Abstract
The intrinsic stabilization of therapeutic proteins by N-glycosylation can endow them with increased shelf and serum half-lives owing to lower populations of misfolded and unfolded states, which are susceptible to aggregation and proteolysis. Conjugation of polyethylene glycol (PEG) oligomers to nucleophilic groups on the surfaces of folded proteins (i.e., PEGylation) is a chemical alternative to N-glycosylation, in that it can also enhance the pharmacologic attributes of therapeutic proteins. However, the energetic consequences PEGylation are currently not predictable. We find that PEGylation of an Asn residue in reverse turn 1 of the Pin WW domain is intrinsically stabilizing in several sequence contexts, unlike N-glycosylation, which is only stabilizing in a particular sequence context. Our thermodynamic data are consistent with the hypothesis that PEGylation destabilizes the protein denatured state ensemble via an excluded volume effect, whereas N-glycosylation-associated stabilization results primarily from native state interactions between the N-glycan and the protein.
Chemical conjugation of polyethylene glycol (PEG) oligomers to nucleophilic groups on the surfaces of folded proteins (i.e., PEGylation) (1) is a widely used strategy for enhancing shelf and serum half-life and decreasing immunogenicity in a number of FDA-approved biotherapeutics (2, 3). However, the energetic consequences of PEGylation are difficult to predict: PEGylation can increase (4–8), decrease (9, 10), or have no effect (11–13) on protein thermodynamic stability, depending on the protein, the number and locations of the conjugation sites, and the molecular weight of the PEG oligomer utilized. Creating engineering guidelines for achieving reliable PEG-associated increases in thermodynamic stability (accompanied by decreases in the populations of aggregation-prone misfolded states and proteolytically-susceptible unfolded states) would be highly desirable for research and therapeutic applications.
N-glycosylation can also intrinsically stabilize proteins, endowing therapeutics with enhanced storage and pharmacokinetic properties (14, 15). N-glycosylation occurs cotranslationally, as the nascent polypeptide chain is inserted into the endoplasmic reticulum. The oligosaccharyl transferase enzyme attaches the Glc3Man9GlcNAc2 oligosaccharide (where Glc is glucose, Man is mannose, and GlcNAc is N-acetylglucosamine) to the Asn side-chain amide nitrogen within the canonical Asn-Xxx-Ser/Thr sequon (16). N-glycosylation is reported to promote secondary structure formation (17–21), accelerate protein folding (22), and increase protein stability (22–24). Moreover, we recently showed that placing a Phe residue prior to an N-glycosylated Asn-Xxx-Thr sequon within specific reverse turn contexts consistently results in stabilizing native-state interactions between the Phe side chain, the N-glycan, and the Thr side chain, in so-called enhanced aromatic sequons (25, 26). We wondered whether similar engineering guidelines might also allow PEGylation to reliably stabilize proteins, although we recognize PEG oligomers differ significantly from N-glycans in terms of their size, flexibility, and structure (3).
With the goal of generating such guidelines, we herein replace the N-glycan of a previously characterized N-glycosylated enhanced aromatic sequon (25, 26) with a short PEG chain, in the host context of the WW domain of the human protein Pin 1 (hereafter called WW). The small size of the WW domain facilitates the preparation of pure, defined N-glycosylated and PEGylated variants via chemical synthesis. Here we show that PEGylation of an Asn residue in reverse turn 1 of the Pin WW domain is intrinsically stabilizing in several sequence contexts, unlike N-glycosylation, which is only stabilizing in a particular sequence context. Our thermodynamic data are consistent with the hypothesis that PEGylation stabilizes the native states of proteins by denatured state destabilization, whereas N-GlcNAcylation- associated stabilization results primarily from native state interactions between N-GlcNAc and the protein, although larger glycans might destabilize the denatured state as well.
