SUMMARY
Cancer cells simultaneously harbor global losses and gains in DNA methylation. We demonstrate that inducing cellular oxidative stress by treatment with hydrogen peroxide, recruits DNA methyltransferase 1 (DNMT1) to damaged chromatin. DNMT1 becomes part of a complex(es) containing DNMT3B and members of Polycomb Repressive Complex 4. Hydrogen peroxide treatment causes translocalization of these proteins from non-GC-rich to GC-rich areas. Key components are similarly enriched at gene promoters in an in vivo colitis model. While high expression genes enriched for members of the complex have histone mark and nascent transcription changes, CpG island-containing low expression genes gain promoter DNA methylation. Thus, oxidative damage induces formation and localization of a silencing complex that may explain cancer-specific aberrant DNA methylation and transcriptional silencing.
INTRODUCTION
Elevated levels of reactive oxygen species (ROS) arising from alterations in cellular metabolism and inflammatory responses constitute a key risk state for increased cancer susceptibility (Federico et al., 2007). The major forms of oxidative DNA damage are nonbulky lesions such as 8-oxo-2′deoxyguanosine (8-oxo-dG) and thymine glycol that are repaired predominantly by base excision repair (BER) (Reardon et al., 1997).
The above DNA repair requires dynamic changes in surrounding chromatin including changes in nucleosome positioning and histone modifications. The best characterized chromatin alteration in DNA repair is the phosphorylation of the histone variant H2AX (γ-H2AX) by DNA damage response protein kinases (Rogakou et al., 1998). This modification helps stabilize the interaction of repair factors with the break sites, leading to further chromatin alterations. Histone acetylases and deacetylases also localize to sites of DNA damage to facilitate repair by increasing access of repair proteins to the break site, repressing transcription at sites of damage, restoring the local chromatin environment after repair is complete, and turning off the DNA damage response (Tamburini and Tyler, 2005). In this regard, (Sirtuin-1) SIRT1 is a NAD+-dependent class III histone deacetylase that plays a role in gene silencing in cancer cells (Pruitt et al., 2006) and has been implicated in DNA damage repair in both yeast and mammalian cells. SIRT1 is recruited to sites of DNA damage and interacts with and deacetylates other proteins involved in the DNA damage (For review, see (Fan and Luo, 2010)). After DNA repair, DNA methylation also needs to be reestablished, possibly by the recruitment of the DNA methyltransferases (DNMTs) that catalyze CpG methylation, including DNMT1 which plays a role in methylating newly replicated DNA (Leonhardt et al., 1992), and DNMT3A and DNMT3B which are mostly responsible for de novo DNA methylation (Okano et al., 1999).
The above epigenetic players have been linked to patterns of cancer-related gene transcriptional silencing, in association with promoter CpG island DNA hypermethylation. We, and others, have shown that a large fraction of the genes that undergo promoter CpG Island DNA hypermethylation in cancer are unmethylated in embryonic stem and progenitor cells and held in low/poised states of transcription by polycomb group (PcG) proteins (Ohm et al., 2007; Schlesinger et al., 2007; Widschwendter et al., 2007). Importantly, SIRT1 has been described as part of a transformation specific PcG complex, PRC4, which is found in embryonic and adult stem cells and cancer cells (Kuzmichev et al., 2005). In addition to SIRT1, the PRC4 complex contains the PcG proteins, Enhancer of Zeste protein-2 (EZH2) which catalyzes the trimethylation of lysine 27 of histone H3 and a specific isoform of EED (EED2) that is absent from previously identified PRC complexes. SIRT1 has also been shown to interact with DNMT1 (Espada et al., 2007). The DNMTs have been linked to PcG proteins in the context of epigenetic gene silencing. Both DNMT1 and DNMT3B interact with EZH2, which in turn facilitates the binding of the DNMTs to EZH2 target promoters (Vire et al., 2006).
In the present study, we investigate epigenetic alterations induced by the ROS, hydrogen peroxide (H2O2), and by inflammation in mouse tissue. We examine changes in the interaction and chromatin binding of the epigenetic proteins discussed above and the functional consequences of these changes. This work attempts to determine a mechanism by which cancer risk states, such as chronic inflammation, can contribute to cancer-related abnormal gene silencing and shifts in DNA methylation.
RESULTS
DNMT1 and SIRT1 Become Tightly Bound to Chromatin after H2O2 Treatment
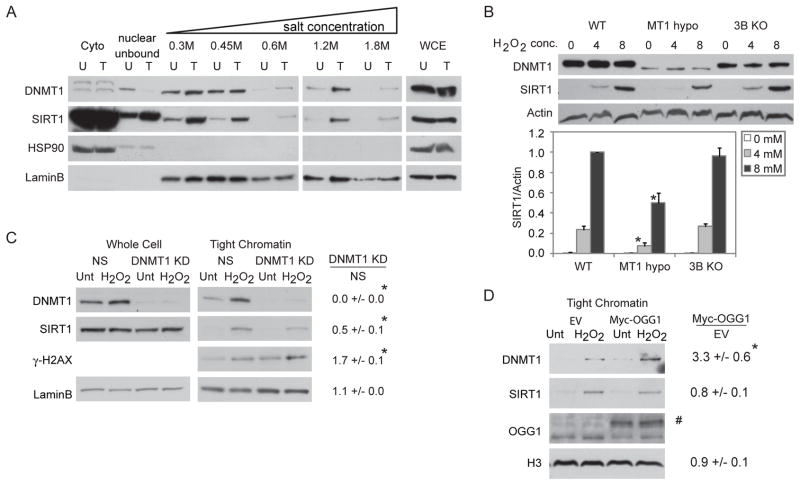
Previously, we have demonstrated that SIRT1 and DNMT1 are rapidly recruited to an induced double strand break in an exogenous promoter CpG island construct (O’Hagan et al., 2008). In this regard, SIRT1, similar to other proteins involved in DNA repair, is known to become more tightly bound to chromatin after oxidative stress (Oberdoerffer et al., 2008). We now find, by examining resistance of the proteins to salt gradient extraction, that both SIRT1 and DNMT1 bind more tightly to chromatin in H2O2-treated human embryonic carcinoma cells (NCCIT) despite their unchanged whole cell levels. As evidence of this tightening, after H2O2 treatment, a portion of SIRT1 is redistributed from the cytoplasmic fraction to the soluble nuclear fraction and is present in all higher salt fractions (Figure 1A). Basally, as has been previously demonstrated, nuclear DNMT1 is loosely bound to the chromatin, being extracted by 0.3 and 0.45 M NaCl (Jeong et al., 2009). However, after H2O2 treatment, DNMT1 is also eluted in salt fractions of 0.6 M, 1.2 M, and 1.8 M NaCl (Figure 1A). HSP90 and LaminB serve as cytoplasmic and nuclear controls, respectively, for the extraction.
Figure 1. DNMT1 and SIRT1 Become Tightly Bound to Chromatin after Treatment with H2O2.
