Abstract
The idea that we could build molecular communications systems can be advanced by investigating how actual molecules from living organisms function. Information theory provides tools for such an investigation. This review describes how we can compute the average information in the DNA binding sites of any genetic control protein and how this can be extended to analyze its individual sites. A formula equivalent to Claude Shannon’s channel capacity can be applied to molecular systems and used to compute the efficiency of protein binding. This efficiency is often 70% and a brief explanation for that is given. The results imply that biological systems have evolved to function at channel capacity, which means that we should be able to build molecular communications that are just as robust as our macroscopic ones.
Keywords: Information theory, Sequence logo, Sequence walker, Channel capacity, Molecular efficiency, Nanotechnology
1. An approach to constructing molecular communications
The fundamental step in communications, including communications at the molecular level, is the accurate reception of a signal. As is well known in communications engineering fields, the mathematical foundation for obtaining good reception was developed by Claude Shannon in 1948 and 1949 [55–57]. How can we apply these ideas to the construction of molecular communications? One approach is to first find out how biomolecules interact with each other and how they set their states. With some changes in perspective from conventional biochemistry, the states and patterns of molecules can be measured by using information theory and the field of study can be called molecular information theory [39,45,47].
So how can we apply Shannon’s theory to molecules found in living organisms? A good starting point is to study the interactions between DNA and proteins that control the expression of DNA. Controlling a gene requires that the protein bind to specific points on the nucleic acid to either prevent or activate expression. There are only four nucleotide bases in DNA, named A, C, G and T, so the DNA to which a protein binds can be represented by the pattern of these letters. A protein is a finite molecule, which means that it can contact only a segment of a DNA at one time, typically 10–20 base pairs. The pattern that one protein searches for and subsequently binds is not always the same string of the four bases; the patterns vary. Some of this variation simply doesn’t matter to the biological system, while other variations can be used by the protein to do different jobs.
2. Sequence logos show binding site information
We can use information theory to measure how much pattern is in a set of binding sites [54]. As an example, consider the Fis protein. In a starving bacterial cell there are below 100 molecules of Fis, but when the cell encounters nutrients, the numbers increase to over 50,000 molecules [4] and the Fis molecules then control many genes in the cell [19]. Fig. 1 shows several experimentally proven Fis sites from the front of the Fis gene itself. When there is not much Fis in the cell, the Fis gene is active, making more Fis molecules. Eventually those molecules occupy the sites at the Fis gene and shut the synthesis of Fis down as a negative feedback loop. How does Fis find these locations on the genome? From inspecting the figure, it is clear that the sequences are quite different, but the central region (around zero) has many A and T bases, while position −7 is almost always a G and +7 is almost always a C. The logo shows that different parts of the site are conserved by different amounts.
Fig. 1.
Aligned listing (top) and sequence logo (bottom) for DNA binding sites of the Fis protein from the bacterium Escherichia coli. The bar of numbers (‘numbar’) on the top is to be read vertically and it shows the range from −10 to +10 for positions across the site. Below the numbar are 6 Fis sites and their complementary sequences. Both are given, since Fis binds as a dimer. On the right is the individual information for each sequence. The sequence logo on the bottom shows the sequence conservation in the complete data set which consists of 60 Fis sites and their complements [19]. The height of each letter is proportional to the frequency of that base at that position and the letters are sorted. The height of the entire stack of letters is the information, measured in bits. The possible variation of the height from small sample effects is shown by error bars. The peak of the sine wave shows where the major groove of DNA faces the protein [30,11,44,29]. It can be used to infer some aspects of how the protein contacts the DNA [40].
How can we characterize the sites precisely? We know that before Fis has bound to the DNA it can see all four possible bases at one position. So, following Shannon, we can say that the protein is uncertain as to what base it will see and that uncertainty can be measured as log2 4 = 2 bits [31,48].
In contrast, once the protein has bound to a Fis site, the uncertainty of what it is touching in different cases is lower. It is nearly just one base in positions −7 and +7 and so there the uncertainty is near log2 1 = 0 bits. However, that is only an approximation since there are other bases at those positions. So the uncertainty is not zero. Fortunately, Shannon worked out how to compute the uncertainty from the frequency of symbols [55]:
| (1) |
where fb,l are the frequencies of the bases b ∈ {A, C, G, T} at a position l in the sequence alignment.
There are several caveats to note at this point. First, since we don’t have infinite numbers of sequences, as Shannon’s theory would require, we substituted the frequencies of bases for the probabilities. This requires making a correction for small sample size [54]. Another point is that the uncertainty is sometimes called the ‘Shannon entropy’, but if we say that then there can be confusion later when we have to discuss the actual entropy changes in the molecular binding process. Finally, it is important to be clear that the uncertainty given by Eq. (1) is not the information, as we discuss below.
Before binding to a site, the Fis molecule is somewhere on the DNA and sees 2 bits of uncertainty. After binding, it has lower uncertainty, H(l). Shannon realized that the receiver of a message will get less information because of noise in the signal, and he showed that the information R received is reduced from the transmitted uncertainty H(x):
| (2) |
He said “The conditional entropy Hy(x) will, for convenience, be called the equivocation. It measures the average ambiguity of the received signal” [55].