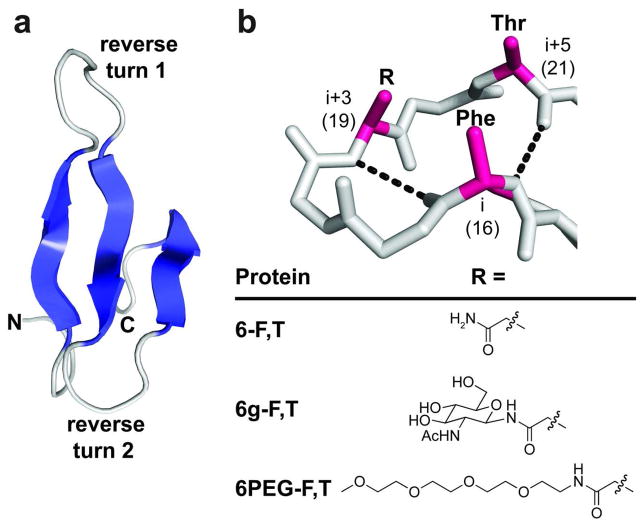
The WW domain is an extensively-characterized (27, 28) glycosylation-naïve protein in which three antiparallel β-strands are connected by two reverse turns (Figure 1, panel a) (29). The first of these reverse turns adopts an unusual “turn-within-a-turn” conformation, featuring a type II β-turn within a six-residue hydrogen-bonded loop (Figure 1, panel b). This turn-within-a-turn projects the side chains at the i, i+3, and i+5 positions (positions 16, 19, and 21 of WW) onto the same side of the loop (29). We previously installed the enhanced aromatic sequon in this turn context by placing Phe, Asn, and Thr at positions 16, 19, and 21, respectively, affording protein 6-F,T (6 for six-residue turn, F for Phe at position 16, and T for Thr at position 21) (25, 26). We showed that chemical N-glycosylation of 6-F,T with a single GlcNAc residue at Asn19 generated glycoprotein 6g-F,T (Figure 1, panel b; g for GlcNAcylated), which is −0.7 kcal mol−1 more stable than 6-F,T at 65 °C (25, 26). N-GlcNAcylation at Asn19 is dramatically less stabilizing when Phe16 is replaced with Ser, or when Thr21 is replaced with Arg (Ser and Arg occupy positions 16 and 21, respectively, in the parent WW sequence), indicating that Phe16, Asn19-GlcNAc, and Thr21 engage in a favorable tripartite interaction that stabilizes 6g-F,T relative to 6-F,T, a conclusion supported by a published thermodynamic cycle analysis (25, 26).
Figure 1.
(a) Ribbon diagram of the WW domain from the human protein Pin 1 (PDB: 1PIN, ref 45). β-strands are shown in blue, reverse turns in gray. (b) Stick representation of reverse turn 1 in the WW domain, main-chain hydrogen bonds represented by black dashes; positions where we previously incorporated components of the enhanced aromatic sequon are highlighted in red. Protein 6-F,T, glycoprotein 6g-F,T (refs. 25, 26) and N-PEGylated protein 6PEG-F,T differ in the group attached to the side-chain amide nitrogen of Asn19 as indicated (see Figure 2, panel a for the complete sequence of 6PEG-F,T). All structures were rendered in Pymol.
We wondered whether similar interactions might allow N-PEGylation to stabilize WW. To test this hypothesis, we replaced GlcNAc in 6g-F,T with a short linear PEG chain comprising four ethylene oxide units (Figure 1, panel b). As with GlcNAc in 6g-F,T, the PEG chain was attached to the side-chain amide nitrogen of Asn19, affording 6PEG-F,T (Figure 1, panel b and Figure 2, panel a; Supplementary Figures S1–S8 and Supplementary Tables S1–S2 provide analytical data for the Fmoc-protected Asn-linked PEG derivative used to synthesize 6PEG-F,T; Supplementary Table S3 summarizes the mass spectrometry characterization data for 6PEG-F,T and its derivatives). The PEG chain shown in Figure 1, panel b has a similar number of carbons and heteroatoms as GlcNAc, but its flexible, weakly amphipathic structure differs substantially from the ring-constrained GlcNAc, which has a distinct hydrophobic α-face. We wondered whether these differences would impact the energetic influence of PEGylating an enhanced aromatic sequon in the six-residue “turn-within-a-turn” context.
Figure 2.