(A) NCCIT cells were untreated (U) or treated with 1 mM H2O2 for 30 minutes (T). Cell pellets were extracted sequentially using cytoplasmic extraction buffer (Cyto), soluble nuclear buffer (nuclear unbound), and buffers with increasing NaCl concentration. Whole cell lysates were prepared separately (WCE). (B) HCT116 (WT), HCT116 hypomorphic DNMT1 (MT1 hypo), and HCT116 DNMT3B KO (3B KO) cells were treated with H2O2 at the indicated concentrations in mM for 30 minutes and total nuclear protein was collected. y-axis is SIRT1 over Actin levels relative to 8 mM treated WT cells. The data presented is the mean of three independent experiments +/− SEM. * p < 0.05 by t-test. (C) NCCIT cells were infected with non-specific shRNA (NS) or DNMT1 shRNA. After 72 hours, they were untreated (Unt) or treated with 1 mM H2O2 for 30 minutes (H2O2). Tight chromatin is the remaining protein in the chromatin pellet after extraction with 0.45 M NaCl buffer. Band densitometry values are displayed as the ratio of DNMT1 knockdown over NS knockdown for protein levels in H2O2 treated cells. The data presented is the mean of three independent experiments +/− SEM. * p < 0.05 by one-tail t-test. (D) NCCIT cells were transiently transfected with empty vector (EV) or c-Myc-tagged OGG1 (Myc-OGG1) plasmids for 48 hours followed by 1 mM H2O2 treatment and analysis as in (C). # Myc-OGG1. * p < 0.05 by one-tail t-test. See also Figure S1.
DNMT1 Functions Upstream of SIRT1 Recruitment to Chromatin Following Oxidative Damage
We next queried the interdependency of the tightening of SIRT1 and DNMT1 to chromatin after H2O2 treatment. SIRT1 knockdown or inhibition causes an increase in the fraction of DNMT1 tightly bound to chromatin after H2O2 treatment relative to non-specific shRNA or mock treated cells, respectively (Figure S1A–C). To examine these dynamics further, we utilized HCT116 cells genetically rendered hypomorphic for DNMT1 or fully deleted for DNMT3B (Rhee et al., 2002; Spada et al., 2007). While H2O2 treatment recruits SIRT1 to the nucleus in the WT and DNMT3B KO lines, there is a significant reduction in nuclear SIRT1 in the DNMT1 hypomorph cells, both by biochemical fractionation (Figure 1B) and immunofluorescence (Figure S1D). However, the residual increase in nuclear SIRT1 in DNMT1 hypomorph cells suggests that additional DNMT1-independent mechanisms may exist for changes in SIRT1 localization after H2O2 treatment. Furthermore, shRNA knockdown of DNMT1 in NCCIT cells significantly reduces the amount of SIRT1 that becomes tightly bound to chromatin after H2O2 treatment to 0.5 fold of control while simultaneously leading to an increase in γ-H2AX levels (Figure 1C). While these results suggest that tightening of DNMT1 and SIRT1 binding to chromatin after H2O2 treatment are dependent on each other, DNMT1 appears to be necessary for the increase in binding of SIRT1 to chromatin.
Because ROS induces DNA damage in the form of base damage, single strand breaks, and double strand breaks, we next examined other types of DNA damaging agents and found that neither ionizing radiation nor ultraviolet light increase the tightness of binding of DNMT1 or SIRT1 to chromatin (Figure S1E&F). Additionally, inhibition of poly (ADP-ribose) polymerase (PARP), an enzyme involved in DNA repair of single and double strand breaks, or knockdown of key components of NER does not affect DNMT1 or SIRT1 recruitment to chromatin after H2O2 treatment (data not shown). Interestingly, we demonstrate that H2O2 treatment induces a significantly higher level of DNMT1 in the tight chromatin fraction in cells over-expressing c-Myc-tagged OGG1, the DNA glycosylase responsible for excising 8-oxo-dG during BER, compared to empty vector cells (Figure 1D). However, OGG1 overexpression does not affect the tightness of SIRT1 binding to chromatin possibly because the chromatin-bound levels of this protein are saturated after H2O2 treatment and therefore cannot be increased further by more DNMT1 recruitment.
DNMTs, SIRT1, and Polycomb Members Interact, as Part of a Large Multi-Protein Complex(es), after H2O2 Treatment
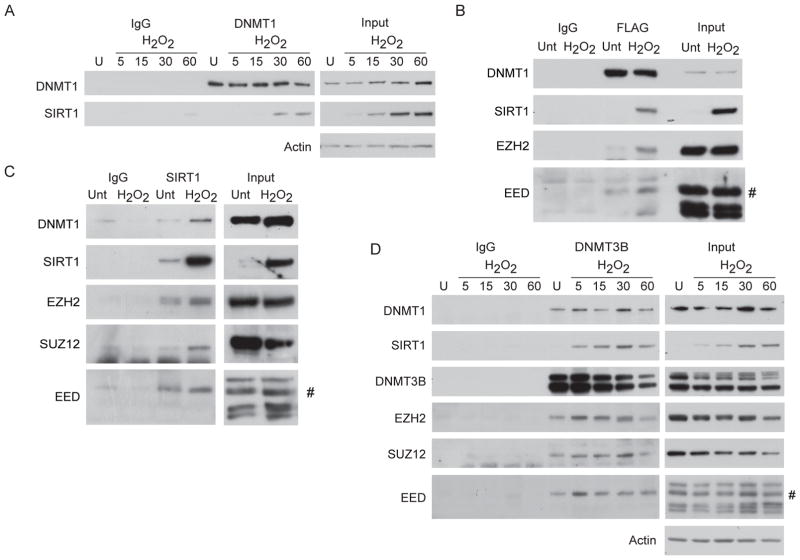
Having demonstrated that H2O2 treatment induces a DNMT1-influenced recruitment of SIRT1 to chromatin, we now queried whether such treatment might facilitate interactions between the two proteins and with other partners. We first observe, using DNMT1 co-immunoprecipitations from NCCIT cells after H2O2 exposure, a time-dependent interaction between endogenous DNMT1 and endogenous SIRT1 30 and 60 minutes after treatment (Figure 2A). We further validate this interaction by expressing a FLAG-tagged full length DNMT1 in the DNMT1 hypomorph HCT116 cell line and finding that H2O2 treatment results in the interaction between the tagged DNMT1 protein and endogenous SIRT1 (Figure 2B).
Figure 2. Oxidative Damage Induces the Interaction between SIRT1, DNMTs, and PcG Components.
(A) NCCIT cells were untreated or treated with 2 mM H2O2 and collected at the indicated time points in minutes after addition of H2O2 to the media. Co-immunoprecipitations were performed with control IgG or anti-DNMT1 antibodies. (B) HCT116 DNMT1 hypomorph cells expressing FLAG-DNMT1 were treated with 8 mM H2O2 for 30 minutes. Co-immunoprecipitations were performed using control IgG or anti-FLAG antibodies. # isoform 2 of EED. (C) NCCIT cells were treated with 2 mM H2O2 for 30 minutes and co-immunoprecipitations were performed using control IgG or anti-SIRT1 antibodies. # isoform 2 of EED. (D) NCCIT cells were treated as in (A) and co-immunoprecipitations were performed using control IgG or anti-DNMT3B antibodies. # isoform 2 of EED. See also Figure S2.
We broadened our search for interacting proteins based on the previously mentioned association between DNA hypermethylated genes and PcG marks. In this regard, we observe that H2O2 treatment increases the interaction of DNMT1 with EZH2 and EED (Figure 2B). Further evidence for SIRT1 in this complex is demonstrated by immunoprecipitated endogenous nuclear SIRT1 pulling down increased levels of DNMT1, EZH2, SUZ12 and the PRC4 specific isoform of EED, EED2, after H2O2 treatment (Figures 2C & S2). Total cellular levels of the above proteins do not change after treatment (Figure 1A and data not shown). Initially, our entrée towards recognition of these interactions was based upon our previous demonstration that there is an acute recruitment of DNMT3B to an induced double strand break which is dependent on SIRT1 (O’Hagan et al., 2008). We now find that H2O2 treatment of NCCIT cells results in an endogenous interaction between SIRT1 and DNMT3B, as analyzed by immunoprecipitation of DNMT3B. This interaction is detectable within five minutes and increases up until 30 minutes after treatment (Figure 2D). As has been shown previously, DNMT3B and DNMT1 also interact (Kim et al., 2002) and this interaction does not change with treatment (Figure 2D). Our DNMT3B co-immunoprecipitation also reveals that the above interactions occur in the context of PRC4 because there is strong interaction of DNMT3B with EZH2, SUZ12 and EED2, both before and after H2O2 treatment (Figure 2D).
