Abstract
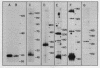
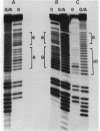
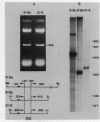
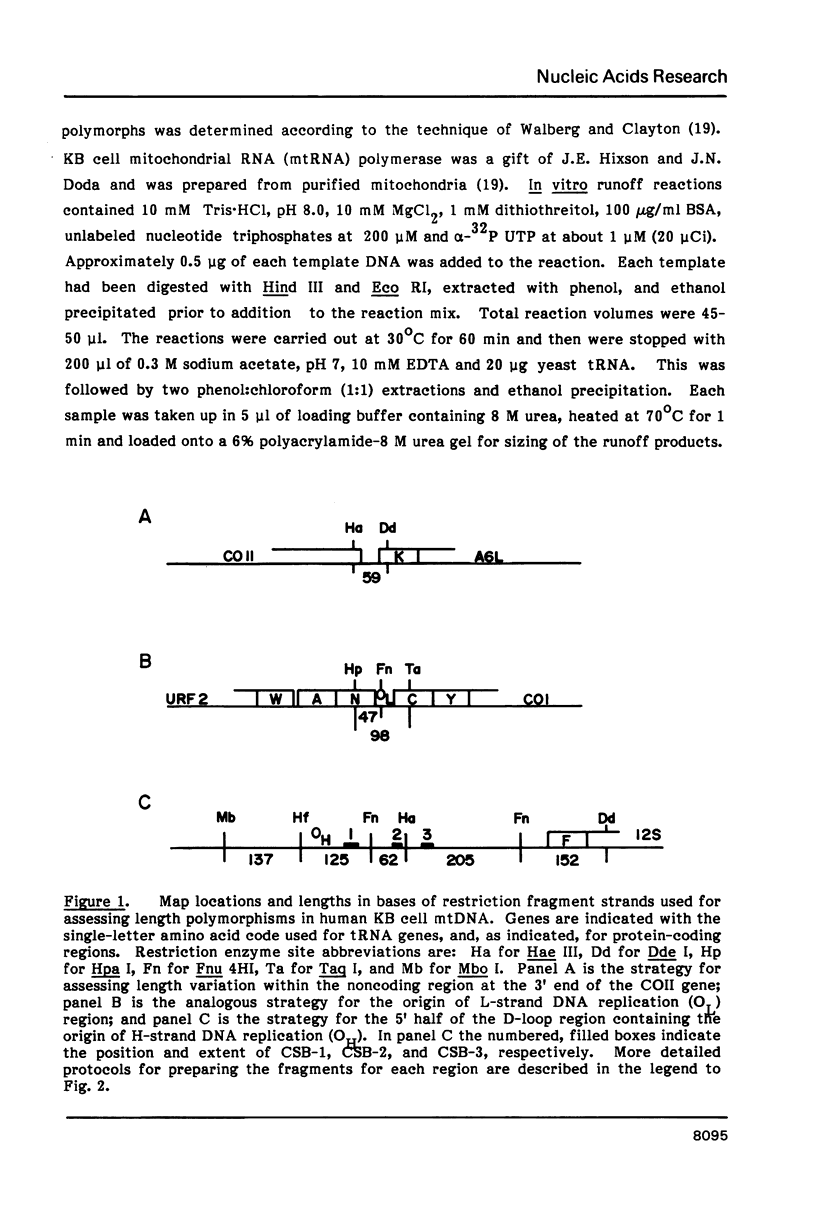
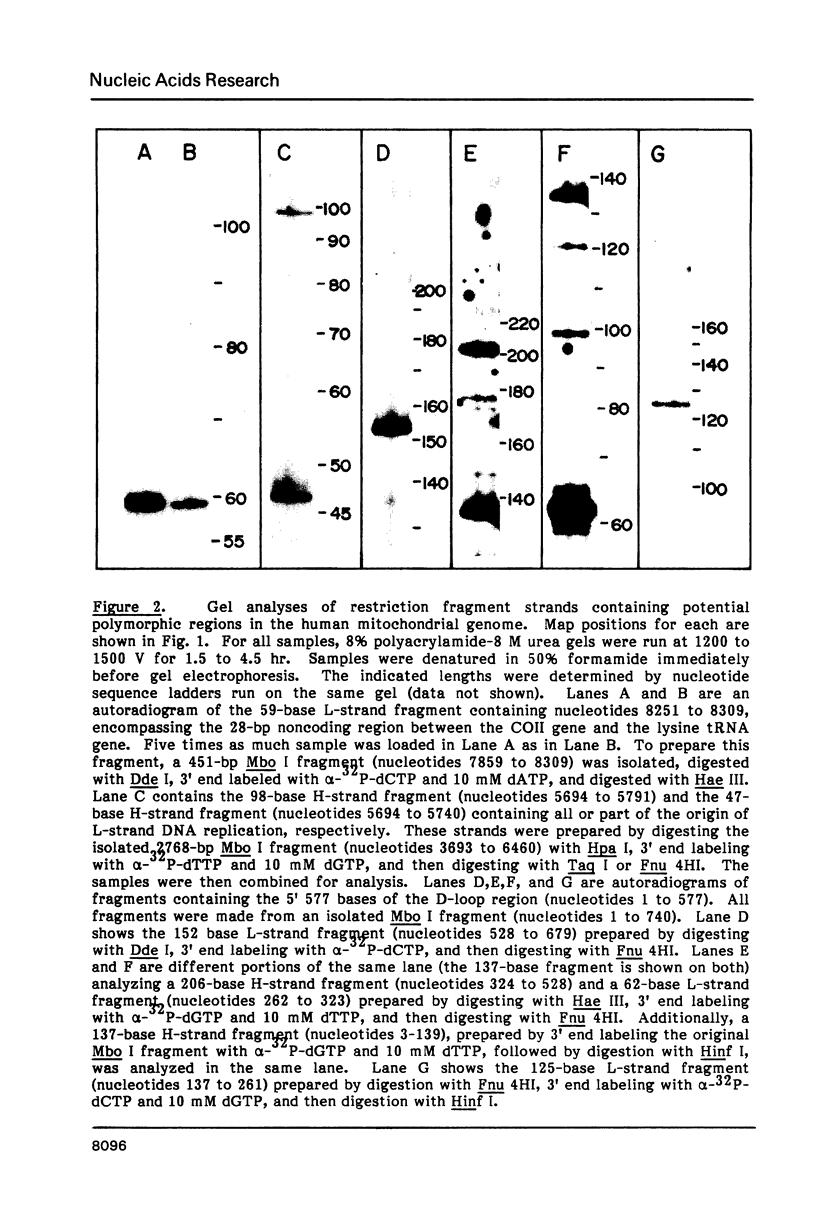
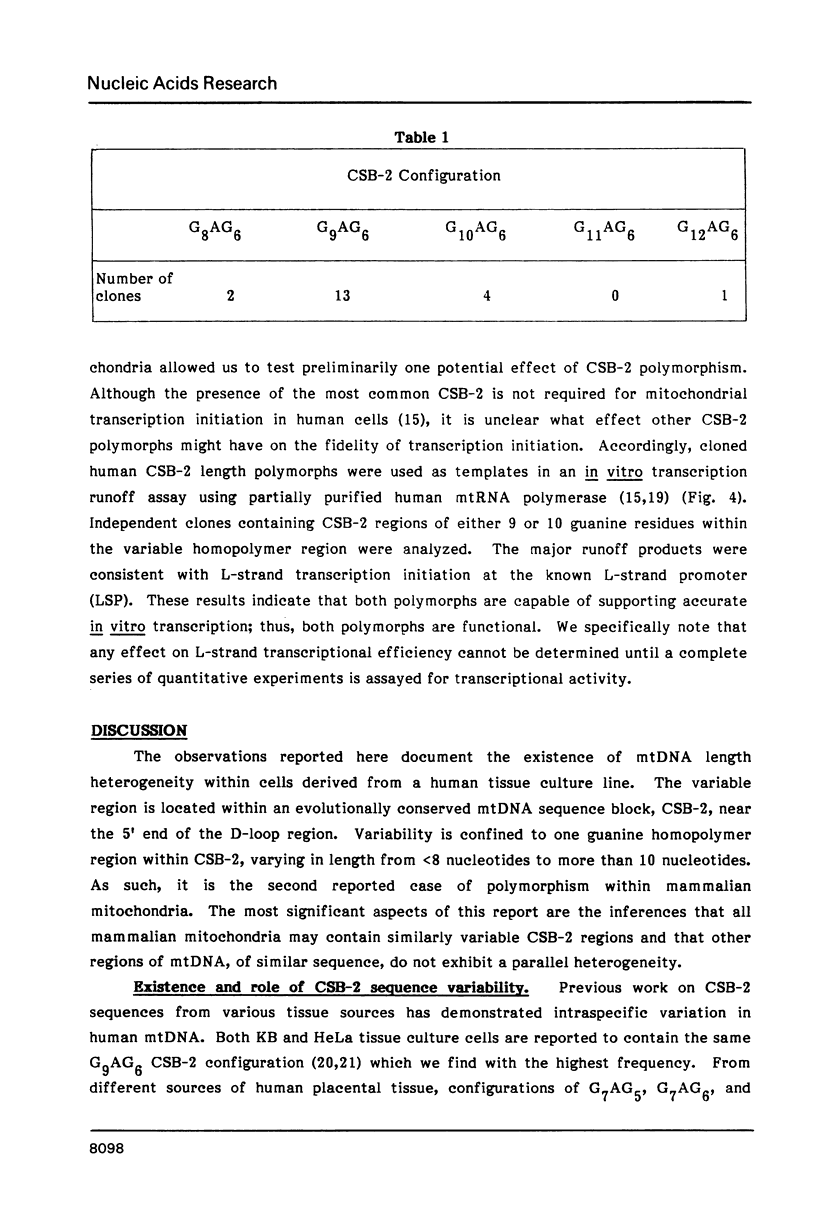
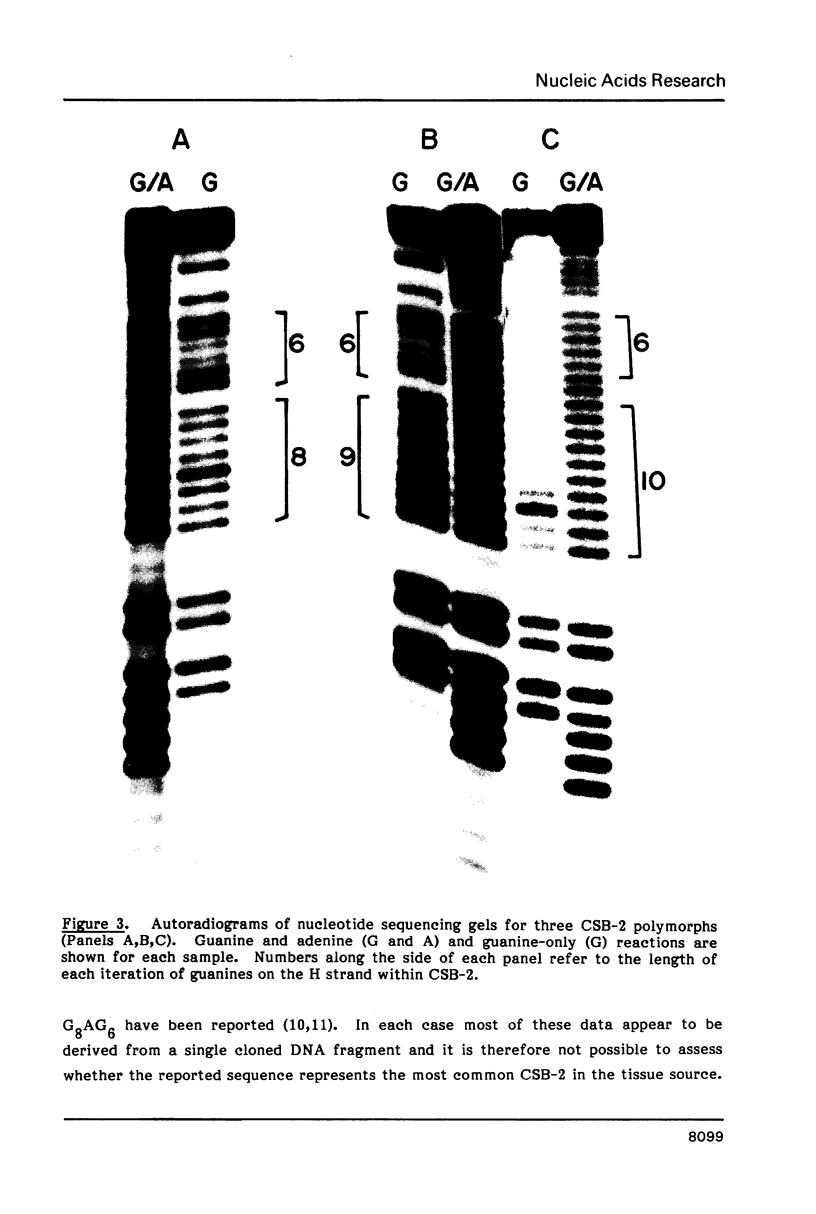
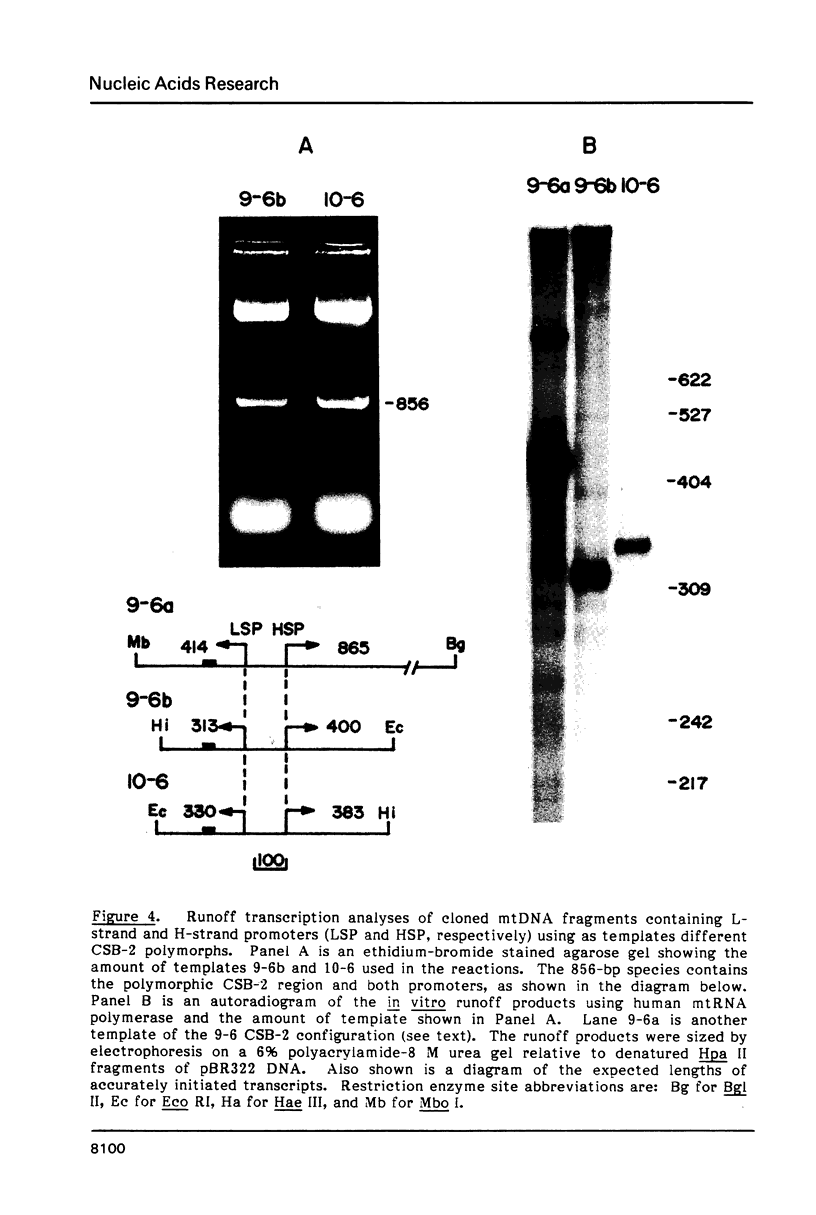
Mitochondrial DNA from human tissue culture cells contains heterogeneous sequences located within a previously identified, evolutionarily conserved region termed CSB-2. CSB-2 is located near the origin of heavy-strand mitochondrial DNA synthesis and the major transcriptional promoters for each strand of human mitochondrial DNA. Nucleotide sequence analysis of cloned mitochondrial DNA and electrophoretic analysis of appropriate small fragments from cellular mitochondrial DNA show that the variability is limited to a homopolymer sequence which can range in length from 6 to 12 residues. In vitro transcriptional analyses, using several of these cloned length polymorphs as templates and partially purified human mitochondrial RNA polymerase, demonstrate that the most common polymorphs