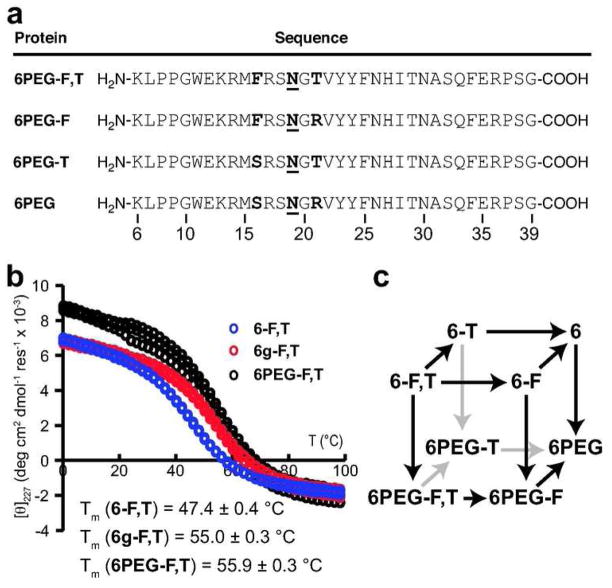
(a) Amino acid sequences of N-PEGylated protein 6PEG-F,T and its derivatives. N = Asn- PEG (see Figure 1, panel b for PEG structure). (b) Variable temperature CD data for protein 6-F,T, glycoprotein 6g-F,T (ref. 26), and N-PEGylated protein 6PEG-F,T at a protein concentration of 10 μM in 20 mM sodium phosphate buffer (pH 7). Data are shown for each of three replicate experiments on each protein. (c) Triple mutant cycle cube formed by proteins 6-F,T, and 6PEG-F,T, and their derivatives.
We used variable temperature far-UV circular dichroism spectropolarimetry (CD) to analyze the thermodynamic stability of 6PEG-F,T (Figure 2, panel b). The melting temperature of 6PEG-F,T is 8.5 ± 0.6 °C higher than that of the unmodified 6-F,T, corresponding to a difference in folding free energy ΔΔGf of −0.81 ± 0.10 kcal mol−1 at 65 °C (Table 1). Surprisingly, 6PEG-F,T is approximately as stable as GlcNAc-containing 6g-F,T (ΔΔGf relative to 6-F,T is −0.70 ± 0.10 kcal mol−1 at 65 °C) (25, 26), suggesting that N-PEGylation of the enhanced aromatic sequon stabilizes WW as effectively as does N-GlcNAcylation (Figure 2, panel b, Table 1).
Table 1.
Melting temperatures and folding free energies of N-PEGylated WW variants and their corresponding non-PEGylated counterparts.a
| Protein | Tm (°C) | ΔTm (°C) | ΔGf (kcal/mol) | ΔΔGf (kcal/mol) |
|---|---|---|---|---|
| 6-F,Tb | 47.4 ± 0.4 | 8.5 ± 0.6 | 1.72 ± 0.09 | −0.81 ± 0.10 |
| 6PEG-F,T | 55.9 ± 0.3 | 0.91 ± 0.05 | ||
| 6-Fb | 51.0 ± 0.3 | 10.9 ± 0.4 | 1.45 ± 0.06 | −1.13 ± 0.07 |
| 6PEG-F | 61.9 ± 0.3 | 0.32 ± 0.03 | ||
| 6-Tb | 52.5 ± 0.3 | 5.3 ± 0.4 | 1.22 ± 0.05 | −0.55 ± 0.06 |
| 6PEG-T | 57.8 ± 0.2 | 0.68 ± 0.03 | ||
| 6b | 56.2 ± 0.3 | 8.0 ± 0.4 | 0.95 ± 0.04 | −0.86 ± 0.05 |
| 6PEG | 64.3 ± 0.2 | 0.09 ± 0.03 |
Tabulated data are given as mean ± standard error at 65 °C for 10 μM solutions of WW variants in 20 mM sodium phosphate buffer (pH 7). CD spectra and variable temperature CD data from which these parameters were derived appear in Figures S17–S20 of the supporting information.
Data for these proteins are from ref. 26.
