Abstract
Bacteriophage T4 alpha- and beta-glucosyltransferases link glucosyl units to the 5-HMdC residues of its DNA. The monoglucosyl group in alpha-linkage predominates over the one in beta linkage. Having recently reported on the nucleotide sequence of gene alpha gt (1) we now determined the nucleotide sequence of gene beta gt. The genes were each cloned on a high expression vector under the control of the lambda pL promoter. After thermo-induction the proteins were isolated and purified to homogeneity. To verify that the translational starting sites and the proposed reading frames are effective in vivo the sequence of the first 31 amino acid residues from gp alpha gt and the first 30 amino acid residues from gp beta gt were determined by Edman degradation. The primary structures of the two proteins seem to have only limited structural similarities. The results are discussed comparing secondary structure predictions and homologies with other proteins from the protein sequence database of the Protein Identification Resource.
Full text
PDF









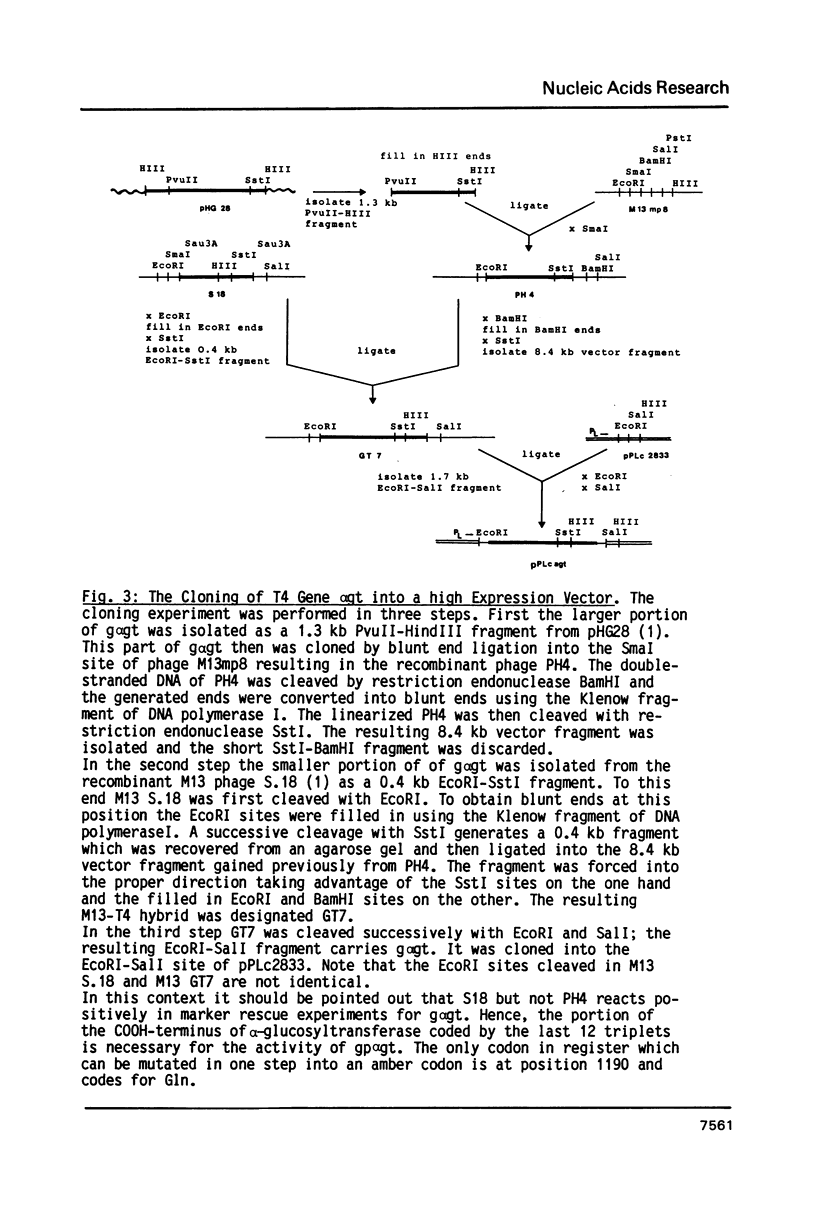

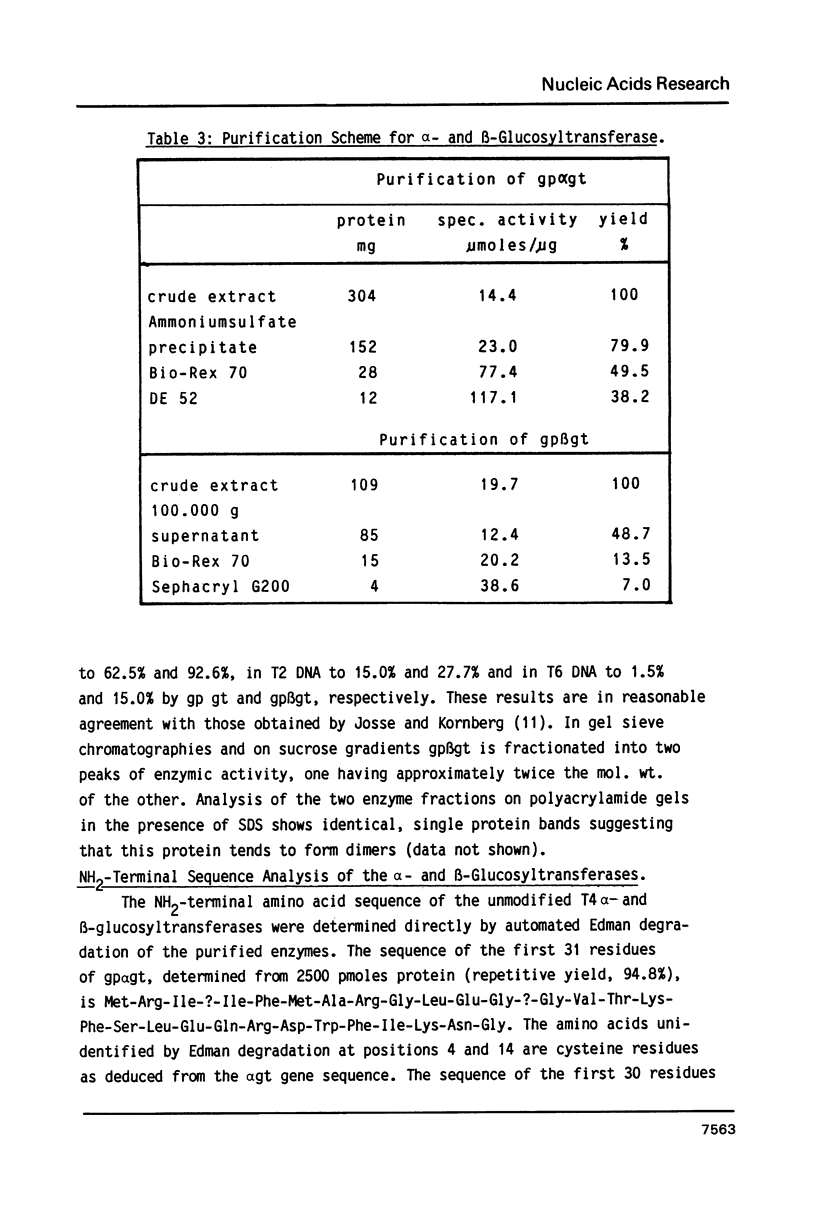





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. R., Lasser G. W., Goldman D. A., Booth J. W., Mathews C. K. T4 phage deoxyribonucleotide-synthesizing enzyme complex. Further studies on enzyme composition and regulation. J Biol Chem. 1983 May 10;258(9):5746–5753. [PubMed] [Google Scholar]
- Baughman G., Nomura M. Localization of the target site for translational regulation of the L11 operon and direct evidence for translational coupling in Escherichia coli. Cell. 1983 Oct;34(3):979–988. doi: 10.1016/0092-8674(83)90555-x. [DOI] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. M., Frampton J., Goelet P., Karn J. Sensitive detection of RNA using strand-specific M13 probes. Gene. 1982 Dec;20(2):139–144. doi: 10.1016/0378-1119(82)90032-4. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of beta-turns. Biophys J. 1979 Jun;26(3):367–383. doi: 10.1016/S0006-3495(79)85259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cox G. S., Conway T. W. Template properties of glucose-deficient T-even bacteriophage DNA. J Virol. 1973 Dec;12(6):1279–1287. doi: 10.1128/jvi.12.6.1279-1287.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Dharmalingam K., Goldberg E. B. Restriction in vivo. III. General effects of glucosylation and restriction on phage T4 gene expression and replication. Virology. 1979 Jul 30;96(2):393–403. doi: 10.1016/0042-6822(79)90097-7. [DOI] [PubMed] [Google Scholar]