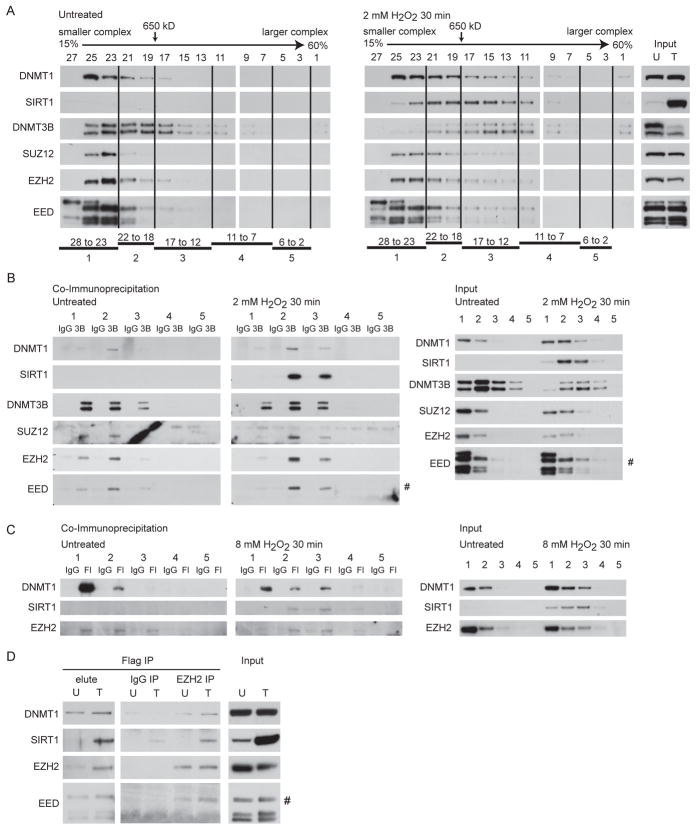
The triggering of multiple, individual, interactions suggests the H2O2 induced generation of a possible mega-complex(es). To examine this hypothesis, we separated nuclear protein complexes by size using sucrose gradient centrifugation from untreated and H2O2 treated cells. After H2O2 treatment, DNMT1, SIRT1 and DNMT3B and to a lesser extent SUZ12, EZH2, and EED proteins all migrate in the gradient in regions of higher molecular mass (greater than 650 kDa) than in untreated cells (Figure 3A). When we perform DNMT3B co-immunoprecipitation on pooled fractions from the gradient in untreated cells, the majority of the PcG proteins (EZH2, SUZ12, and EED2) and DNMT1 co-immunoprecipitate with DNMT3B in lower molecular weight pools 1 and 2 (smaller than 650 kDa – left panel - Figure 3B). Importantly, after treatment, the co-immunoprecipitation between DNMT3B and the PcG members and DNMT1 now shifts towards pools 2 and 3 indicating interaction of the proteins in a much larger complex(es) (middle panel - Figure 3B). After treatment, SIRT1 interacts with DNMT3B in the pools 2 and 3 (middle panel - Figure 3B), suggesting that all DNMT3B interacting members are present in the same size large complex(es) after H2O2 treatment.
Figure 3. H2O2 Treatment Induces the Formation of a Large Complex(es) Containing DNA Methyltransferases, SIRT1, and Polycomb Group Proteins.
(A) Nuclear extracts from untreated NCCIT cells or cells treated with 2 mM H2O2 for 30 minutes were added to a 15 to 60% sucrose gradient and fractions were assayed by immunoblotting. Fraction numbers and 650 kDa molecular mass standard are across the top. Larger fraction numbers indicate smaller molecular weight of the complex(es). (B) Fractions from (A) were pooled into 5 groups as indicated at the bottom of (A). Co-immunoprecipitations for control IgG or anti-DNMT3B (3B) antibodies were performed from the pooled fractions. Right panels are inputs from the pooled fractions. # isoform 2 of EED. (C) HCT116 DNMT1 hypomorph cells expressing FLAG-DNMT1 were untreated or treated with 8 mM H2O2 for 30 minutes. Nuclear extracts, sucrose gradients, and pooling of fractions were performed as in (A). Co-immunoprecipitations for control IgG or anti-FLAG (Fl) antibodies were performed from pooled fractions. Right panels are inputs from the pooled fractions. (D) Flag co-immunoprecipitations were performed in HCT116 DNMT1 hypomorph cells expressing FLAG-DNMT1 that were either untreated (U) or treated with 8 mM H2O2 for 30 minutes (T). After elution with flag peptide (elute) a second immunoprecipitation was done using IgG or EZH2 antibodies. # isoform 2 of EED.
To examine the above complex formation further, we performed similar gradient analyses and co-immunoprecipitations of DNMT1 in the DNMT1 hypomorph HCT116 cell line exogenously expressing FLAG-tagged full length DNMT1. The input protein analyses for this co-IP indicate that there is more nuclear DNMT1, SIRT1, and EZH2 in the higher molecular weight gradient fractions after H2O2 treatment than before treatment (right panel - Figure 3C). In untreated cells, the majority of FLAG-DNMT1 is immunoprecipitated from lower molecular weight gradient pools 1 and 2 and interacts with EZH2 in pools 1, 2, and 3 (left panel – Figure 3C). As expected, SIRT1 is not seen prior to treatment. After H2O2 treatment, FLAG-DNMT1 is now immunoprecipitated more prominently from pool 3 in addition to pools 1 and 2, peak interaction of EZH2 with FLAG-DNMT1 is now in pool 3, and interaction is seen with SIRT1 in pools 2 and 3 (middle panel – Figure 3C). Thus, as for DNMT3B, DNMT1 interacts with EZH2 and SIRT1 as part of a large multi-protein complex(es) after H2O2 treatment. To demonstrate that DNMT1, EZH2, and SIRT1 were indeed all bound together in these large complexes we performed sequential co-immunoprecipitation in the FLAG-DNMT1 cells. First, we immunoprecipitated DNMT1 containing complexes. After eluting these complexes, we immunoprecipitated EZH2 interacting proteins. As demonstrated by the SIRT1 band in the EZH2 IP lane in the DNMT1 elute from the treated cells, EZH2 that is bound to DNMT1 after H2O2 treatment is also bound to SIRT1 (Figure 3D). In total, this data suggests that oxidative damage induces the formation of a large complex(es) containing the DNMTs and PRC4 members.