We previously used triple mutant cycle analysis to probe the sequence dependence of the energetic effect of N-GlcNAcylating the enhanced aromatic sequon (25, 26). Here, we used a similar strategy to ask whether the stabilizing effect of N-PEGylating an enhanced aromatic sequon in the context of the six-residue “turn-within-a-turn” described above depends on stabilizing interactions with Phe and Thr (25, 26). We prepared three variants of 6PEG-F,T (Figure 2, panel a) in which we replaced Phe16 and Thr21 with Ser16 and Arg21, respectively, in every possible combination (mass spectrometry and analytical HPLC data for 6PEG-F,T and its derivatives appear in Supplementary Figures S9–16, and are summarized in Supplementary Table S3). As we have done previously (25, 26), we chose Ser and Arg because these residues occupy positions 16 and 21, respectively, in the parent WW protein from which 6PEG-F,T is derived. Ser occupies position 16 in proteins 6PEG and 6PEG-T, whereas Phe occupies position 16 in 6PEG-F. Arg occupies position 21 in PEGylated proteins 6PEG and 6PEG-F whereas Thr occupies position 21 in 6PEG-T.
We compared the thermodynamic stabilities of proteins 6PEG, 6PEG-F and 6PEG-T with the thermodynamic stabilities of the corresponding unmodified proteins 6, 6-F, and 6-T, which we prepared and characterized previously (25, 26) (Table 1, Supplementary Figures S17–S20 show complete variable temperature CD data for 6PEG-F,T and its derivatives). The stabilizing effect of N-PEGylation at Asn19 (compare 6PEG-F,T with 6-F,T: ΔΔGf = −0.81 ± 0.10 kcal mol−1 at 65 °C) is still substantial when Phe16 is replaced by Ser16 (compare 6PEG-T with 6-T: ΔΔGf = −0.55 ± 0.6 kcal mol−1 at 65 °C), when Thr21 is replaced by Arg21 (compare 6PEG-F with 6-F: ΔΔGf = −1.13 ± 0.07 kcal mol−1 at 65 °C), or when both modifications are made (compare 6PEG with 6: ΔΔGf = −0.86 ± 0.05 kcal mol−1 at 65 °C). In contrast, as reported previously (25, 26), the stabilizing effect of N-GlcNAcylation at Asn19 (compare 6g-F,T with 6-F,T; ΔΔGf = −0.70 ± 0.10 kcal mol−1 at 65 °C) decreases substantially when Phe16 is replaced by Ser16 (compare 6g-T and 6-T; ΔΔGf = −0.04 ± 0.07 kcal mol−1 at 65 °C), when Thr21 is replaced by Arg21 (compare 6g-F and 6-F; ΔΔGf = −0.17 ± 0.08 kcal mol−1 at 65 °C), or when both modifications are made (compare 6g and 6; ΔΔGf = 0.21 ± 0.06 kcal mol−1 at 65 °C). Thus, the stabilizing influence of N-PEGylation is much less dependent on the identity of nearby residues than N-GlcNAcylation.(25, 26)
WW proteins 6, 6-F, 6-T, and 6-F,T, and N-PEGylated proteins 6PEG, 6PEG-F, 6PEG-T, and 6PEG-F,T enable a triple mutant cycle analysis (Figure 2, panel c), which reveals information about the two- and three-way interactions between Phe16, Asn19-PEG, and Thr21 in 6PEG-F,T. As we have done previously (25, 26), we used least-squares regression of the thermodynamic data for each variant to extract this information, according to the following equation:
| (1) |
In equation (1), ΔGf is the folding free energy of a given variant of 6; ΔGf° is the average folding free energy of 6; CS→F, CN→N, and CR→T describe the intrinsic energetic consequences of the Ser16 to Phe16, Asn19 to Asn19-PEG, and Arg21 to Thr21 mutations, respectively; CF,N, CF,T, and CN,T describe the two-way interaction energies between Phe16 and Asn19-PEG, between Phe16 and Thr21, and between Asn19-PEG and Thr21, respectively; CF,N,T describes the three-way interaction energy between Phe16, Asn19-PEG, and Thr21; WF is 0 when position 16 is Ser or 1 when it is Phe; WN = 0 when position 19 is Asn or 1 when it is Asn-PEG; WT = 0 when position 21 is Arg or 1 when it is Thr. Equation 1 assumes that the Ser16 side chain does not interact with the side chains at positions 19 and 21, and that the Arg21 side chain does not interact with the side chains at positions 19 and 21, assumptions which are consistent to a first approximation with the available structural data (26, 29). The results of this analysis are summarized in Figure 3 and in Supplementary Table S4.
Figure 3.