- Fleischman R. A., Cambell J. L., Richardson C. C. Modification and restriction of T-even bacteriophages. In vitro degradation of deoxyribonucleic acid containing 5-hydroxymethylctosine. J Biol Chem. 1976 Mar 25;251(6):1561–1570. [PubMed] [Google Scholar]
- Fuhrman S. A., Deininger P. L., LaPorte P., Friedmann T., Geiduschek E. P. Analysis of transcription of the human Alu family ubiquitous repeating element by eukaryotic RNA polymerase III. Nucleic Acids Res. 1981 Dec 11;9(23):6439–6456. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada K., Abelson J. DNA sequence of a T4 transfer RNA gene cluster. J Mol Biol. 1980 May 25;139(3):377–391. doi: 10.1016/0022-2836(80)90136-9. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Location of glucosyl transferase genes on the genetic map of phage T4. Virology. 1968 Feb;34(2):364–366. doi: 10.1016/0042-6822(68)90250-x. [DOI] [PubMed] [Google Scholar]
- Gram H., Liebig H. D., Hack A., Niggemann E., Rüger W. A physical map of bacteriophage T4 including the positions of strong promoters and terminators recognized in vitro. Mol Gen Genet. 1984;194(1-2):232–240. doi: 10.1007/BF00383522. [DOI] [PubMed] [Google Scholar]
- Gram H., Rüger W. Genes 55, alpha gt, 47 and 46 of bacteriophage T4: the genomic organization as deduced by sequence analysis. EMBO J. 1985 Jan;4(1):257–264. doi: 10.1002/j.1460-2075.1985.tb02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Guthrie C., McClain W. H. Rare transfer ribonucleic acid essential for phage growth. Nucleotide sequence comparison of normal and mutant T4 isoleucine-accepting transfer ribonucleic acid. Biochemistry. 1979 Aug 21;18(17):3786–3795. doi: 10.1021/bi00584a023. [DOI] [PubMed] [Google Scholar]
- HATTMAN S., FUKASAWA T. HOST-INDUCED MODIFICATION OF T-EVEN PHAGES DUE TO DEFECTIVE GLUCOSYLATION OF THEIR DNA. Proc Natl Acad Sci U S A. 1963 Aug;50:297–300. doi: 10.1073/pnas.50.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hood L. E. Analysis of phenylthiohydantoins by ultrasensitive gradient high-performance liquid chromatography. Methods Enzymol. 1983;91:486–493. doi: 10.1016/s0076-6879(83)91045-5. [DOI] [PubMed] [Google Scholar]
