Abstract
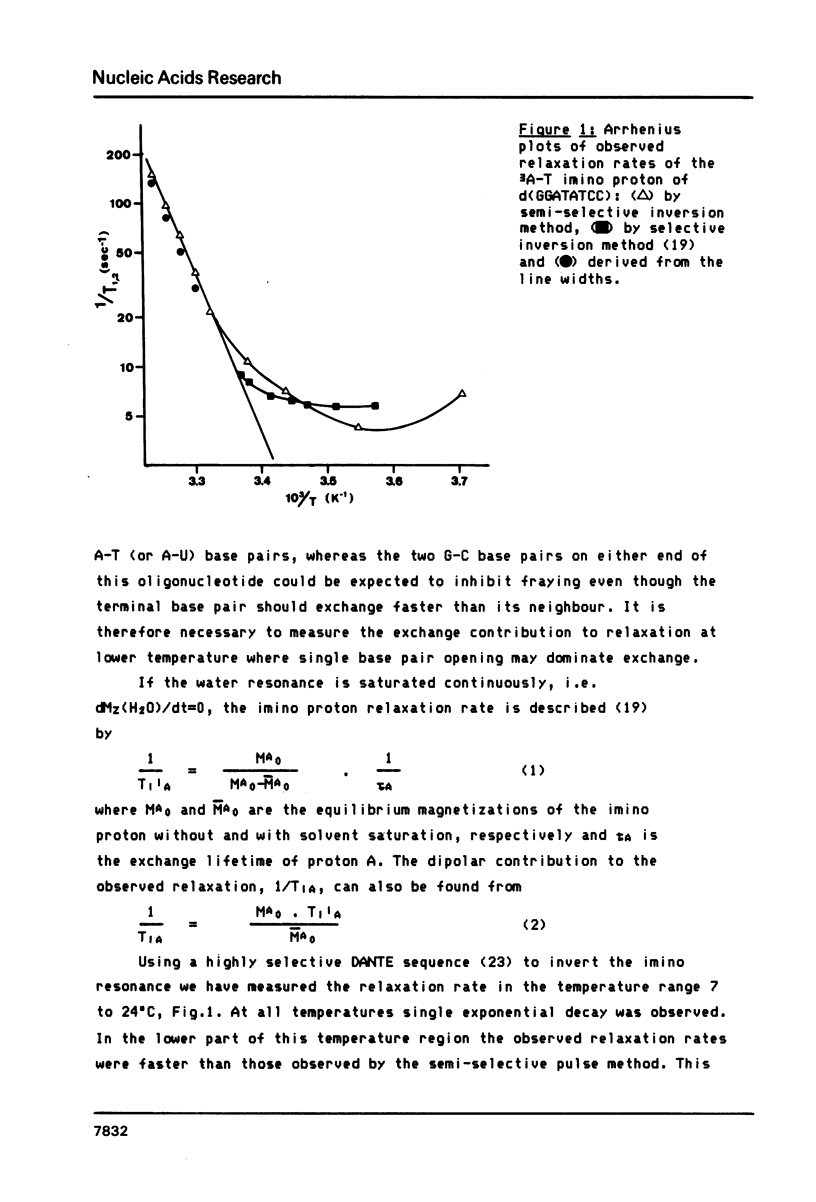
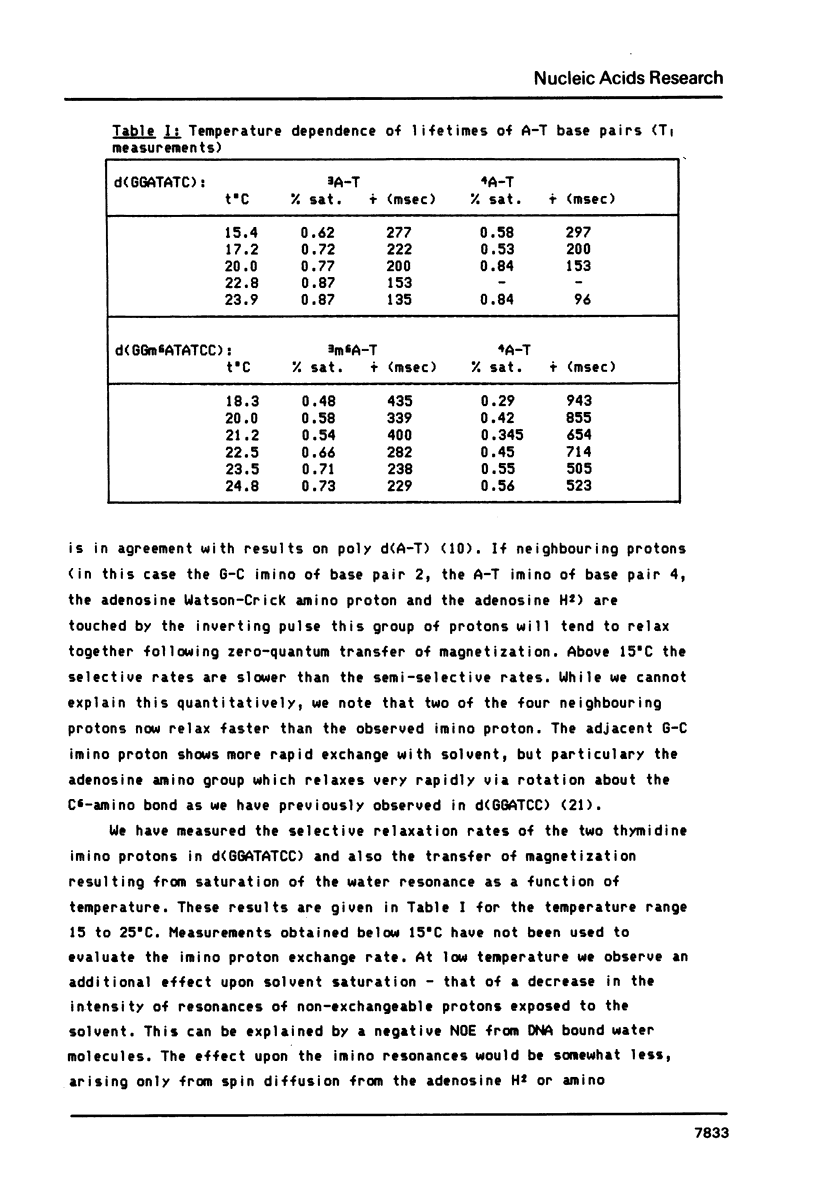
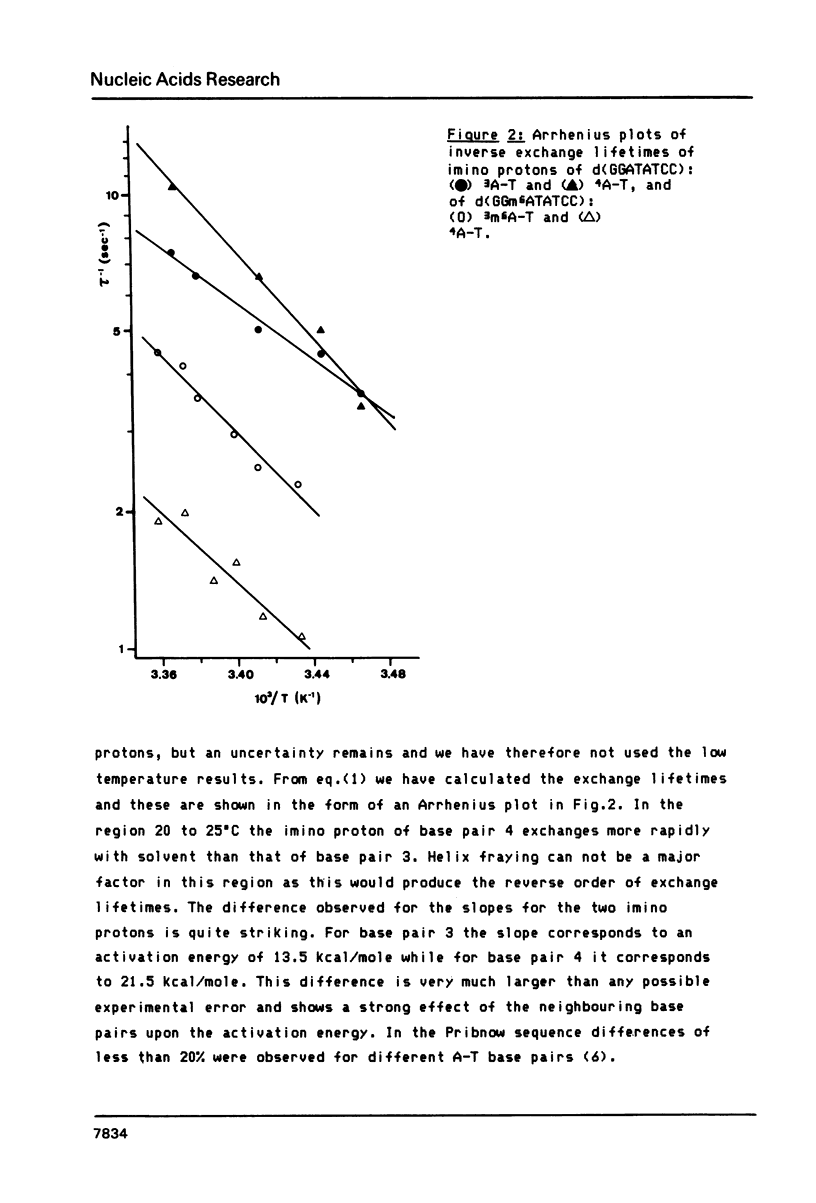
We report relaxation time measurements by semi-selective and totally selective NMR techniques on the thymidine imino protons of d(GGATATCC) and d(GGm6ATATCC). For these oligonucleotides helix fraying, rather than single base pair opening, is the major exchange mechanism even 25 degrees C below the Tm. We have therefore applied a new saturation transfer technique to measure exchange rates at temperatures where fraying has a very small or negligible contribution. Measurements of exchange rates as a function of temperature give significantly different activation energies for base pairs 3 and 4 in d(GGATATCC). Adenine methylation results in a slowing down of the opening rate for the m6A-T base pair but surprisingly has an even greater effect upon the adjacent non-methylated A-T base pair.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Assa-Munt N., Granot J., Behling R. W., Kearns D. R. 1H NMR relaxation studies of the hydrogen-bonded imino protons of poly(dA-dT). Biochemistry. 1984 Feb 28;23(5):944–955. doi: 10.1021/bi00300a023. [DOI] [PubMed] [Google Scholar]
- Calladine C. R. Mechanics of sequence-dependent stacking of bases in B-DNA. J Mol Biol. 1982 Oct 25;161(2):343–352. doi: 10.1016/0022-2836(82)90157-7. [DOI] [PubMed] [Google Scholar]