will support accurate transcriptional initiation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Avise J. C., Lansman R. A., Shade R. O. The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. I. Population structure and evolution in the genus Peromyscus. Genetics. 1979 May;92(1):279–295. doi: 10.1093/genetics/92.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Applegate E. F., Yoza B. K. Identification of a promoter for transcription of the heavy strand of human mtDNA: in vitro transcription and deletion mutagenesis. Cell. 1984 Apr;36(4):1105–1113. doi: 10.1016/0092-8674(84)90061-8. [DOI] [PubMed] [Google Scholar]
- Brown G. G., DesRosiers L. J. Rat mitochondrial DNA polymorphism: sequence analysis of a hypervariable site for insertions/deletions. Nucleic Acids Res. 1983 Oct 11;11(19):6699–6708. doi: 10.1093/nar/11.19.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984 Mar;36(3):635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985 Jan;82(2):351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Hauswirth W. W., Clayton D. A. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985 Jun;4(6):1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. Replication of animal mitochondrial DNA. Cell. 1982 Apr;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Blackler A. W. Maternal and cytoplasmic inheritance of mitochondrial DNA in Xenopus. Dev Biol. 1972 Oct;29(2):152–161. doi: 10.1016/0012-1606(72)90052-8. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Giles R. E., Blanc H., Cann H. M., Wallace D. C. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg B. D., Newbold J. E., Sugino A. Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene. 1983 Jan-Feb;21(1-2):33–49. doi: 10.1016/0378-1119(83)90145-2. [DOI] [PubMed] [Google Scholar]
- Hauswirth W. W., Laipis P. J. Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4686–4690. doi: 10.1073/pnas.79.15.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauswirth W. W., Van de Walle M. J., Laipis P. J., Olivo P. D. Heterogeneous mitochondrial DNA D-loop sequences in bovine tissue. Cell. 1984 Jul;37(3):1001–1007. doi: 10.1016/0092-8674(84)90434-3. [DOI] [PubMed] [Google Scholar]