Likewise, H(l) is the ambiguity ‘observed’ by the DNA binding protein once it has bound to a site, so the information in the binding site is the uncertainty before binding less that after binding:
| (3) |
To show this graphically, we can create a sequence logo [52], as shown on the bottom of Fig. 1. We plot Rsequence(l) across the binding site and use that to vary the heights of stacks of letters representing the relative abundances of bases at each position. Sequence logos are now widely used in molecular biology to present patterns in DNA, RNA or protein.
Variation of one part of a binding site is generally independent of other parts [6], so the information values at all of the positions in a binding site can be added together to find the total information in a binding site,
| (4) |
This is the ‘area’ under the logo, found by adding the heights of the letter stacks together.
3. Evolution of information
The significance of Rsequence was found by comparing it to another measure of information. In many cases (but not Fis) the number of binding sites on the genome is known. So the problem facing the DNA binding protein is to locate a number of sites, γ, from the entire genome of size G. In information theory terms, the uncertainty before being bound to one of the sites is log2 G, while after being bound it reduces to log2 γ. So, as with the computation of the information in the binding sites, the information required to find the binding sites is
| (5) |
Natural binding sites have Rsequence close to Rfrequency [54]. In other words, the information found in binding sites is just sufficient to locate the binding sites on the genome. Since the genome size and number of sites are more or less fixed by the environment, the information at the binding sites, Rsequence, has to evolve towards that required, Rfrequency, and a computer model called Ev has verified this prediction [43]. A Java version that you can run on your own computer is now available at http://alum.mit.edu/www/toms/papers/ev/. Using this, Evj, which was written by Paul C. Anagnostopoulos, one can watch the sequence logo evolve from scratch.
4. Sequence walkers show individual information of binding sites
The significance of Rsequence is that it reflects how the genetic control system evolves to meet the demands of the environment as represented by Rfrequency. If we inspect how Rsequence is computed from Eqs. (1), (3) and (4), we can see that it relies on the probability-weighted sum in Eq. (1). That is, Rsequence is an average of the function −log2 fb,l. But what does this average represent? It turns out that it can be expressed as an average of the information of individual sequences by adding together weights of the form Ri(b, l) = 2 + log2 fb,l for a sequence [41,51,17]. The computed values for each binding site sequence in Fig. 1 are on the right side of the figure.
Like a sequence logo, the information of a single binding site can be presented graphically [42]. For example, Fig. 2 shows a portion of the Fis promoter region using sequence walkers. The figure reveals the marvelous but previously unsuspected complexity of the genetic control system.
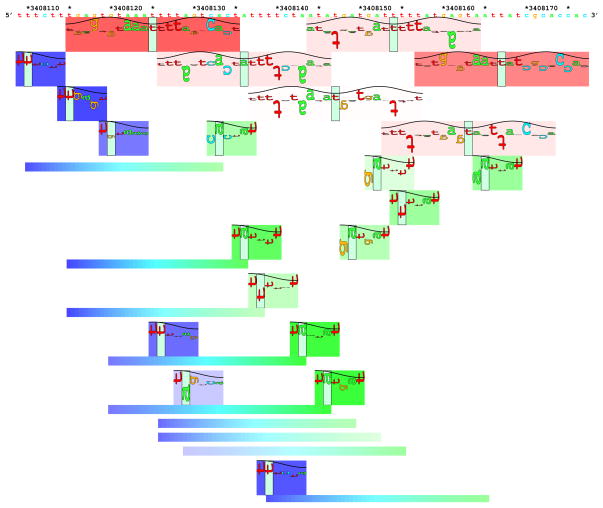
Fig. 2.
Sequence walkers show a map of DNA binding sites of the Fis protein on part of the Fis promoter on the genome of the bacterium Escherichia coli [19]. The DNA sequence from GenBank entry Accession NC_000913.2 is given on the top with every 5th and 10th base marked with an asterisk (*) and every 10th base numbered. A sequence walker, much like a sequence logo, is a set of adjacent letters with varying heights, but there is only one letter per position. A walker represents the location of where a protein binds. As in Fig. 1, sine waves show how the protein is oriented on the DNA. For example, directly below the DNA sequence are red and pink rectangles (called ‘petals’) each with a green bar in the center that marks the location of the binding site (zero coordinate of the logo in Fig. 1). Sequence walkers for Fis sites are shown superimposed on these red rectangles. The letters of each walker correspond to the DNA sequence directly above. The height of a letter is how well that base is conserved in the original data set of binding sites, measured in bits. The scale runs from 2 bits at the top of each rectangle to−3 bits at the bottom. Preferred bases go upwards while bases that are bad for binding go downwards. The total information for each site is shown by the saturation of the color, so a red rectangle means that the site has more information than a pink or white one. A total of six Fis sites (two red, three pink and one white) are predicted to be in this piece of DNA. The two red sites are numbers 3 and 5 of Fig. 1. The bottom of this map also shows a set of promoters for making RNA from this region. The σ70 promoters shown have two parts, a ‘−35’, shown in blue rectangles on the left and a ‘−10’ region, shown by green rectangles on the right [58]. These parts are connected together by a variable-length horizontal bar below the rectangles [59]. The color of the connecting bar shades from one side to the other to help identify the corresponding sequence walkers at their zero coordinates, indicated by the green bars. Because Fis binds as a dimer, the binding sites are symmetrical and the sequence walker letters are vertical. Since promoters are asymmetrical, the letters in these sequence walkers are turned sideways so that reading ‘down’ the sequence of letters indicates the direction that transcription will take place. The map shows that since the gene for Fis is ‘downstream’ (to the right) of this region, Fis protein binds to its own promoter to control its own synthesis in a negative feedback loop.