DNMT1, SIRT1, and EZH2 Form DNA Damage Foci
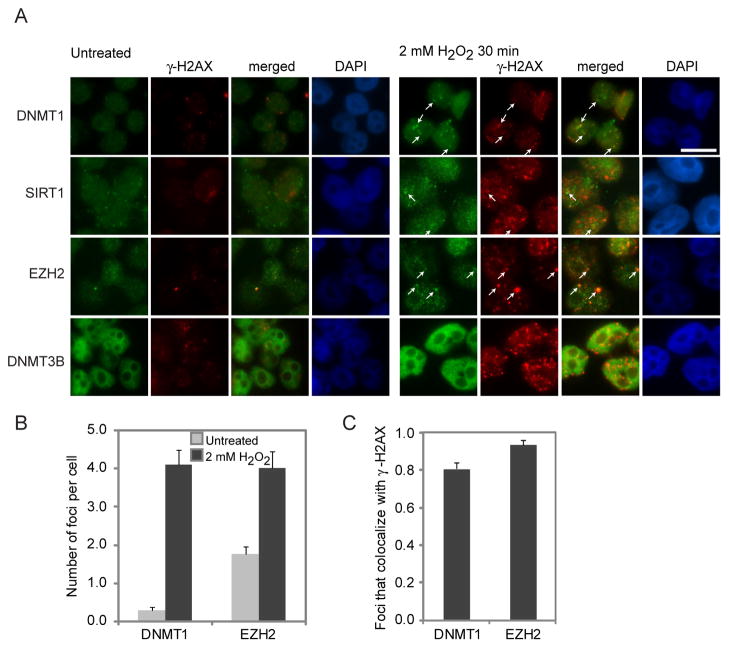
To determine the possible interactions of DNMTs and members of PRC4 directly with DNA damage sites, we performed co-immunofluorescence of key interacting proteins with γ-H2AX in NCCIT cells. We used a paraformaldehyde fixation method that does not visualize DNA replication foci, since DNMT1 is constitutively present at these sites in S-phase (Figure S3A). After H2O2 treatment, the chromatin bound protein, LaminB, does not co-localize with γ-H2AX foci (Figure S3B). In contrast, there is a increase in total nuclear DNMT1, and in DNMT1 foci (0.3 foci in untreated and 4.1 in treated cells – Figure 4B), the majority of which co-localize with γ-H2AX (Figure 4C). The PcG member, EZH2, behaves similarly to DNMT1 (1.8 to 4.0 foci per cell) (Figure 4A–C). SIRT1, also, exhibits occasional larger foci after H2O2 treatment that co-localize with γ-H2AX (Figure 4A) but the considerable increase in nuclear staining makes precise quantitation difficult. Dense and widely distributed nuclear staining of DNMT3B before and after treatment did not allow visualization of foci after treatment. Overall, it appears that the tight binding of DNMT1 and SIRT1 to chromatin induced by H2O2, and the interaction between these two proteins and other PcG components, occurs, at least in part, at DNA damage sites.
Figure 4. DNMT1 and EZH2 Form Nuclear Foci after H2O2 Treatment that Co-localize with γ-H2AX.
(A) NCCIT cells were untreated or treated with 2 mM H2O2 for 30 minutes. Immunofluorescence analysis was performed using the indicated antibodies. White arrows indicate examples of foci. White scale bar is 5 μm. (B) More than 50 nuclei from cells in (A) were scored per antibody in at least two independent experiments. Graphs represent the sample mean +/− SEM. Grey and black bars are untreated and H2O2 treated cells, respectively. (C) More than 50 nuclei from cells in (A) were scored per antibody in at least two independent experiments. Graphs represent the sample mean +/− SEM. Black bars are H2O2 treated cells. See also Figure S3.
Oxidative Damage Recruits Members of the H2O2 Induced Silencing Protein Complex to Promoter CpG Islands
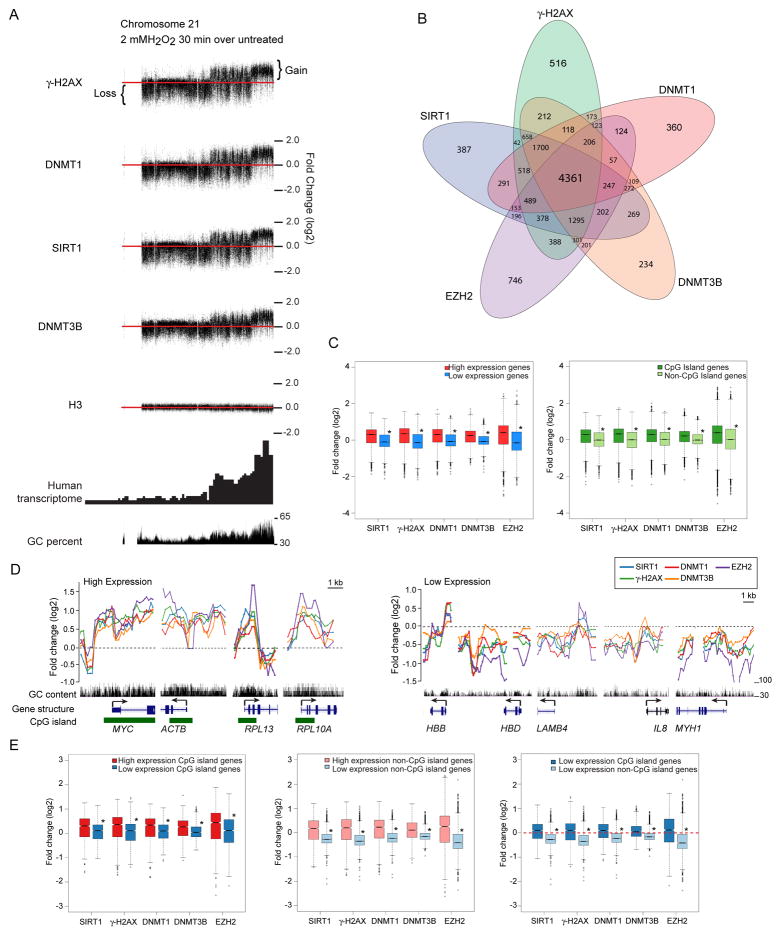
We next explored the genomic regions to which the oxidative damage-induced complexes may localize using the SW480 colon carcinoma cell line, where we have observed similar tightening of DNMT1 and SIRT1 to chromatin after H2O2 treatment (Figure S4A). We first find, utilizing customized ChIP-chip arrays for chromosome 18, 19, and 21 and histone H3 as a control, damage-induced concurrent shifts for DNMT1, DNMT3B, SIRT1 and γ-H2AX after H2O2 treatment (Figure 5A & S4B). In each case, the chromosome regions with enrichment constitute those previously mapped by others (Folle et al., 2010) to harbor high gene transcription activity and GC content. This enrichment shift is particularly observable for chromosome 21 where the sub-telomeric regions of the chromosome harbor most of the gene transcription and GC content (Figure 5A). Importantly, concomitant to the co-enrichments above, there is a notable loss of the same silencing proteins from the transcriptionally inactive and GC-poor regions (Figure 5A).
Figure 5. Oxidative Damage Induces Recruitment of Silencing Proteins to the Promoters of Actively Transcribed Genes and/or High GC Content Regions.
(A) SW480 cells were either untreated or treated with 2 mM H2O2 for 30 minutes, followed by ChIP-Chip using the whole chromosome array of chromosomes 18, 19, and 21. Differences in log2 ratios of IP signals over input signals between treated and untreated samples were plotted along chromosome 21. Red line represents the zero (no change) line. Signals above and below the red line represent gain and loss, respectively, of corresponding marker. (B) Venn diagram for ChIP enriched genes for each antibody in treated over untreated samples. SW480 cells were either untreated or treated with 2 mM H2O2 for 30 minutes. ChIP samples were hybridized to the 244K promoter array. (C) For box plots, the y-axis depicts the differences in log2 ratios of IP signals over input signals between treated and untreated samples. For the left panel, red and blue represent the top 1000 genes with high and low expression, respectively, from expression array data. In the right panel, dark and light green represent groups of CpG island and non-CpG island genes, respectively. * p < 2×10−10 by two-tail t-test. (D) Plots of ChIP-chip signals in treated over untreated samples for individual genes. y-axis is the same as in (C). Black dash line represents the no change line. Black vertical bars indicate GC content, ranging from 30% to 100%. Blue lines-boxes represent the position and construction of genes, with the boxes indicating the position of exons, lines indicating the position of introns, and arrow indicating the direction of transcription. Green boxes represent the position of CpG islands. The names of genes are indicated at the bottom of the plots. (E) Values were plotted as in (C). In the left panel, red and blue represent the top 100 CpG island genes with high and low expression, respectively. In the middle panel, pink and light blue represent the top 100 non-CpG island genes with high and low expression, respectively. In the right panel, blue and light blue represent the top 100 CpG island and non-CpG island genes, respectively, with similar levels of low expression. Red dash line represents the no change line. See also Figure S4.
