficient of variation (CV) values of the dual-energy X-ray absorptionmetry (DXA) measurements for spine and hip BMDs were approximately 1.01% and 1.33%, respectively.
Caucasian sample
The Caucasian sample consisted of 2,286 unrelated adults. All of the subjects were US Caucasians of Northern European origin living in Midwestern area. The exclusion criteria were the same as with Chinese samples. BMD at hip and spine were measured using the same model Hologic 4500 W machines (Hologic Inc., Bedford, MA, USA) under the same strict protocols used in the Chinese sample. The CV values of the DXA measurements for spine and hip BMDs were approximately 1.98% and 1.87%, respectively.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using standard protocols. For all the three samples, SNP genotyping was performed using Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA), according to the Affymetrix protocol. Only samples with a minimum call rate of 95% were included. Due to efforts of repeat experiments, all samples met this criteria and the final mean call rate reached a high level of 98.93% for Caucasian sample and 98.96% for Chinese sample. SNPs that deviated from Hardy-Weinberg equilibrium (HWE, P<0.0001) and had a minor allele frequency (MAF)<0.01 were discarded in each sample set. Thus, 141 SNPs in FTO were included for subsequent association analyses. The basic characteristics of these SNPs are summarized in Table S1.
Statistical analyses
Before association analyses, principal component analysis implemented in EIGENSTRAT [22] was used to adjust for potential population stratification that may lead to spurious association results. The first ten principal components emerging from the EIGENSTRAT analyses, along with sex, age, height, weight and BMI, were used as covariates to adjust the raw BMD values in each sample. The residues were used for association analyses. Linear regression implemented in PLINK [23] was used to test for association under the additive inheritance model.
Meta-analysis statistics were generated using METAL software package (http://genome.sph.umich.edu/wiki/METAL_Documentation) taking into account sample size and direction of effect. SNAP was used to characterize linkage disequilibrium (LD) and depict the regional association plot [24]. A raw P value of <0.05 in our study was considered nominally significant, which were further subjected to a false discovery rate (FDR) of Benjamini and Hochberg procedure [25] to account for multiple comparisons.
Supporting Information
Properties of FTO SNPs tested in this study
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from National Natural Science Foundation of China (81000363, 31000554), the PhD. Programs Foundation of Ministry of Education of China (20100201120058), the Fundamental Research Funds for the Central Universities, Shanghai Leading Academic Discipline Project (S30501), and startup funds from University of Shanghai for Science and Technology and Xi'an Jiaotong University. The study was also funded by grants from National Institutes of Health (NIH R01 AR050496, R21 AG027110, R01 AG026564, R01 AR057049-01A1 and R21 AA015973) and a SCOR (Specialized Center of Research) grant (P50 AR055081). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Toth E, Ferenc V, Meszaros S, Csupor E, Horvath C. [Effects of body mass index on bone mineral density in men]. Orv Hetil. 2005;146:1489–1493. [PubMed] [Google Scholar]
- 2.Rosen CJ, Bouxsein ML. Mechanisms of disease: is osteoporosis the obesity of bone? Nat Clin Pract Rheumatol. 2006;2:35–43. doi: 10.1038/ncprheum0070. [DOI] [PubMed] [Google Scholar]
- 3.Zhao LJ, Liu YJ, Liu PY, Hamilton J, Recker RR, et al. Relationship of obesity with osteoporosis. J Clin Endocrinol Metab. 2007;92:1640–1646. doi: 10.1210/jc.2006-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gimble JM, Robinson CE, Wu X, Kelly KA. The function of adipocytes in the bone marrow stroma: an update. Bone. 1996;19:421–428. doi: 10.1016/s8756-3282(96)00258-x. [DOI] [PubMed] [Google Scholar]
- 5.Deng FY, Lei SF, Li MX, Jiang C, Dvornyk V, et al. Genetic determination and correlation of body mass index and bone mineral density at the spine and hip in Chinese Han ethnicity. Osteoporos Int. 2006;17:119–124. doi: 10.1007/s00198-005-1930-4. [DOI] [PubMed] [Google Scholar]
- 6.Zhao LJ, Guo YF, Xiong DH, Xiao P, Recker RR, et al. Is a gene important for bone resorption a candidate for obesity? An association and linkage study on the RANK (receptor activator of nuclear factor-kappaB) gene in a large Caucasian sample. Hum Genet. 2006;120:561–570. doi: 10.1007/s00439-006-0243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Bradfield JP, Li M, Zhang H, Mentch FD, et al. BMD-associated variation at the Osterix locus is correlated with childhood obesity in females. Obesity (Silver Spring) 2011;19:1311–1314. doi: 10.1038/oby.2010.324. [DOI] [PubMed] [Google Scholar]
- 8.Liu YZ, Pei YF, Liu JF, Yang F, Guo Y, et al. Powerful bivariate genome-wide association analyses suggest the SOX6 gene influencing both obesity and osteoporosis phenotypes in males. PLoS One. 2009;4:e6827. doi: 10.1371/journal.pone.0006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dina C, Meyre D, Gallina S, Durand E, Korner A, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 10.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cha SW, Choi SM, Kim KS, Park BL, Kim JR, et al. Replication of genetic effects of FTO polymorphisms on BMI in a Korean population. Obesity (Silver Spring) 2008;16:2187–2189. doi: 10.1038/oby.2008.314. [DOI] [PubMed] [Google Scholar]
- 13.Chang YC, Liu PH, Lee WJ, Chang TJ, Jiang YD, et al. Common variation in the fat mass and obesity-associated (FTO) gene confers risk of obesity and modulates BMI in the Chinese population. Diabetes. 2008;57:2245–2252. doi: 10.2337/db08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hotta K, Nakata Y, Matsuo T, Kamohara S, Kotani K, et al. Variations in the FTO gene are associated with severe obesity in the Japanese. J Hum Genet. 2008;53:546–553. doi: 10.1007/s10038-008-0283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubacek JA, Bohuslavova R, Kuthanova L, Kubinova R, Peasey A, et al. The FTO gene and obesity in a large Eastern European population sample: the HAPIEE study. Obesity (Silver Spring) 2008;16:2764–2766. doi: 10.1038/oby.2008.421. [DOI] [PubMed] [Google Scholar]
- 16.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Church C, Lee S, Bagg EA, McTaggart JS, Deacon R, et al. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet. 2009;5:e1000599. doi: 10.1371/journal.pgen.1000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 19.Gao X, Shin YH, Li M, Wang F, Tong Q, et al. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stratigopoulos G, Padilla SL, LeDuc CA, Watson E, Hattersley AT, et al. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1185–1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Tan LJ, Lei SF, Yang TL, Chen XD, et al. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6:e1000806. doi: 10.1371/journal.pgen.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Properties of FTO SNPs tested in this study
(DOC)