As with Fis in Fig. 2, by using sequence walkers we find a similar complexity of potential promoters and ribosome binding sites between known genes [18]. The method produces a wealth of experimental predictions and can also be used to understand human diseases. Many genetic changes alter a single base at a time (single nucleotide polymorphism, SNP) and about 15% of those that cause genetic disease affect RNA splicing. The SNP changes can be analyzed using individual information and sequence walkers to predict whether the change is likely to be the cause of a disease [32]. Likewise, more than 50% of tumors in some types of cancer have mutations in the DNA binding p53 protein, so it is important for understanding cancer biology. We built an information theory model of how p53 binds to DNA, predicted that 16 genes should be under p53 control and then demonstrated experimentally that 15 of the 16 are indeed under p53 control [28]. Since 11 of the 16 sites had not been identified by other methods, information theory provides another tool for understanding the genetics of diseases.
5. Information and energy
Having determined practical measures of information in biological systems, the question arises, how is this information related to the binding energy? Surprisingly, the answer comes from two apparently different directions [38].
The first approach is from the Second Law of Thermo-dynamics expressed as the Clausius inequality [10,66,3]:
| (6) |
Here S is the total entropy of a system and it has units of joules per kelvin since Q is heat and T is the absolute temperature. The Boltzmann–Gibbs entropy of a physical system is
| (7) |
in which kB is Boltzmann’s constant, pi is the probability of a microstate and Ω is the number of microstates. Within binding sites for a protein there are only four possible states, represented by the individual bases, and these therefore correspond to the ‘microstates’ of the system. So we can equate the estimated probabilities of Eq. (1) with the probabilities of Eq. (7). This allows us to find the relationship between the uncertainty and the entropy:
| (8) |
Applying this to the Second Law, one can show that when the temperature is constant (as it is before and after a molecule functions since it equilibrates with the surroundings in a few picoseconds)
| (9) |
where εmin is the minimum energy that must be dissipated out of a system for that system to gain one bit of information [38]. This is another form of the Second Law of Thermodynamics [22], and so it provides a hard bound on what a system can do.
The other approach comes from Shannon’s channel capacity equation. Shannon’s equation describes the maximum bits that can be sent through a communications channel given the bandwidth, the power dissipated at the receiver and the thermal noise at the receiver. An equivalent equation was developed for molecules [37], in which the ‘machine capacity’ is the maximum bits that a molecular machine can select amongst, given that it has a number of independently acting parts dspace (equivalent to bandwidth) and that it dissipates energy Py (equivalent to power for a single molecular binding or selection event) in the presence of thermal noise Ny:
| (10) |
This equation relates bits to energy so one can define the actual joules used (i.e. dissipated) per bit gained as:
| (11) |
Taking the limit of ε as Py goes to zero gives Eq. (9) again [38].
εmin is not only a version of the Second Law of Thermodynamics (at constant temperature) but can also be used as an ideal conversion factor between energy and information. So we are now in a position to compare the energy dissipated when a DNA binding protein binds to DNA to the information it gains in doing so. The best example to use for this is the protein EcoRI.
Found in strains of the bacterium Escherichia coli, EcoRI acts as a molecular defense system. When a virus attacks a bacterium, the virus injects its DNA into the bacterial cell and the DNA contains signals that cause it to take over the cell metabolism to generate new virus particles. However, if the cell has EcoRI, the EcoRI scans along the viral DNA until it finds the sequence GAATTC, which it then cuts between the G and first A. This happens on both strands of the DNA since GAATTC on one side of the DNA reads the same in the opposite direction on the complementary strand. With both strands cut, the viral DNA falls apart and the virus is stopped. But why don’t the bacteria destroy their own DNA? Because they have another enzyme that protects the GAATTC sequences by putting a methyl group on the second A [35]. Since the virus doesn’t have that protection, they are attacked.
A sequence logo for EcoRI looks just like GAATTC since there is essentially no variation to the pattern that EcoRI binds. So to define the first position, the G, requires a choice of 1 in 4 or 2 bits. There are 6 positions so the total information to define GAATTC is 6 × 2 = 12 bits.
How much energy is dissipated when EcoRI binds? This has been carefully measured [15] and using εmin we find that it is equivalent to 17.3 bits. That is, for the energy dissipation that occurs during binding, EcoRI could make Renergy = 17.3 bits of choices but from the DNA sequence we see that it only does Rsequence = 12 bits. It is inefficient, and we can measure its efficiency compared to the ideal as Rsequence/Renergy = 12/17.3 = 69% [49]. Many other genetic systems are around 70% efficient (in preparation). What causes this effect?
An answer to why DNA binding proteins are 70% efficient comes from the machine capacity equation. A sketch of the result is given here and details can be found in the original paper [49]. εmin from Eq. (9) is the best a molecule can do to make decisions by using energy since it gives the minimum joules dissipated to get one bit. In contrast, ε from Eq. (11) is what a molecule actually does. So we can define a theoretical efficiency as
| (12) |
Substituting into Eq. (12) Eqs. (9), (11) and (10), and then using Ny = dspacekBT [38] we find the elegant form
| (13) |
It can be shown that this is an upper bound on the molecular efficiency since it comes from the channel capacity, which is an upper bound. The efficiency is unitless and has the range from 0 to 1. Intriguingly, if Py = Ny, then ϵt = ln 2 = 0.69. So we can understand the occurrence of 70% efficiencies if we understand why Py would equal Ny.