- Chou S. H., Wemmer D. E., Hare D. R., Reid B. R. Sequence-specific recognition of DNA: NMR studies of the imino protons of a synthetic RNA polymerase promoter. Biochemistry. 1984 May 8;23(10):2257–2262. doi: 10.1021/bi00305a026. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Cole P. E., Hilbers C. W., Shulman R. G. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974 Jul 25;87(1):63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. Base sequence and helix structure variation in B and A DNA. J Mol Biol. 1983 May 25;166(3):419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Hillen W., Wells R. D. A 300- and 600-MHz proton nuclear magnetic resonance investigation of a 12 base pair deoxyribonucleic acid restriction fragment: relaxation behavior of the low-field resonances in water. Biochemistry. 1981 Jun 23;20(13):3756–3764. doi: 10.1021/bi00516a014. [DOI] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Hillen W., Wells R. D. A 300-MHz proton nuclear magnetic resonance investigation of deoxyribonucleic acid restriction fragments: dynamic properties. Biochemistry. 1981 Jun 23;20(13):3764–3769. doi: 10.1021/bi00516a015. [DOI] [PubMed] [Google Scholar]
- Engel J. D., von Hippel P. H. Effects of methylation on the stability of nucleic acid conformations: studies at the monomer level. Biochemistry. 1974 Sep 24;13(20):4143–4158. doi: 10.1021/bi00717a013. [DOI] [PubMed] [Google Scholar]
- Feigon J., Denny W. A., Leupin W., Kearns D. R. Proton nuclear magnetic resonance investigation of the conformation and dynamics in the synthetic deoxyribonucleic acid decamers d(ATATCGATAT) and d(ATATGCATAT). Biochemistry. 1983 Dec 6;22(25):5930–5942. doi: 10.1021/bi00294a037. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Patel D. J. Proton nuclear magnetic resonance investigations of the nucleation and propagation reactions associated with the helix-coil transition of d-ApTpGpCpApT in H2O solution. Biochemistry. 