- JOSSE J., KORNBERG A. Glucosylation of deoxyribonucleic acid. III. alpha- and beta-Glucosyl transferases from T4-infected Escherichia coli. J Biol Chem. 1962 Jun;237:1968–1976. [PubMed] [Google Scholar]
- Kabsch W., Sander C. How good are predictions of protein secondary structure? FEBS Lett. 1983 May 8;155(2):179–182. doi: 10.1016/0014-5793(82)80597-8. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. On the use of sequence homologies to predict protein structure: identical pentapeptides can have completely different conformations. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1075–1078. doi: 10.1073/pnas.81.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf K. W. Simple isolation method and assay for T4 DNA ligase and characterization of the purified enzyme. Eur J Biochem. 1977 Feb 15;73(1):33–38. doi: 10.1111/j.1432-1033.1977.tb11289.x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LEHMAN I. R., PRATT E. A. On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J Biol Chem. 1960 Nov;235:3254–3259. [PubMed] [Google Scholar]
- LUNT M. R., NEWTON E. A. GLUCOSYLATED NUCLEOTIDE SEQUENCES FROM T-EVEN BACTERIOPHAGE DEOXYRIBONUCLEIC ACIDS. Biochem J. 1965 Jun;95:717–723. doi: 10.1042/bj0950717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mattson T., Van Houwe G., Bolle A., Selzer G., Epstein R. Genetic identification of cloned fragments of bacteriophage T4 DNA and complementation by some clones containing early T4 genes. Mol Gen Genet. 1977 Sep 9;154(3):319–326. doi: 10.1007/BF00571289. [DOI] [PubMed] [Google Scholar]
- McNicol L. A., Goldberg E. B. An immunochemical characterization of glucosylation in bacteriophage T4. J Mol Biol. 1973 May 15;76(2):285–301. doi: 10.1016/0022-2836(73)90392-6. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- Nagano K. Triplet information in helix prediction applied to the analysis of super-secondary structures. J Mol Biol. 1977 Jan 15;109(2):251–274. doi: 10.1016/s0022-2836(77)80033-8. [DOI] [PubMed] [Google Scholar]
- Niggemann E., Green I., Meyer H. P., Rüger W. Physical mapping of bacteriophage T4. Mol Gen Genet. 1981;184(2):289–299. doi: 10.1007/BF00272920. [DOI] [PubMed] [Google Scholar]