What would make the energy dissipated from a molecule (Py) exactly equal the thermal noise flowing through the molecule at the same time (Ny)? This is almost a koan in its simplicity, but a distinct answer can be found by looking at the origins of the machine/channel capacity. (Readers who wish a challenge may stop at this point, read Refs. [37,38] but not [49] and see if they can find a solution. It took the author 6 years.)
6. Coding theory explains molecular efficiency
In Shannon’s model of communications, a series of D independent voltage pulses sent over a wire is represented by a point in a D dimensional space [56]. Although the pulses are initially distinct values, thermal noise distorts them by the time they reach the receiver. Essentially each pulse undergoes a drunkard’s walk, which means there will be a Gaussian variation around each signal pulse. The noise on the pulses is also independent and a combination of independent Gaussian distributions forms a sphere in the high dimensional space [56,37,46].
Starting from the center of a sphere in high dimensional space (the initial pure message) and moving away from that point because of thermal noise, there are so many ways to go that most of the received points will be on the surface of the sphere [8,9,37]. One may think of the sphere as similar to a well-defined ping-pong ball—the continuous thermal noise is almost certain to put the received point on the surface. So for a transmitted point (the sphere center) the received point will almost certainly be somewhere on the surface of a sphere whose radius is determined by the thermal noise.
Each message is a point, but what is received is a point on a sphere around the message point. The receiver’s job is to determine the original message from the received point, which means to find the nearest possible transmitted message point. This is possible as long as the spheres do not intersect. An arrangement of message points that avoids sphere intersections is called a coding, and finding the nearest sphere center is called ‘decoding’. The total space in which spheres can reside is determined by the power and it is also spherical but larger than the thermal noise spheres. Shannon noted that the maximum possible number of distinct messages M can be determined by dividing the volume of the large sphere by the volume of a smaller sphere. Then log2 M in time t is the number of bits per second, the channel capacity.
Shannon proved an important theorem about the capacity. First, one cannot transmit data at a rate greater than the channel capacity C; at most C bits per seconds will get through. Conversely, so long as one transmits at a rate R less than or equal to the channel capacity, one may have as few errors as desired. This result is possible by appropriate coding. After more than 60 years to develop codes near Shannon’s limit and the creation of computer chips to compute the algorithms for those codes we now have mobile cell phones that take advantage of this theorem. The result explains why our communications are so good.
An equivalent model can be built for molecules [37]. In this model the individual pulses are replaced by independently moving parts of the molecule. These parts are called ‘pins’ because they are analogous to the independently moving pins in a lock. Thermal noise impacts on the pins from all directions causing them to gain and lose energy either as potential energy or as a transient velocity. The series of random impacts means that the distribution of velocities for each pin is Gaussian. As with the communications model, the set of independent Gaussian distributions are represented by a sphere in a high dimensional space. A theorem equivalent to the channel capacity theorem says that so long as they do not exceed the machine capacity, living molecular machines may make as few errors as necessary for survival.
With several more concepts we can determine (if not fully understand) why molecular efficiencies are near 70%. The model for molecular machines is of spheres in a high dimensional space. A sphere represents the possible thermal vibrations of a molecule residing in one state. For example, the EcoRI protein bound to GAATTC would be a state. Another state would be EcoRI bound incorrectly to a similar sequence, say TAATTC. If these two states were not distinct for EcoRI, then EcoRI would be able to bind to TAATTC and cut there. This would be a disaster for the bacterium because the TAATTC sequences are not protected by methyl groups, the genome would be chopped up, and the cell would die. Obviously binding to GAATTC and to TAATTC should be different physically, so to go from one to the other requires changes in the positions of atoms. In the high dimensional coding space, this is a vector between two spheres.
When EcoRI has a lot of energy it slides along the DNA. At every position it can move closer to the DNA [67] and if it is at a GAATTC sequence, the molecular surfaces will match and EcoRI will bind, dissipating energy to stick there [68,27]. So before binding, the state of EcoRI is in a large ‘before’ sphere and after binding it is in a smaller ‘after’ sphere. If EcoRI were not to discriminate between sequences, it would end up in a sphere at the center of the large sphere. This sphere, called the ‘degenerate sphere’ represents binding to any sequence without discrimination. That would be fatal. So EcoRI must bind to a sphere representing GAATTC that is some distance away from the central degenerate sphere, the ‘forward’ sphere.
To get away from the central degenerate sphere to the forward GAATTC sphere requires dissipating energy. EcoRI must move outward by at least the radius of the degenerate sphere. As soon as the molecule has gone that far, it can decode into the GAATTC sphere. That is, the center of motion of the molecule can switch from one to the other state by a rearrangement of the atoms. Once the rearrangement has occurred, the molecule will ‘orbit’ around the new state. This continuous motion around a central state is the equivalent of ‘decoding’ in a communications system. (See the appendix of [49] for more details of this model.)
Getting from the degenerate sphere center to the radius requires expending energy Py that is at least equal to Ny (which determines the thermal noise radius) and so Py > Ny. Plugging this into Eq. (13) gives an efficiency of no more than 69%, as we observe. While this result makes mathematical sense, unfortunately it is not intuitive.