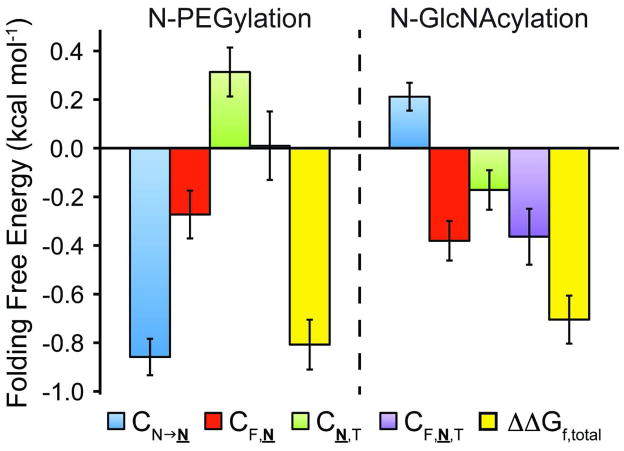
The increase in stability of 6-F,T upon N-PEGylation (ΔΔGf,total, yellow bars) is the sum of the energetic effects of (1) changing Asn19 to Asn19-PEG (CN→N, blue bars); (2) the two-way interaction between Phe16 and Asn19-PEG (CF,N, red bars); (3) the two-way interaction between Asn19-PEG and Thr21 (CN,T, green bars); and (4) the three-way interaction between Phe16, Asn19- PEG, and Thr21, (CF,N,T, purple bars). Error bars represent parameter standard errors. Analogous data for N-glycosylation (ref. 26) are shown for comparison.
The parameters obtained via regression of equation 1 provide insights into why PEGylation stabilizes 6PEG-F,T relative to 6-F,T. According to equation 1, ΔΔGf = ΔGf(6PEG-F,T) − ΔGf(6-F,T) = CN→N + CF,N + CN,T + CF,N,T. Thus, the differences in how N-glycosylation and N-PEGylation stabilize WW (Figure 3) are depicted by the differences between the values of CN→N, CF,N, CN,T, and CF,N,T for N-PEGylation and for N-GlcNAcylation of 6-F,T, which were measured previously (26).
As reported previously, the stabilizing effect of N-GlcNAcylation on 6g-F,T (Figure 3) comes predominantly from the favorable three-way interaction between Phe16, Asn19-GlcNAc, and Thr21 (CF,N,T = −0.36 ± 0.11 kcal mol−1) and the favorable two-way interaction between Phe16 and Asn19- GlcNAc (CF,N = −0.38 ± 0.08 kcal mol−1), offset by the intrinsically unfavorable effect of changing Asn19 to Asn19-GlcNAc (CN→N = 0.21 ± 0.06 kcal mol−1). In contrast, the stabilizing effect of N-PEGylation on 6PEG-F,T (Figure 3) depends strongly on the intrinsically favorable effect of changing Asn19 to Asn19-PEG (CN→N = −0.86 ± 0.08 kcal mol−1) and to a lesser extent on a favorable two-way interaction between Phe16 and Asn19-PEG (CF,N = −0.27 ± 0.10 kcal mol−1), offset by an unfavorable two-way interaction between Asn19-PEG and Thr21 (CN,T = 0.31 ± 0.10 kcal mol−1). Notably, there is no significant three-way interaction between Phe16, Asn19-PEG, and Thr21 (CF,N,T, see Figure 3).
These results imply fundamental differences in the mechanisms by which N-glycosylation and N-PEGylation stabilize proteins. N-GlcNAcylation of the enhanced aromatic sequon in the six-residue reverse turn of WW is stabilizing due to favorable native-state tripartite interactions between GlcNAc and the nearby Phe and Thr side chains; without these side chains N-GlcNAcylation of Asn19 destabilizes WW (25, 26). In contrast, N-PEGylation of Asn19 in the six-residue reverse turn context of WW is strongly intrinsically stabilizing and not as dependent on the identities of side chains at positions 16 and 21 as is N-GlcNAcylation. Indeed, N-PEGylating an isolated Asn residue (compare 6PEG and 6; ΔΔGf = −0.86 ± 0.05 kcal mol−1 at 65 °C, Table 1) is approximately as favorable as N-PEGylating an Asn residue within an enhanced aromatic sequon context (compare 6PEG-F,T with 6-F,T: ΔΔGf = − 0.81 ± 0.10 kcal mol−1 at 65 °C, Table 1). It remains to be seen whether N-PEGylation of reverse turns provides more stabilization than N-PEGylation of other secondary structures and whether the turn type and exact position of PEGylation within the reverse turn are important.