- Rand K. N., Gait M. J. Sequence and cloning of bacteriophage T4 gene 63 encoding RNA ligase and tail fibre attachment activities. EMBO J. 1984 Feb;3(2):397–402. doi: 10.1002/j.1460-2075.1984.tb01819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Rawlings N., Ashman K., Wittmann-Liebold B. Computerised version of the Chou and Fasman protein secondary structure predictive method. Int J Pept Protein Res. 1983 Nov;22(5):515–524. doi: 10.1111/j.1399-3011.1983.tb02124.x. [DOI] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Remaut E., Tsao H., Fiers W. Improved plasmid vectors with a thermoinducible expression and temperature-regulated runaway replication. Gene. 1983 Apr;22(1):103–113. doi: 10.1016/0378-1119(83)90069-0. [DOI] [PubMed] [Google Scholar]
- Robson B., Suzuki E. Conformational properties of amino acid residues in globular proteins. J Mol Biol. 1976 Nov 5;107(3):327–356. doi: 10.1016/s0022-2836(76)80008-3. [DOI] [PubMed] [Google Scholar]
- Russell R. L. Comparative genetics of the T-even bacteriophages. Genetics. 1974 Dec;78(4):967–988. doi: 10.1093/genetics/78.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüger W. Transcription of bacteriophage T4 DNA in vitro: selective initiation with dinucleotides. Eur J Biochem. 1978 Jul 17;88(1):109–117. doi: 10.1111/j.1432-1033.1978.tb12427.x. [DOI] [PubMed] [Google Scholar]
- SHEDLOVSKY A., BRENNER S. A CHEMICAL BASIS FOR THE HOST-INDUCED MODIFICATION OF T-EVEN BACTERIOPHAGES. Proc Natl Acad Sci U S A. 1963 Aug;50:300–305. doi: 10.1073/pnas.50.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spicer E. K., Nossal N. G., Williams K. R. Bacteriophage T4 gene 44 DNA polymerase accessory protein. Sequences of gene 44 and its protein product. J Biol Chem. 1984 Dec 25;259(24):15425–15432. [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait R. C., Rodriguez R. L., West R. W., Jr The rapid purification of T4 DNA ligase from a lambda T4 lig lysogen. J Biol Chem. 1980 Feb 10;255(3):813–815. [PubMed] [Google Scholar]
- WYATT G. R., COHEN S. S. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953 Dec;55(5):774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Molecular cloning of the DNA ligase gene from bacteriophage T4. I. Characterisation of the recombinants. J Mol Biol. 1979 Aug 15;132(3):471–491. doi: 10.1016/0022-2836(79)90270-5. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waard A., Ubbink T. E., Beukman W. On the specificity of bacteriophage-induced hydroxymethylcytosine glucosyltransferases. II. Specificities of hydroxymethylcytosine alphaand beta-glucosyltransferases induced by bacteriophage T4. Eur J Biochem. 1967 Oct;2(3):303–308. doi: 10.1111/j.1432-1033.1967.tb00139.x. [DOI] [PubMed] [Google Scholar]