In any case, we can draw several important lessons from it. First, the result implies that the molecule EcoRI is functioning at machine/channel capacity because to get this result it must be operating on the efficiency curve. To some people this is a surprising result – how could a tiny molecule approach this ultimate limit? – but to a biologist it should not be surprising. If there is an optimal solution, so long as there are no barriers in the way, we expect an evolutionary process to find the solution.
All the codes people have devised for communications systems potentially have applications in biology—we ‘just’ have to look at the biological data to see where they might apply. The efficiency result is telling us the reverse too. All the sophisticated mechanisms of biology must be based on codes—we ‘just’ have to learn what those codes are so that we can apply them to our own technologies. This result is highly encouraging since it means that we too could build molecules that have error rates as low as we may desire.
Communications at the molecular level will be limited by the machine/channel capacity. One can build a hybrid model for molecular machines that receive parallel signals and decode them both in space and in time as described in the appendix of [37]. Likewise, we can envision building single molecules that use coding systems to reliably transmit molecular states to the outside world. An example of such a device is the patent pending Medusa™ sequencer [50]. This molecule consists of a DNA or RNA polymerase that reads along a DNA or RNA and produces a series of light pulses that represent the DNA sequence. It is possible to code the output spectrum with a Hamming-like code so that damage to the Medusa™ sequencer will be reported, in which case its signals can be ignored.
7. Information theory in biology
We have used molecular information theory to investigate many biological systems across the ‘central dogma’ and beyond:
DNA replication initiation by bacteriophage P1 RepA and other proteins [30,11,44,29]
RNA polymerases including those in T7 and related phages [54,53,13,14,59]
splice junctions [62] and mutations in splice junctions causing human disease [34,32,1,64], including the cancer-causing xeroderma pigmentosum [24,16,26, 25,21]
RNA folding [7]
protein structure [23]
These papers only represent ones from my lab; hundreds of others have been published by many other groups, but a fair review of them would be far beyond the scope of this paper. Why is information theory so applicable to biology? Shannon embedded into his mathematics a single idea that does not appear in physics or thermodynamics: that states must not intersect. As discussed above, in communications we demand that messages not be confused, so the spheres in high dimensional space must not touch. Because it is a high dimensional space, this is possible, and Shannon took advantage of it to create the channel capacity formula. There is nothing in physics or thermodynamics that demands this separation. Likewise, molecular states in biological systems must also be distinct—for survival, and Shannon’s theorem and Darwinian evolution guarantee that a solution can be found. This inevitably leads biological systems that make choices between two or more states to have 70% efficiency. Claude Shannon probably never realized that his work on the mathematics of communications was about biology [46].
Acknowledgments
I thank Don Court, Amar Klar, Ryan Shultzaberger, Rose Chiango and Carrie Paterson for reading and commenting on the manuscript. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Biography
Thomas D. Schneider is a Research Biologist in the Gene Regulation and Chromosome Biology Laboratory, National Cancer Institute, a part of the National Institutes of Health. Dr. Schneider received a B.S. in biology at MIT in 1978 and received his Ph.D. in 1984 from the University of Colorado, Department of Molecular, Cellular and Developmental Biology. His thesis was on applying Shannon’s information theory to DNA and RNA binding sites (Schneider1986). He is continuing this work at NIH as a tenured research biologist. A permanent web link is: http://alum.mit.edu/www/toms.

References
- 1.Allikmets R, Wasserman WW, Hutchinson A, Smallwood P, Nathans J, Rogan PK, Schneider TD, Dean M. Organization of the ABCR gene: analysis of promoter and splice junction sequences. Gene. 1998;215:111–122. doi: 10.1016/s0378-1119(98)00269-8. http://alum.mit.edu/www/toms/papers/abcr/ [DOI] [PubMed]
- 2.Arvidson DN, Youderian P, Schneider TD, Stormo GD. Automated kinetic assay of β-galactosidase activity. BioTechniques. 1991;11:733–738. [PubMed] [Google Scholar]
- 3.Atkins PW. The Second Law. W.H. Freeman and Co; NY: 1984. [Google Scholar]
- 4.Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrick D, Villanueba K, Childs J, Kalil R, Schneider TD, Lawrence CE, Gold L, Stormo GD. Quantitative analysis of ribosome binding sites in E. coli. Nucleic Acids Res. 1994;22:1287–1295. doi: 10.1093/nar/22.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benos PV, Bulyk ML, Stormo GD. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bindewald E, Schneider TD, Shapiro BA. CorreLogo: an online server for 3D sequence logos of RNA and DNA alignments. Nucleic Acids Res. 2006;34:w405–w411. doi: 10.1093/nar/gkl269. http://alum.mit.edu/www/toms/papers/correlogo/ [DOI] [PMC free article] [PubMed]