For all three chromosomes examined changes in ChIP signals are most prominent around the transcription start sites (TSS) of genes (example for DNMT1 signals in Figure S4C). To explore this finding in more depth, we used promoter arrays to examine the co-localization of the proteins at gene promoters across the genome. In these promoter studies we included, one of the key PcG members, EZH2. We also matched the results to previously obtained genome wide expression array data (Easwaran et al., 2010). These studies not only confirm targeting of all the tested proteins, after damage, to transcriptionally active GC-rich promoter regions, but also extend our findings in key ways.
First, DNMT1, DNMT3B, SIRT1, EZH2 and γ-H2AX enrichment that is lost from transcriptionally poor and low GC-content regions is translocalized to the promoters of genes that contain CpG islands and are highly expressed, with a high degree of overlap of enrichment between the different proteins (Figure 5B–E). The direct targeting to promoter CpG islands can be appreciated for example genes, MYC, ACTB, RPL13, and RPL10A (Figure 5D-left panel). The loss of enrichment for low expression, non-CpG island promoter genes is well appreciated for the example genes, HBB, HBD, LAMB4, IL8, and MYH1 (Figure 5D-right panel). These genomic analyses were confirmed by local ChIP and quantitative PCR (Figure S4D).
Second, transcriptional activity is associated with targeting of the members of the complex separately from the presence of CpG islands (Figure 5E & S4E–F). We find that high expression genes gain more enrichment than low expression genes in groups of genes both with and without CpG islands (Figure 5E, left and middle panels). However, the presence of CpG islands is still important as targeting is increased in CpG island versus non-CpG island genes with similar low basal expression (Figure 5E, right panel & Figure S4F). Further scrutiny of the pattern of translocalization revealed that the position of peaks also correlates with areas of high GC content, including but not limited to CpG islands (Figure 5D [note gene ACTB] & Figure S4G), indicating that GC content, in addition to the presence of CpG islands, is a contributing factor to targeting. Altogether, this data suggests that members of the complex undergo H2O2-induced enrichment at gene promoters with high expression and/or high GC-content, including those with CpG islands.
Functional Consequences of Enrichment of Members of the Oxidative Damage-Induced Complex
The fact that oxidative damage-induces proteins involved in gene silencing to form a complex and be enriched at CpG island-containing promoters, suggests a potential functional role of this complex in transcriptional changes accompanying DNA damage and/or cancer-specific, abnormal gene silencing. Examination of changes in histone marks, transcription, and DNA methylation supports this hypothesis.
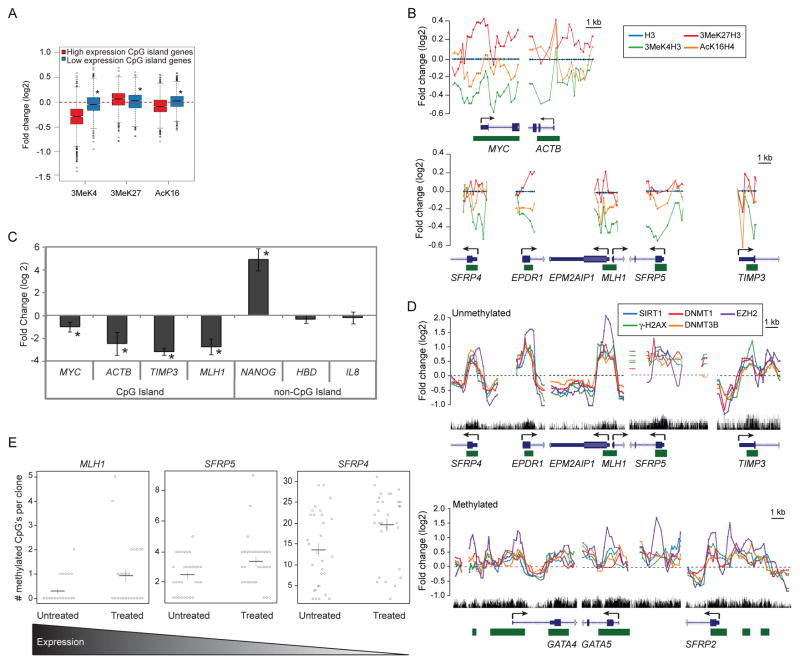
We first examined histone modifications at CpG island-containing gene promoters most targeted by members of the complexes. Using genome-wide promoter arrays and histone H3 to normalize for nucleosome positions, we observed, as expected, marked relative enrichment for the active transcription marks, 3MeK4H3 and AcK16H4, and low amounts of the PcG repressive mark, 3MeK27H3, at the promoter of high expression genes in untreated cells (Figure S5A). After treatment, there is reduction in the active marks 3MeK4H3 and AcK16H4, the later is consistent with the deacetylation activity of SIRT1, and enrichment of the H3K27me3 mark, which can be catalyzed by the PcG component EZH2 (Figure 6A). These global changes are verified by examining patterns at the CpG island-containing promoters of genes, including, MYC, ACTB, SFRP4, MLH1, SFRP5, and TIMP3 (Figure 6B) and by local ChIP studies (Figure S5B). We see similar changes, although with less magnitude, for high expression non-CpG island genes (Figure S5C left panel). Interestingly, there appears to be a slight gain in 3MeK4H3 and loss of 3MeK27H3 (Figure S5C, right panel) at low expression non-CpG island gene promoters where there is relative decrease in the complex constituents (Figure 5E, right panel).
Figure 6. Gene Promoters with Oxidative Damage-Induced Enrichment of the Members of the Silencing Complex have Reduced Levels of Nascent Transcription and/or Increased CpG Methylation.
(A) ChIP samples were hybridized to the 1M promoter array. Plots are of ChIP-chip signals from SW480 cells treated with 2 mM H2O2 for 30 minutes over untreated samples. Box plots are constructed as in Figure 5C except ChIP over input signals are normalized to H3. Red and blue represent the top 100 CpG island genes with high and low expression, respectively. * p< 2×10−10 by two-tail t-test. (B) Plots are constructed as in Figure 5D with ChIP signals normalized to H3. (C) SW480 cells were either untreated or treated with 2 mM H2O2 for 30 minutes and the nascent RNA was labeled concurrently. Quantitative RTPCR data are presented as the mean of the log 2 ratio of the treated over the untreated values for three independent biological replicates +/− SEM. * p < 0.05 by one-sided t-test. (D) Plots are constructed as in Figure 5D. (E) Bisulfite sequencing was performed on DNA from SW480 cells that were untreated or treated with 2 mM H2O2 for 30 minutes. Results are shown as a combination of three independent biological replicates with at least eight clones per experiment. Circles are the individual clones. Black horizontal lines are the mean for all clones with the vertical line representing the standard error. p-values by one-sided Welch’s t-test for MLH1, SFRP5, and SFRP4 are 0.014, 0.012, and 0.005, respectively. The gradient bar at the bottom depicts the relative log 2 basal expression levels, by expression array, which are 11.13, 6.82, and 6.40 for MLH1, SFRP5, and SFRP4, respectively. See also Figure S5.