1975 Jun 17;14(12):2656–2660. doi: 10.1021/bi00683a015. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Borer P. N., Ts'o P. O. Conformation and interaction of short nucleic acid double-stranded helices. II. Proton magnetic resonance studies on the hydrogen-bonded NH-N protons of ribosyl ApApGpCpUpU helix. Biochemistry. 1975 Nov 4;14(22):4864–4869. doi: 10.1021/bi00693a013. [DOI] [PubMed] [Google Scholar]
- Mirau P. A., Kearns D. R. Sequence and conformational effects on imino proton exchange in A.T- and A.U-containing DNA and RNA duplexes. Biopolymers. 1985 Apr;24(4):711–724. doi: 10.1002/bip.360240410. [DOI] [PubMed] [Google Scholar]
- Pardi A., Martin F. H., Tinoco I., Jr Comparative study of ribonucleotide, deoxyribonucleotide, and hybrid oligonucleotide helices by nuclear magnetic resonance. Biochemistry. 1981 Jul 7;20(14):3986–3996. doi: 10.1021/bi00517a007. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of imino protons in the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix and in two similar helices that contain a G . T base pair, d(C-G-T-G-A-A-T-T-C-G-C-G), and an extra adenine, d(C-G-C-A-G-A-A-T-T-C-G-C-G). Biochemistry. 1982 Dec 7;21(25):6567–6574. doi: 10.1021/bi00268a038. [DOI] [PubMed] [Google Scholar]
- Pardi A., Tinoco I., Jr Kinetics for exchange of imino protons in deoxyribonucleic acid, ribonucleic acid, and hybrid oligonucleotide helices. Biochemistry. 1982 Sep 14;21(19):4686–4693. doi: 10.1021/bi00262a026. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. Nuclear magnetic resonance studies of the helix-coil transition of poly (dA-dT) in aqueous solution. Proc Natl Acad Sci U S A. 1976 Mar;73(3):674–678. doi: 10.1073/pnas.73.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Hilbers C. W. Proton nuclear magnetic resonance investigations of fraying in double-stranded d-ApTpGpCpApT in H2O solution. Biochemistry. 1975 Jun 17;14(12):2651–2656. doi: 10.1021/bi00683a014. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Ikuta S., Kozlowski S., Itakura K. Sequence dependence of hydrogen exchange kinetics in DNA duplexes at the individual base pair level in solution. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2184–2188. doi: 10.1073/pnas.80.8.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Hare D. R., Reid B., Ikuta S., Lander N., Itakura K. Conformation, dynamics, and structural transitions of the TATA box region of self-complementary d[(C-G)n-T-A-T-A-(C-G)n] duplexes in solution. Biochemistry. 1985 Feb 12;24(4):926–935. doi: 10.1021/bi00325a018. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Hare D. R., Reid B., Ikuta S., Lander N., Itakura K. Conformation, dynamics, and structural transitions of the TATA box region of self-complementary d[(C-G)n-T-A-T-A-(C-G)n] duplexes in solution. Biochemistry. 1985 Feb 12;24(4):926–935. doi: 10.1021/bi00325a018. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Broka C., Rice J. A., Itakura K., Breslauer K. J. Premelting and melting transitions in the d(CGCGAATTCGCG) self-complementary duplex in solution. Biochemistry. 1982 Feb 2;21(3):428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]