- 8.Brillouin L. Science and Information Theory. Academic Press, Inc; New York: 1962. [Google Scholar]
- 9.Callen HB. Thermodynamics and an Introduction to Thermostatistics. John Wiley & Sons, Ltd; NY: 1985. [Google Scholar]
- 10.Castellan GW. Physical Chemistry. Addison-Wesley Publishing Company; Reading, Mass: 1971. [Google Scholar]
- 11.Chattoraj DK, Schneider TD. Replication control of plasmid P1 and its host chromosome: the common ground. Prog Nucleic Acid Res Mol Biol. 1997;57:145–186. doi: 10.1016/s0079-6603(08)60280-9. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, Storz G, Schneider TD. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 2007;35:6762–6777. doi: 10.1093/nar/gkm631. http://alum.mit.edu/www/toms/papers/fur/ [DOI] [PMC free article] [PubMed]
- 13.Chen Z, Schneider TD. Information theory based T7-like promoter models: classification of bacteriophages and differential evolution of promoters and their polymerases. Nucleic Acids Res. 2005;33:6172–6187. doi: 10.1093/nar/gki915. http://alum.mit.edu/www/toms/papers/t7like/ [DOI] [PMC free article] [PubMed]
- 14.Chen Z, Schneider TD. Comparative analysis of tandem T7-like promoter containing regions in enterobacterial genomes reveals a novel group of genetic islands. Nucleic Acids Res. 2006;34:1133–1147. doi: 10.1093/nar/gkj511. http://alum.mit.edu/www/toms/papers/t7island/ [DOI] [PMC free article] [PubMed]
- 15.Clore GM, Gronenborn AM, Davies RW. Theoretical aspects of specific and non-specific equilibrium binding of proteins to DNA as studied by the nitrocellulose filter binding assay: co-operative and non-co-operative binding to a one-dimensional lattice. J Mol Biol. 1982;155:447–466. doi: 10.1016/0022-2836(82)90481-8. [DOI] [PubMed] [Google Scholar]
- 16.Emmert S, Schneider TD, Khan SG, Kraemer KH. The human XPG gene: gene architecture, alternative splicing and single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:1443–1452. doi: 10.1093/nar/29.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erill I, O’Neill MC. A reexamination of information theory-based methods for DNA-binding site identification. BMC Bioinformatics. 2009;10:57. doi: 10.1186/1471-2105-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemm MR, Paul BJ, Schneider TD, Storz G, Rudd KE. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol Microbiol. 2008;70:1487–1501. doi: 10.1111/j.1365-2958.2008.06495.x. http://alum.mit.edu/www/toms/papers/smallproteins/ [DOI] [PMC free article] [PubMed]
- 19.Hengen PN, Bartram SL, Stewart LE, Schneider TD. Information analysis of Fis binding sites. Nucleic Acids Res. 1997;25:4994–5002. doi: 10.1093/nar/25.24.4994. http://alum.mit.edu/www/toms/papers/fisinfo/ [DOI] [PMC free article] [PubMed]
- 20.Hengen PN, Lyakhov IG, Stewart LE, Schneider TD. Molecular flip-flops formed by overlapping Fis sites. Nucleic Acids Res. 2003;31:6663–6673. doi: 10.1093/nar/gkg877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inui H, Oh KS, Nadem C, Ueda T, Khan SG, Metin A, Gozukara E, Emmert S, Slor H, Busch DB, Baker CC, Digiovanna JJ, Tamura D, Seitz CS, Gratchev A, Wu WH, Chung KY, Chung HJ, Azizi E, Woodgate R, Schneider TD, Kraemer KH. Xeroderma Pigmentosum-variant patients from America, Europe, and Asia. J Investig Dermatol. 2008;128:2055–2068. doi: 10.1038/jid.2008.48. http://alum.mit.edu/www/toms/papers/xpv/ [DOI] [PMC free article] [PubMed]
- 22.Jaynes ET. The evolution of Carnot’s principle. In: Erickson GJ, Smith CR, editors. Maximum-Entropy and Bayesian Methods in Science and Engineering. Vol. 1. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1988. pp. 267–281. http://bayes.wustl.edu/etj/articles/ccarnot.ps.gz, http://bayes.wustl.edu/etj/articles/ccarnot.pdf. [Google Scholar]
- 23.Kannan N, Schneider TD, Vishveshwara S. Logos for amino acid preferences in different backbone packing density regions of protein structural classes. Acta Crystallogr Sect D. 2000;56:1156–1165. doi: 10.1107/s0907444900008969. [DOI] [PubMed] [Google Scholar]
- 24.Khan SG, Levy HL, Legerski R, Quackenbush E, Reardon JT, Emmert S, Sancar A, Li L, Schneider TD, Cleaver JE, Kraemer KH. Xeroderma Pigmentosum Group C splice mutation associated with mutism and hypoglycinemia—a new syndrome? J Investig Dermatol. 1998;111:791–796. doi: 10.1046/j.1523-1747.1998.00391.x. [DOI] [PubMed] [Google Scholar]
- 25.Khan SG, Metin A, Gozukara E, Inui H, Shahlavi T, Muniz-Medina V, Baker CC, Ueda T, Aiken JR, Schneider TD, Kraemer KH. Two essential splice lariat branchpoint sequences in one intron in a xeroderma pigmentosum DNA repair gene: mutations result in reduced XPC mRNA levels that correlate with cancer risk. Hum Mol Genet. 2004;13:343–352. doi: 10.1093/hmg/ddh026. http://hmg.oxfordjournals.org/cgi/content/full/13/3/343. [DOI] [PubMed]
- 26.Khan SG, Muniz-Medina V, Shahlavi T, Baker CC, Inui H, Ueda T, Emmert S, Schneider TD, Kraemer KH. The human XPC DNA repair gene: arrangement, splice site information content and influence of a single nucleotide polymorphism in a splice acceptor site on alternative splicing and function. Nucleic Acids Res. 2002;30:3624–3631. doi: 10.1093/nar/gkf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Grable JC, Love R, Greene PJ, Rosenberg JM. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990;249:1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- 28.Lyakhov I, Annangarachari K, Schneider TD. Discovery of novel tumor suppressor p53 response elements using information theory. Nucleic Acids Res. 2008;36:3828–3833. doi: 10.1093/nar/gkn189. http://alum.mit.edu/www/toms/papers/p53/ [DOI] [PMC free article] [PubMed]