Second, the change in 3MeK4H3 levels suggests that enrichment of members of the complex may also rapidly induce repressive transcriptional changes mainly in CpG island-containing genes with significant basal expression. Indeed, the CpG island-containing genes, MYC, ACTB, TIMP3, and MLH1, all have reduced nascent transcription levels within 30 minutes after treatment (Figure 6C). In contrast, the non-CpG island-containing genes, NANOG, HBD, and IL8, either gain or have no change in their nascent transcription levels suggesting that the reduction in transcription for high expression genes is not solely due to a global DNA damage-induced decrease in transcription (Figure 6C).
Third, we find important correlates for genes that develop cancer-specific, abnormal, CpG island DNA hypermethylation. Genes with a high frequency for this change in colon cancer that are unmethylated or hypermethylated in SW480 cells, have damage-induced enrichment of the silencing proteins (Figure 6D & Figure S5D).
Fourth, we examined DNA methylation by bisulfite sequencing for representative genes. We did not observe any CpG methylation in the promoter CpG island of the high expression gene, MYC, which does not become hypermethylated in cancer (data not shown). However, at the short time point examined we find increases in DNA methylation for the MLH1, SFRP5, and SFRP4 genes, which are frequently hypermethylated in colon cancer but are unmethylated in SW480 cells (Figure 6E). These three genes are basally expressed to varying degrees in SW480 cells and the level of observed increases in DNA methylation are inverse to this expression (Figure 6E). This finding fits well with emerging data that cancer specific, promoter DNA methylation mostly targets genes, which normally have low basal expression (Hahn et al., 2008). This data suggests that localization of members of the complex to gene promoters has functional consequences, including histone mark changes, reduction in nascent transcription, and/or increases in DNA methylation.
Inflammation Induced Changes in EZH2, SIRT1, and DNMT1 in a Mouse Model of Colitis
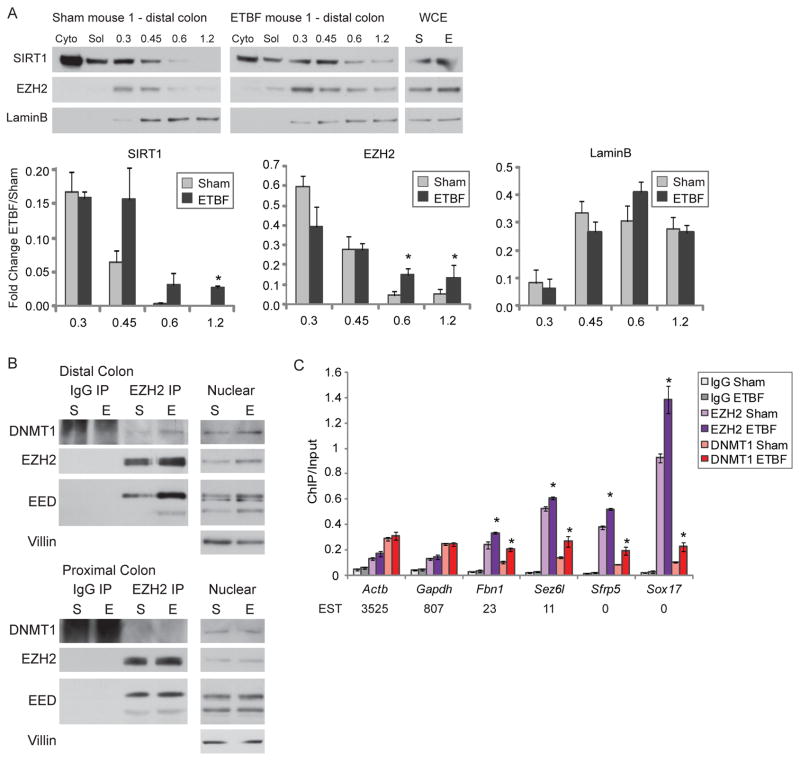
To examine whether our in vitro findings are applicable to the cancer risk state of inflammation, we studied an in vivo model of colitis in which infection with the human commensal enterotoxigenic Bacteroides fragilis (ETBF) induces inflammation and tumorigenesis primarily in the distal colons of Multiple intestinal neoplasia (Min) mice which are heterozygous for loss of Adenomatous polyposis coli (Apc) (Rhee et al., 2009; Wu et al., 2009). Importantly, in vitro treatment of colon cancer cells with purified B. fragilis toxin induces an increase in γ-H2AX and ROS suggesting that this model provides an in vivo scenario to assess the endogenous impact of the oxidative damage we examined in our in vitro model (Goodwin et al., 2011). First, we examined whether any of the players involved in our complex become more tightly bound to chromatin in the inflamed distal tissue (Figure S6A). Similar levels of Villin, an epithelial marker, establish that cells harvested by scraping from ETBF and mock inoculated (sham) mice have similar epithelial content, though this method does not obtain pure populations of epithelial cells (Figure S6B). Using the salt gradient extraction employed in Figure 1A, while we detect no change in DNMT1 (data not shown), we demonstrate that both SIRT1 and EZH2 are more tightly bound to chromatin in distal, but not proximal, colon epithelial cells from ETBF mice than sham mice two days after inoculation (Figure 7A, S6B & S6C). These results suggest the change in binding is due to the specific high level of inflammation that occurs during the acute phase of the ETBF model.
Figure 7. In a Mouse Model of Acute Colonic Inflammation, Members of the Silencing Complex Become Enriched at Promoter CpG Islands of Low Expression Genes.
(A) Mice were sham-inoculated (sham) or inoculated with ETBF. Two days post-inoculation, colon epithelial cells were extracted sequentially using cytoplasmic extraction buffer (Cyto), soluble nuclear buffer (Sol), and buffers with increasing NaCl concentration. Whole cell lysate was prepared separately (WCE). Blots from one set of representative mice are depicted. The value calculated for each fraction is the ratio of that fraction over the total of all fractions. The graphs represent the mean values for 3 separate mice +/− SEM. * p-value < 0.05 by one-tail t-test. (B) Co-immunoprecipitations for control IgG or anti-EZH2 antibodies were performed in nuclear lysates of colon epithelial cells from sham-inoculated mice (S) or ETBF-inoculated mice (E). Blots from one set of representative mice are depicted. (C) Using distal colon epithelial cells, ChIP was performed for IgG, EZH2, or DNMT1 and analyzed by quantitative PCR. The data presented is the mean of ChIP performed in samples from 3 sham and 3 ETBF mice +/− SEM. * p < 0.05 by one-sided t-test for the difference between the means. Values below the gene names are the expressed sequence tag (EST) counts for mouse intestine from the Unigene database. See also Figure S6.
Secondly, we examined whether any of the proteins in our complex interact in the inflamed tissue. We performed co-immunoprecipitations using anti-EZH2 antibodies in proximal or distal colon epithelial cells from two separate pairs of sham and ETBF mice. While in all cases EZH2 co-immunoprecipitates predominantly isoform 1 of EED (isoform is determined by comparison to the four isoforms present in mouse embryonic carcinoma cells - Figure S6D), co-immunoprecipitation of DNMT1 by EZH2 is more prominent in the tissue from the distal colon of the ETBF mice (Figure 7B & S6D) suggesting that this interaction is increased by the inflammation in this tissue.
Thirdly, we performed local ChIP for EZH2 and DNMT1 in epithelial cells from the distal colon of ETBF and sham mice. Interestingly, unlike the in vitro model, we do not see enrichment of EZH2 and DNMT1 at all promoter CpG islands (Figure 7C). High expression housekeeping genes such as Actb and Gapdh have no change in EZH2 or DNMT1 enrichment between the ETBF and sham mice. However, lower expression genes, such as Fbn1, Sez6l, Sfrp5, and Sox17, that have higher basal levels of EZH2 than the high expression genes, all have more enrichment of EZH2 and DNMT1 at their promoter CpG islands in inflamed distal epithelial cells from ETBF mice compared to sham mice. Interestingly, three of these genes, Fbn1, Sez6l, and Sox17, have been demonstrated to undergo inflammation and tumor-specific DNA methylation in a model of intestinal inflammation and all four are methylated in human cancers (Hahn et al., 2008). These data suggest that, in this model, where changes in ROS are likely less dramatic and induced over a longer time frame than for our in vitro model, the recruitment of members of the silencing complex is most persistent at promoter CpG island-containing genes with lower basal expression. These genes are the most likely to be targets of cancer-specific DNA methylation (Hahn et al., 2008).