The intrinsic PEG-associated stabilization of WW in a variety of sequence contexts is consistent with the hypothesis that PEG acts as an excluded-volume conjugate that destabilizes the WW denatured state ensemble by restricting its conformational entropy (24). In a recently-described native topology model of WW glycosylation (30), Asn-linked GlcNAc was modeled as an excluded-volume conjugate. This model incorrectly predicted that N-GlcNAcylation of Asn19 within the six-residue turn of WW (without any other sequence modifications) would be stabilizing. In fact, N-GlcNAcylation of Asn19 is destabilizing (ΔΔGf = +0.21 ± 0.06 kcal mol−1 at 65 °C) (25, 26, 30), whereas N-PEGylation of Asn19 is stabilizing (ΔΔGf = −0.86 ± 0.05 kcal mol−1 at 65 °C), perhaps indicating that PEG more closely resembles an excluded-volume conjugate than does GlcNAc (24, 30). It is possible that N-glycan-based excluded volume effects become important only for the larger N-glycans present in secreted glycoproteins.
In addition, our results show that PEG can interact favorably with nearby side chains, though the energetic impact of these interactions is smaller than the large intrinsic energetic impact of PEGylation. PEG, like GlcNAc, interacts favorably with Phe16 (CF,N = −0.27 ± 0.10 kcal mol−1 for PEG, −0.38 ± 0.08 kcal mol−1 for GlcNAc), which is consistent with displacement of ordered water molecules from the Phe side-chain surface by PEG (8). In contrast, PEG and Thr21 do not interact favorably (CN,T = 0.31 ± 0.10 kcal mol−1), whereas GlcNAc and Thr21 do (CN,T = −0.17 ± 0.08 kcal mol−1), possibly because of favorable hydrogen bonding between the side chains of Asn19-GlcNAc and Thr21, but not between Asn19-PEG and Thr21. Alternatively, PEG may interact more favorably with Arg21 than with Thr21, though the nature of this potential interaction is unclear. In any case, our results indicate that favorable interactions with nearby side-chains can add to the already large and sequence-context-independent stabilization of WW by PEGylation. Other side chains besides Ser and Phe at position 16 and Arg and Thr at position 21 may be able to interact more favorably with PEG, a hypothesis we are currently investigating.
PEGylation has been used for more than three decades to safely improve the shelf-life and pharmacokinetic properties of proteins (1–3). PEGylation can increase protein thermodyamic stability (4–8), but the molecular basis for this effect is poorly understood, due, in part, to PEGylation methodologies that generate mixtures of protein-PEG conjugates that differ in the number of attached PEGs and/or the location(s) of the PEGylation site(s). Herein, chemical synthesis of uniformly N-PEGylated WW variants facilitates a quantitative assessment of N-PEGylation as a function of sequence context. Unlike N-GlcNAcylation, N-PEGylation of the six-residue reverse turn within WW is intrinsically stabilizing–possibly due to denatured state destabilization via an excluded volume effect. Future kinetic studies on the folding and unfolding on 6PEG and its analogs, on variants thereof where the MW of the PEG is altered, and on WW variants where the location of N-PEGylation is varied will allow us to further scrutinize this mechanistic hypothesis. Previous energetic results emerging from N-GlcNAcylation of the enhanced aromatic sequon in the WW domain have translated readily to N-glycosylation in more complex proteins (25). We expect the same to be true for applying energetic principles obtained from N-PEGylating the WW domain. Because N-PEGylation of Asn residues and N-glycosylation of enhanced aromatic sequons in certain reverse turn contexts appear to increase protein stability by distinct mechanisms, it should also be possible to use both approaches additively or synergistically to stabilize a protein of interest.