- 29.Lyakhov IG, Hengen PN, Rubens D, Schneider TD. The P1 phage replication protein RepA contacts an otherwise inaccessible thymine N3 proton by DNA distortion or base flipping. Nucleic Acids Res. 2001;29:4892–4900. doi: 10.1093/nar/29.23.4892. http://alum.mit.edu/www/toms/papers/repan3/ [DOI] [PMC free article] [PubMed]
- 30.Papp PP, Chattoraj DK, Schneider TD. Information analysis of sequences that bind the replication initiator RepA. J Mol Biol. 1993;233:219–230. doi: 10.1006/jmbi.1993.1501. [DOI] [PubMed] [Google Scholar]
- 31.Pierce JR. An Introduction to Information Theory: Symbols, Signals and Noise. 2nd edition. Dover Publications, Inc; NY: 1980. [Google Scholar]
- 32.Rogan PK, Faux BM, Schneider TD. Information analysis of human splice site mutations. Hum Mutat. 1998;12:153–171. doi: 10.1002/(SICI)1098-1004(1998)12:3<153::AID-HUMU3>3.0.CO;2-I. http://alum.mit.edu/www/toms/papers/rfs/ [DOI] [PubMed]; Hum Mutat. 1999;13(1):82. erratum.
- 33.Rogan PK, Salvo JJ, Stephens RM, Schneider TD. Visual display of sequence conservation as an aid to taxonomic classification using PCR amplification. In: Pickover CA, editor. Visualizing Biological Information. World Scientific; Singapore: 1995. pp. 21–32. [Google Scholar]
- 34.Rogan PK, Schneider TD. Using information content and base frequencies to distinguish mutations from genetic polymorphisms in splice junction recognition sites. Hum Mutat. 1995;6:74–76. doi: 10.1002/humu.1380060114. http://alum.mit.edu/www/toms/papers/colonsplice/ [DOI] [PubMed]
- 35.Rubin RA, Modrich P. EcoRI methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1977;252:7265–7272. [PubMed] [Google Scholar]
- 36.Rudd KE, Schneider TD. Compilation of E. coli ribosome binding sites. In: Miller JH, editor. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1992. pp. 17.19–17.45. [Google Scholar]
- 37.Schneider TD. Theory of molecular machines. I. Channel capacity of molecular machines. J Theoret Biol. 1991;148:83–123. doi: 10.1016/s0022-5193(05)80466-7. http://alum.mit.edu/www/toms/papers/ccmm/ [DOI] [PubMed]
- 38.Schneider TD. Theory of molecular machines. II. Energy dissipation from molecular machines. J Theoret Biol. 1991;148:125–137. doi: 10.1016/s0022-5193(05)80467-9. http://alum.mit.edu/www/toms/papers/edmm/ [DOI] [PubMed]
- 39.Schneider TD. Sequence logos, machine/channel capacity, Maxwell’s demon, and molecular computers: a review of the theory of molecular machines. Nanotechnology. 1994;5:1–18. http://alum.mit.edu/www/toms/papers/nano2/
- 40.Schneider TD. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. http://alum.mit.edu/www/toms/papers/oxyr/ [DOI] [PubMed]
- 41.Schneider TD. Information content of individual genetic sequences. J Theoret Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. http://alum.mit.edu/www/toms/papers/ri/ [DOI] [PubMed]
- 42.Schneider TD. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res. 1997;25:4408–4415. doi: 10.1093/nar/25.21.4408. http://alum.mit.edu/www/toms/papers/walker/ [DOI] [PMC free article] [PubMed]; Nucleic Acids Res. 1998;26(4):1135. erratum. [Google Scholar]
- 43.Schneider TD. Evolution of biological information. Nucleic Acids Res. 2000;28:2794–2799. doi: 10.1093/nar/28.14.2794. http://alum.mit.edu/www/toms/papers/ev/ [DOI] [PMC free article] [PubMed]
- 44.Schneider TD. Strong minor groove base conservation in sequence logos implies DNA distortion or base flipping during replication and transcription initiation. Nucleic Acids Res. 2001;29:4881–4891. doi: 10.1093/nar/29.23.4881. http://alum.mit.edu/www/toms/papers/baseflip/ [DOI] [PMC free article] [PubMed]
- 45.Schneider TD. Some lessons for molecular biology from information theory. In: Karmeshu, editor. Entropy Measures, Maximum Entropy Principle and Emerging Applications. Vol. 119. Springer-Verlag; New York: 2003. pp. 229–237. Special Series on Studies in Fuzziness and Soft Computing. (Festschrift Volume in honour of Professor J.N. Kapour, Jawaharlal Nehru University, India) http://alum.mit.edu/www/toms/papers/lessons2003/ [Google Scholar]
- 46.Schneider TD. Claude Shannon: biologist. IEEE Eng Med Biol Mag. 2006;25:30–33. doi: 10.1109/memb.2006.1578661. http://alum.mit.edu/www/toms/papers/shannonbiologist/ [DOI] [PMC free article] [PubMed]
- 47.Schneider TD. Biol Theory; Twenty years of delila and molecular information theory: the Altenberg–Austin workshop in theoretical biology biological information, beyond metaphor: causality, explanation, and unification Altenberg; Austria. 11–14 July 2002; 2006. pp. 250–260. http://alum.mit.edu/www/toms/papers/schneider2006/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider TD. Information theory primer. 2010 Published on the web at: http://alum.mit.edu/www/toms/papers/primer/
- 49.Schneider TD. 70% efficiency of bistate molecular machines explained by information theory, high dimensional geometry and evolutionary convergence. Nucleic Acids Res. 2010;38:5995–6006. doi: 10.1093/nar/gkq389. http://alum.mit.edu/www/toms/papers/emmgeo/ [DOI] [PMC free article] [PubMed]
- 50.Schneider TD, Lyakhov I, Needle D. Probe for nucleic acid sequencing and methods of use. 2010 US patent claims allowed; European patent number 1960550 granted on 2010 September 15. US patent number 7,871,777 granted on 2011 January 18.