DISCUSSION
In the present study, we link several proteins involved in transcriptional repression to the DNA damage response. We provide evidence for a role for DNMT1 in the response of cells to H2O2 treatment. This enzyme becomes more tightly bound to chromatin after H2O2 treatment in the context of damage-induced foci that co-localize with γ-H2AX. Moreover, DNMT1 appears responsible for the tightening of the PRC4 component, SIRT1, to chromatin after H2O2 treatment. SIRT1 has been implicated in the response to DNA damage and transcriptional repression in many ways as previously discussed. While in inflamed mouse tissue we do not see an increase in binding of DNMT1 to chromatin, we see an increase in binding of the PRC4 components, SIRT1 and EZH2. Increases in binding of DNMT1 to chromatin may occur at an earlier time point than the one we studied, as the in vivo model time point is two days after infection compared to 30 minutes after treatment for our in vitro model. However, we do implicate DNMT1 in the in vivo response to colonic inflammation by demonstrating interaction between EZH2 and DNMT1 and enrichment of both of these proteins at the CpG islands of low expression genes in this inflamed tissue, in a manner consistent with our in vitro results.
Our observations presented here suggest that tightening of DNMT1 and SIRT1 to chromatin after H2O2 treatment is actively associated with DNA damage and/or repair. Our evidence by immunofluorescence studies for co-localization of DNMT1, SIRT1, and EZH2 to DNA damage-induced foci marked by γ-H2AX suggests targeting to ongoing DNA damage. Previously, we have implicated DNMT1 in double strand break repair and recently another group has further studied this role for DNMT1 (Ha et al., 2010; O’Hagan et al., 2008). We did not see DNMT1 become more tightly bound to chromatin after IR treatment and the observed narrow localization of γ-H2AX signal at the promoter region by ChIP-chip after H2O2 exposure is not representative of the typical mega base domain enrichment seen after double strand breaks (Rogakou et al., 1999). Thus, our findings suggest that, after H2O2 treatment, double strand breaks are not the dominant trigger for the dynamics we are reporting. Because levels of OGG1 modulate the tightening of DNMT1 binding to chromatin, we suggest that either the specific 8-oxodG base damage or the BER pathway that repairs this type of damage is responsible for recruitment of member of the silencing complex to chromatin. We propose that the promoter targeted γ-H2AX is likely marking sites of base damage and this hypothesis is supported by the enrichment of 8-oxoguanine at GC-rich promoters after H2O2 treatment. We suggest that the enrichment of damage at these areas of high GC content may be because guanine is the most easily oxidized of the four deoxyribonucleosides and therefore may be targeted by oxidative damage (Steenken, 1997). The co-enrichment of DNMT1, SIRT1, DNMT3B, and EZH2 at these sites suggests that these proteins are being localized to sites of base damage induced by H2O2 treatment. While in our in vivo model we have not assessed whether the enrichment of key complex constituents is also occurring at the sites of base damage, it has been demonstrated that purified B. fragilis toxin causes an increase in γ-H2AX and ROS in colonic epithelial cells. This data suggests that our in vivo model involves a similar induction of oxidative damage as our in vitro model (Goodwin et al., 2011).
One of the intriguing implications of our data is the potential role for increased levels of cellular ROS that accompany cancer-risk states such as inflammation in the formation of cancer-specific aberrant patterns of DNA methylation and transcriptional silencing. First, as we have noted, cellular transformation has been associated with the presence of the PRC4 iso-complex (Kuzmichev et al., 2005) that we now link to DNMTs during H2O2 exposure. This complex has altered substrate specificity from the typical PRC2/3 complexes possibly due to the specific isoform of EED that it contains (EED2) (Kim et al., 2007). Since both pre-neoplastic and transformed cells undergo a significant amount of endogenous oxidative damage (Federico et al., 2007), the basal PRC4 complex previously described in transformed cells may be the same as the complex we describe here. Because nuclear SIRT1 levels increase after H2O2 treatment, it is possible that the increase in interaction demonstrated between SIRT1 and the other proteins in Figure 2 is due to higher levels of SIRT1, not to an induced interaction per say. However, either cause has the same outcome, namely higher levels of the complex.
Second, our findings suggest one potential mechanism that might help explain a conundrum in the abnormalities of DNA methylation in human cancer - namely, why cancer cells simultaneously harbor both widespread chromosomal loss of DNA methylation and increased DNA methylation in CpG islands of gene promoters (Jones and Baylin, 2007). In terms of the losses, we find that enzymes that catalyze DNA methylation, DNMT1 and DNMT3B, shift away from non-GC-rich gene and chromosome regions. In a similar manner, it has previously been demonstrated in yeast and mammalian cells that DNA damage leads to a shift in localization of SIRT1 from repressed gene regions to sites of induced DNA damage resulting in transcriptional de-repression of genes that are basally repressed by SIRT1 (Mills et al., 1999; Oberdoerffer et al., 2008). We suggest that when cells are exposed to chronic oxidative damage that is present during all phases of tumorigenesis, the induced shifts in chromosome localization that we demonstrate may be associated with losses of DNA methylation observed in cancer cells.
Finally, our observations may also help explain gains in DNA methylation at gene promoters in cancer cells. By examining histone mark and transcription changes, we demonstrate that enrichment of members of this silencing complex is associated with gene silencing. Importantly, however, in cells progressing towards transformation, the above translocalization would probably be transient at most genes with high basal transcription levels, such as housekeeping genes, and oncogenes, for which silencing would be detrimental to tumor cell growth. This hypothesis is supported by the lack of EZH2 and DNMT1 enrichment at the promoters of high expression CpG island-containing genes in the less harsh, longer time-frame, inflammatory in vivo model studied. In this regard, active transcription may prevent de novo promoter CpG island methylation (Thomson et al., 2010). We demonstrate that genes with a history of frequent, cancer specific, CpG island promoter DNA hypermethylation show damage-induced enrichment for the members of the complex in cell culture and enrichment of EZH2 and DNMT1 in inflamed mouse tissue. In vitro we see an increase in DNA methylation that correlates with the low basal expression level of these genes, which harbor PcG complexes in embryonic stem and progenitor cells (Ohm et al., 2007; Schlesinger et al., 2007; Widschwendter et al., 2007). We hypothesize that such localization of the DNMT-PRC4 complex and increase in DNA methylation at low expression promoter CpG island-containing genes might be more persistent over the course of chronic ROS damage during tumorigenesis setting up a scenario for the expansion of DNA methylation in the CpG islands involved. Our previous work with a promoter CpG island, double strand break DNA damage model, suggests this time-dependent context for the expansion of such DNA hypermethylation (O’Hagan et al., 2008).
EXPERIMENTAL PROCEDURES
Cell culture, Chemicals, Treatments, and Plasmids
Cells were maintained as described in the Supplemental Experimental Procedures. The DNMT1 hypomorph HCT116 cell line was clonally selected for stable expression of exogenous FLAG-tagged full length DNMT1 and maintained in media containing puromycin. For H2O2 exposure, 30% H2O2 (Sigma) was diluted in PBS immediately before adding it to the medium. Time of treatment is the time after H2O2 is added to the media. c-Myc-Nuc-hOGG1 (Chatterjee et al., 2006) was kindly provided by D. Sidransky (Addgene plasmid 18709).