- Hayashi J., Yonekawa H., Gotoh O., Motohashi J., Tagashira Y. Two different molecular types of rat mitochondrial DNAs. Biochem Biophys Res Commun. 1978 Apr 14;81(3):871–877. doi: 10.1016/0006-291x(78)91432-8. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Newbold J. E., Potter S. S., Edgell M. H. Maternal inheritance of mammalian mitochondrial DNA. Nature. 1974 Oct 11;251(5475):536–538. doi: 10.1038/251536a0. [DOI] [PubMed] [Google Scholar]
- Jones C. W., Kafatos F. C. Accepted mutations in a gene family: evolutionary diversification of duplicated DNA. J Mol Evol. 1982;19(1):87–103. doi: 10.1007/BF02100227. [DOI] [PubMed] [Google Scholar]
- Kroon A. M., Pepe G., Bakker H., Holtrop M., Bollen J. E., Van Bruggen E. F., Cantatore P., Terpstra P., Saccone C. The restriction fragment map of rat-liver mitochondrial DNA: a reconsideration. Biochim Biophys Acta. 1977 Sep 20;478(2):128–145. doi: 10.1016/0005-2787(77)90177-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ojala D., Crews S., Montoya J., Gelfand R., Attardi G. A small polyadenylated RNA (7 S RNA), containing a putative ribosome attachment site, maps near the origin of human mitochondrial DNA replication. J Mol Biol. 1981 Aug 5;150(2):303–314. doi: 10.1016/0022-2836(81)90454-x. [DOI] [PubMed] [Google Scholar]
- Olivo P. D., Van de Walle M. J., Laipis P. J., Hauswirth W. W. Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial DNA D-loop. Nature. 1983 Nov 24;306(5941):400–402. doi: 10.1038/306400a0. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Taira M., Yoshida E., Kobayashi M., Yaginuma K., Koike K. Tumor-associated mutations of rat mitochondrial transfer RNA genes. Nucleic Acids Res. 1983 Mar 25;11(6):1635–1643. doi: 10.1093/nar/11.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. In vitro transcription of human mitochondrial DNA. Identification of specific light strand transcripts from the displacement loop region. J Biol Chem. 1983 Jan 25;258(2):1268–1275. [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. F., Ma D. P., Wilson R. K., Roe B. A. DNA sequence of the Xenopus laevis mitochondrial heavy and light strand replication origins and flanking tRNA genes. Nucleic Acids Res. 1983 Jul 25;11(14):4977–4995. doi: 10.1093/nar/11.14.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]