- 51.Schneider TD, Rogan PK. Computational analysis of nucleic acid information defines binding sites. 5867402. United States Patent. 1999 http://alum.mit.edu/www/toms/patent/walker/
- 52.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. http://alum.mit.edu/www/toms/papers/logopaper/ [DOI] [PMC free article] [PubMed]
- 53.Schneider TD, Stormo GD. Excess information at bacteriophage T7 genomic promoters detected by a random cloning technique. Nucleic Acids Res. 1989;17:659–674. doi: 10.1093/nar/17.2.659. [DOI] [PMC free article] [PubMed]
- 54.Schneider TD, Stormo GD, Gold L, Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. http://alum.mit.edu/www/toms/papers/schneider1986/ [DOI] [PubMed] [Google Scholar]
- 55.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423. 623–656. http://tinyurl.com/Shannon1948.
- 56.Shannon CE. Communication in the presence of noise. Proc IRE. 1949;37:10–21. [Google Scholar]
- 57.Shannon CE, Weaver W. The Mathematical Theory of Communication. University of Illinois Press; Urbana: 1949. [Google Scholar]
- 58.Shultzaberger RK, Bucheimer RE, Rudd KE, Schneider TD. Anatomy of Escherichia coli ribosome binding sites. J Mol Biol. 2001;313:215–228. doi: 10.1006/jmbi.2001.5040. http://alum.mit.edu/www/toms/papers/flexrbs/ [DOI] [PubMed]
- 59.Shultzaberger RK, Chen Z, Lewis KA, Schneider TD. Anatomy of Escherichia coli σ70 promoters. Nucleic Acids Res. 2007;35:771–788. doi: 10.1093/nar/gkl956. http://alum.mit.edu/www/toms/papers/flexprom/ [DOI] [PMC free article] [PubMed]
- 60.Shultzaberger RK, Roberts LR, Lyakhov IG, Sidorov IA, Stephen AG, Fisher RJ, Schneider TD. Correlation between binding rate constants and individual information of E. coli Fis binding sites. Nucleic Acids Res. 2007;35:5275–5283. doi: 10.1093/nar/gkm471. http://alum.mit.edu/www/toms/papers/fisbc/, http://dx.doi.org/10.1093/nar/gkm471. [DOI] [PMC free article] [PubMed]
- 61.Shultzaberger RK, Schneider TD. Using sequence logos and information analysis of Lrp DNA binding sites to investigate discrepancies between natural selection and SELEX. Nucleic Acids Res. 1999;27:882–887. doi: 10.1093/nar/27.3.882. http://alum.mit.edu/www/toms/papers/lrp/ [DOI] [PMC free article] [PubMed]
- 62.Stephens RM, Schneider TD. Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J Mol Biol. 1992;228:1124–1136. doi: 10.1016/0022-2836(92)90320-j. http://alum.mit.edu/www/toms/papers/splice/ [DOI] [PubMed]
- 63.Stormo GD, Schneider TD, Gold L. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 1986;14:6661–6679. doi: 10.1093/nar/14.16.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svojanovsky SR, Schneider TD, Rogan PK. Redundant designations of BRCA1 intron 11 splicing mutation; c.4216-2A > G; IVS11-2A > G; L78833, 37698, A > G. Hum Mutat. 2000;16:264. doi: 10.1002/1098-1004(200009)16:3<264::AID-HUMU9>3.0.CO;2-1. http://www3.interscience.wiley.com/cgi-bin/abstract/73001161/START. [DOI] [PubMed]
- 65.Tooley PW, Salvo JJ, Schneider TD, Rogan PK. Phylogenetic inference based on information theory-based PCR amplification. J Phytol. 1998;146:427–430. [Google Scholar]
- 66.Weast RC, Astle MJ, Beyer WH. CRC Handbook of Chemistry and Physics. CRC Press, Inc; Boca Raton, Florida: 1988. [Google Scholar]
- 67.Weber IT, Steitz TA. A model for the non-specific binding of catabolite gene activator protein to DNA. Nucleic Acids Res. 1984;12:8475–8487. doi: 10.1093/nar/12.22.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber IT, Steitz TA. Model of specific complex between catabolite gene activator protein and B-DNA suggested by electrostatic complementarity. Proc Natl Acad Sci USA. 1984;81:3973–3977. doi: 10.1073/pnas.81.13.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]