Methods
Protein Synthesis
WW domain variants were synthesized via solid phase peptide synthesis, using the standard Fmoc protecting group strategy as described previously (30), and in the supporting information. The synthesis of Fmoc-Asn(PEG)-OH (N2-fluorenylmethyoxycarbonyl-N4-{11-methoxy- 3,6,9-trioxaundecyl}-L-asparagine) is also described in the supporting information (characterization data appear in Supplementary Tables S1–S2 and Supplementary Figures S1–S8). Following cleavage from resin, WW variants were purified by reverse-phase HPLC on a C18 column using a linear gradient of water in acetonitrile with 0.2% v/v TFA. The identity of each WW domain was confirmed by matrix-assisted laser desorption/ionization time-of-flight spectrometry (MALDI-TOF, see Supplementary Table S3 and Supplementary Figures S9–S12), and purity was evaluated by analytical HPLC (see Supplementary Figures S13–S16).
CD Measurements
CD spectra and variable temperature CD data were collected using an Aviv 62A DS spectropolarimeter. We fit the variable temperature CD data to obtain Tm and ΔGf values for each protein, as described previously (30), and in the supporting information. CD data for 6PEG-F,T and its derivatives are shown in Supplementary Figures S17–S20.
Supplementary Material
Acknowledgments
We thank D.L. Powers for helpful prepublication discussions. This work was supported in part by NIH grant GM051105 to J.W.K. and E.T.P, the Skaggs Institute for Chemical Biology, and the Lita Annenberg Hazen Foundation. J.L.P was supported in part by an NRSA NIH post-doctoral fellowship F32 GM086039.
Footnotes
Supporting Information Available. Complete experimental methods, compound characterization, and circular dichroism data, and full triple mutant cycle analysis results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Abuchowski A, Vanes T, Palczuk NC, Davis FF. Alteration of Immunological Properties of Bovine Serum-Albumin by Covalent Attachment of Polyethylene-Glycol. J Biol Chem. 1977;252:3578–3581. [PubMed] [Google Scholar]
- 2.Frokjaer S, Otzen DE. Protein drug stability: A formulation challenge. Nat Rev Drug Discov. 2005;4:298–306. doi: 10.1038/nrd1695. [DOI] [PubMed] [Google Scholar]
- 3.Jevsevar S, Kunstelj M, Porekar VG. PEGylation of therapeutic proteins. Biotechnol J. 2010;5:113–128. doi: 10.1002/biot.200900218. [DOI] [PubMed] [Google Scholar]
- 4.Kinstler OB, Brems DN, Lauren SL, Paige AG, Hamburger JB, Treuheit MJ. Characterization and stability of N-terminally PEGylated rhG-CSF. Pharm Res. 1996;13:996–1002. doi: 10.1023/a:1016042220817. [DOI] [PubMed] [Google Scholar]
- 5.Dhalluin C, Ross A, Leuthold LA, Foser S, Gsell B, Muller F, Senn H. Structural and biophysical characterization of the 40 kDa PEG-interferon-alpha(2a) and its individual positional isomers. Bioconjugate Chem. 2005;16:504–517. doi: 10.1021/bc049781+. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Martinez JA, Sola RJ, Castillo B, Cintron-Colon HR, Rivera-Rivera I, Barletta G, Griebenow K. Stabilization of alpha-Chymotrypsin Upon PEGylation Correlates With Reduced Structural Dynamics. Biotechnol Bioeng. 2008;101:1142–1149. doi: 10.1002/bit.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu K, Agoubi LL, Lee I, Limpar MT, Lowe JW, Goh SL. Effects of Polymer Molecular Weight on the Size, Activity, and Stability of PEG-Functionalized Trypsin. Biomacromolecules. 2010;11:3688–3692. doi: 10.1021/bm1006954. [DOI] [PubMed] [Google Scholar]
- 8.Yang C, Lu DN, Liu Z. How PEGylation Enhances the Stability and Potency of Insulin: A Molecular Dynamics Simulation. Biochemistry. 2011;50:2585–2593. doi: 10.1021/bi101926u. [DOI] [PubMed] [Google Scholar]
- 9.So T, Ueda T, Abe Y, Nakamata T, Imoto T. Situation of monomethoxypolyethylene glycol covalently attached to lysozyme. J Biochem. 1996;119:1086–1093. doi: 10.1093/oxfordjournals.jbchem.a021352. [DOI] [PubMed] [Google Scholar]
- 10.Gokarn YR. PhD Dissertation. University of New Hampshire; 2003. Hydrodynamic Behavior and Thermal Stability of a PEGylated Protein: Studies with Hen Egg Lysozyme. [Google Scholar]