Salt Gradient Extraction and Tight Chromatin Fractionation
Cells were collected 30 minutes after H2O2 exposure and subjected to sequential extraction with buffers indicated in the Supplemental Experimental Procedures. Band densitometry for western blots was analyzed using ImageJ software.
shRNA Knockdown
Cells were transduced with the indicated lentiviral particles following the manufacturer’s protocol (Sigma).
Co-Immunoprecipitation
Co-immunoprecipitations were performed from nuclear extracts that were treated with oligoamines to release chromatin bound proteins as described in the Supplemental Experimental Methods.
Sucrose Gradient
Nuclear extracts prepared as for co-immunoprecipitation were applied to a 15–60% (w/v) sucrose gradient, and centrifuged in a SW41 rotor for 20 hours at 40,000 rpm at 4°C. Equivalent volumes from each odd fraction were separated by SDS-PAGE and analyzed by immunoblot. The remaining fractions were pooled as indicated. Buffer was exchanged to modified RIPA using PD-10 columns (GE Healthcare). Co-immunoprecipitations were performed from each pool as in the section above.
Immunofluorescence
Cells grown on coverslips were preextracted and fixed as described in the Supplemental Experimental Methods.
ChIP-chip
Cells were crosslinked using 1% formaldehyde and 0.5 mM DSG. Nuclear extraction was performed using CEBN and CEB, followed by ChIP-chip as previously described (McGarvey et al., 2008) using antibodies indicated in Supplemental Experimental Methods. Samples were either hybridized to the Agilent 1M custom array for human chromosomes 18, 19 and 21 or the human promoter 244K or 1M ChIP-chip arrays from Agilent Technologies.
Nascent Transcription
Nascent transcription assays were performed using the Click-iT Nascent RNA Capture Kit (Invitrogen). Cells were labeled with ethynyl uridine for 30 minutes concurrently with the H2O2 treatment if indicated.
Bisulfite Sequencing
Bisulfite treatment was performed with the EZ DNA Methylation Kit (Zymo). Bisulfite sequencing was performed as previously described (McGarvey et al., 2007).
Mice
C57BL/6J mice were handled and inoculated as in (Rhee et al., 2009). Distal and proximal epithelium was collected by scraping the mucosal surface of the dissected colon, washed three times in PBS, and then subjected to the indicated protocol. Such scraping has been shown by others to be an effective method to obtain samples of intestinal epithelial cells (Ortega-Cava et al., 2006). ChIP from this tissue was performed using the Magna ChIP™ G Tissue Kit (Millipore). All mouse protocols were approved by the Johns Hopkins University Animal Care and Use Committee in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International.
Statistical Analysis
All western blot, immunofluorescence, and local ChIP data are presented as the mean +/− standard error (SEM). These data are evaluated by one-tail t-test and considered statistically significant with a p-value < 0.05.
ChIP-Chip data were analyzed utilizing the limma and Ringo packages from Bioconductor (Smyth and Speed, 2003; Toedling et al., 2007) as described in the Supplemental Experimental Methods.
Supplementary Material
SIGNIFICANCE.
Tumors have aberrant gains and losses in DNA methylation, although the mechanisms establishing these changes are not well understood. Here we demonstrate that oxidative damage induces the formation of a large silencing complex(es) containing DNA methyltransferases and constituents of the polycomb complex, PRC4, including SIRT1. PRC4 is found uniquely in cancer and embryonic and adult stem cells. Key constituents of the damage-induced complex are recruited from transcriptionally poor regions of the genome to GC-rich areas, including promoter CpG islands. Such translocalization causes changes in histone marks, transcription, and DNA methylation. We suggest that this relocalization may be a mechanism by which oxidative damage can be responsible for both promoter CpG island specific hypermethylation and global hypomethylation seen in cancer.
HIGHLIGHTS.
DNMT1 becomes more tightly bound to chromatin after oxidative damage.
Oxidative damage induces formation of a complex containing DNMT1, DNMT3B and PRC4.
DNMT-PRC4 enrichment at CpG islands may explain aberrant gene silencing in cancer.
Promoters enriched for these proteins have histone mark and DNA methylation changes
Acknowledgments
We thank Kathy Bender for manuscript preparation, Dr. D. Sidransky for the c-Myc-nuc-hOGG1 plasmid, and Dr. B. Karim for assessment of H&E stained colon slides.
This work was supported by National Cancer Institute Grant CA043318, National Institute of Environmental Health Sciences grant ES011858, and National Institutes of Health Grant CA116160, all to S.B.B, as well as National Institutes of Health Grants R01DK080817 and R01CA151325 awarded to C.L.S. and National Cancer Institute Grants CA51085 and CA98454 to R.A.C.. C.D.S. is supported by National Institute of Environmental Health Sciences Training grant ES07141.
Footnotes
ACCESSION NUMBERS
Microarray data sets were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) with the accession number GSE32382.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chatterjee A, Mambo E, Zhang Y, Deweese T, Sidransky D. Targeting of mutant hogg1 in mammalian mitochondria and nucleus: effect on cellular survival upon oxidative stress. BMC Cancer. 2006;6:235. doi: 10.1186/1471-2407-6-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran HP, Van Neste L, Cope L, Sen S, Mohammad HP, Pageau GJ, Lawrence JB, Herman JG, Schuebel KE, Baylin SB. Aberrant silencing of cancer-related genes by CpG hypermethylation occurs independently of their spatial organization in the nucleus. Cancer Res. 2010;70:8015–8024. doi: 10.1158/0008-5472.CAN-10-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. Epub 2007 Mar 2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. 2007;121:2381–2386. doi: 10.1002/ijc.23192. [DOI] [PubMed] [Google Scholar]
- Folle GA, Liddle P, Lafon-Hughes L, Di Tomaso MV. Close encounters: RIDGEs, hyperacetylated chromatin, radiation breakpoints and genes differentially expressed in tumors cluster at specific human chromosome regions. Cytogenet Genome Res. 2010;128:17–27. doi: 10.1159/000296072. [DOI] [PubMed] [Google Scholar]
- Goodwin AC, Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K, Lee GE, Palii SS, Brown KD, Takeda Y, Liu K, Bhalla KN, Robertson KD. Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum Mol Genet. 2010;20:126–140. doi: 10.1093/hmg/ddq451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–10289. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Liang G, Sharma S, Lin JC, Choi SH, Han H, Yoo CB, Egger G, Yang AS, Jones PA. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GD, Ni J, Kelesoglu N, Roberts RJ, Pradhan S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. Embo J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Levenson JM, Korsmeyer S, Sweatt JD, Schumacher A. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J Biol Chem. 2007;282:9962–9972. doi: 10.1074/jbc.M608722200. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–5102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, Schuebel KE, Baylin SB. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Ortega-Cava CF, Ishihara S, Rumi MA, Aziz MM, Kazumori H, Yuki T, Mishima Y, Moriyama I, Kadota C, Oshima N, et al. Epithelial toll-like receptor 5 is constitutively localized in the mouse cecum and exhibits distinctive down-regulation during experimental colitis. Clin Vaccine Immunol. 2006;13:132–138. doi: 10.1128/CVI.13.1.132-138.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci U S A. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Rhee KJ, Wu S, Wu X, Huso DL, Karim B, Franco AA, Rabizadeh S, Golub JE, Mathews LE, Shin J, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Langst G, Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol. 2007;176:565–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenken S. Electron transfer in DNA? Competition by ultra-fast proton transfer? Biol Chem. 1997;378:1293–1297. [PubMed] [Google Scholar]
- Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. Ringo--an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.