- 11.Callahan WJ, Narhi LO, Kosky AA, Treuheit MJ. Sodium chloride enhances the storage and conformational stability of BDNF and PEG-BDNF. Pharm Res. 2001;18:261–266. doi: 10.1023/a:1011034425896. [DOI] [PubMed] [Google Scholar]
- 12.Popp MW, Dougan SK, Chuang TY, Spooner E, Ploegh HL. Sortase-catalyzed transformations that improve the properties of cytokines. Proc Natl Acad Sci USA. 2011;108:3169–3174. doi: 10.1073/pnas.1016863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plesner B, Westh P, Nielsen AD. Biophysical characterisation of GlycoPEGylated recombinant human factor VIIa. Int J Pharmaceut. 2011;406:62–68. doi: 10.1016/j.ijpharm.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 14.Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 15.Sola RJ, Griebenow K. Glycosylation of Therapeutic Proteins An Effective Strategy to Optimize Efficacy. Biodrugs. 2010;24:9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornfeld R, Kornfeld S. Assembly of Asparagine-Linked Oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 17.Wormald MR, Wooten EW, Bazzo R, Edge CJ, Feinstein A, Rademacher TW, Dwek RA. The conformational effects of N-glycosylation on the tailpiece from serum IgM. Eur J Biochem. 1991;198:131–139. doi: 10.1111/j.1432-1033.1991.tb15995.x. [DOI] [PubMed] [Google Scholar]
- 18.Imperiali B, Rickert KW. Conformational implications of asparagine-linked glycosylation. Proc Natl Acad Sci USA. 1995;92:97–101. doi: 10.1073/pnas.92.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Live DH, Kumar RA, Beebe X, Danishefsky SJ. Conformational influences of glycosylation of a peptide: A possible model for the effect of glycosylation on the rate of protein folding. Proc Natl Acad Sci USA. 1996;93:12759–12761. doi: 10.1073/pnas.93.23.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connor SE, Imperiali B. Conformational Switching by Asparagine-Linked Glycosylation. J Am Chem Soc. 1997;119:2295–2296. [Google Scholar]
- 21.Bosques CJ, Tschampel SM, Woods RJ, Imperiali B. Effects of Glycosylation on Peptide Conformation: A Synergistic Experimental and Computational Study. J Am Chem Soc. 2004;126:8421–8425. doi: 10.1021/ja049266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson SR, Culyba EK, Hsu TL, Wong CH, Kelly JW, Powers ET. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci USA. 2009;106:3131–3136. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Eufemi M, Turano C, Giartosio A. Influence of the Carbohydrate Moiety on the Stability of Glycoproteins. Biochemistry. 1996;35:7299–7307. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 24.Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: A close look at thermodynamic stabilization. Proc Natl Acad Sci USA. 2008;105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Culyba EK, Price JL, Hanson SR, Dhar A, Wong CH, Gruebele M, Powers ET, Kelly JW. Protein Native-State Stabilization by Placing Aromatic Side Chains in N-Glycosylated Reverse Turns. Science. 2011;331:571–575. doi: 10.1126/science.1198461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price JL, Powers DL, Powers ET, Kelly JW. Glycosylation of the Enhanced Aromatic Sequon is Similarly Stabilizing in Three Distinct Reverse Turn Contexts. Proc Natl Acad Sci USA. 2011;108:14127–14132. doi: 10.1073/pnas.1105880108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jäger M, Dendle M, Kelly JW. Sequence determinants of thermodynamic stability in a WW domain---An all-β-sheet protein. Protein Sci. 2009;18:1806–1813. doi: 10.1002/pro.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jäger M, Deechongkit S, Koepf EK, Nguyen H, Gao J, Powers ET, Gruebele M, Kelly JW. Understanding the Mechanism of β-Sheet Folding From a Chemical and Biological Perspective. Biopolymers. 2008;90:751–758. doi: 10.1002/bip.21101. [DOI] [PubMed] [Google Scholar]
- 29.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 30.Price JL, Shental-Bechor D, Dhar A, Turner MJ, Powers ET, Gruebele M, Levy Y, Kelly JW. Context-Dependent Effects of Asparagine Glycosylation on Pin WW Folding Kinetics and Thermodynamics. J Am Chem Soc. 2010;132:15359–15367. doi: 10.1021/ja106896t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.