Abstract
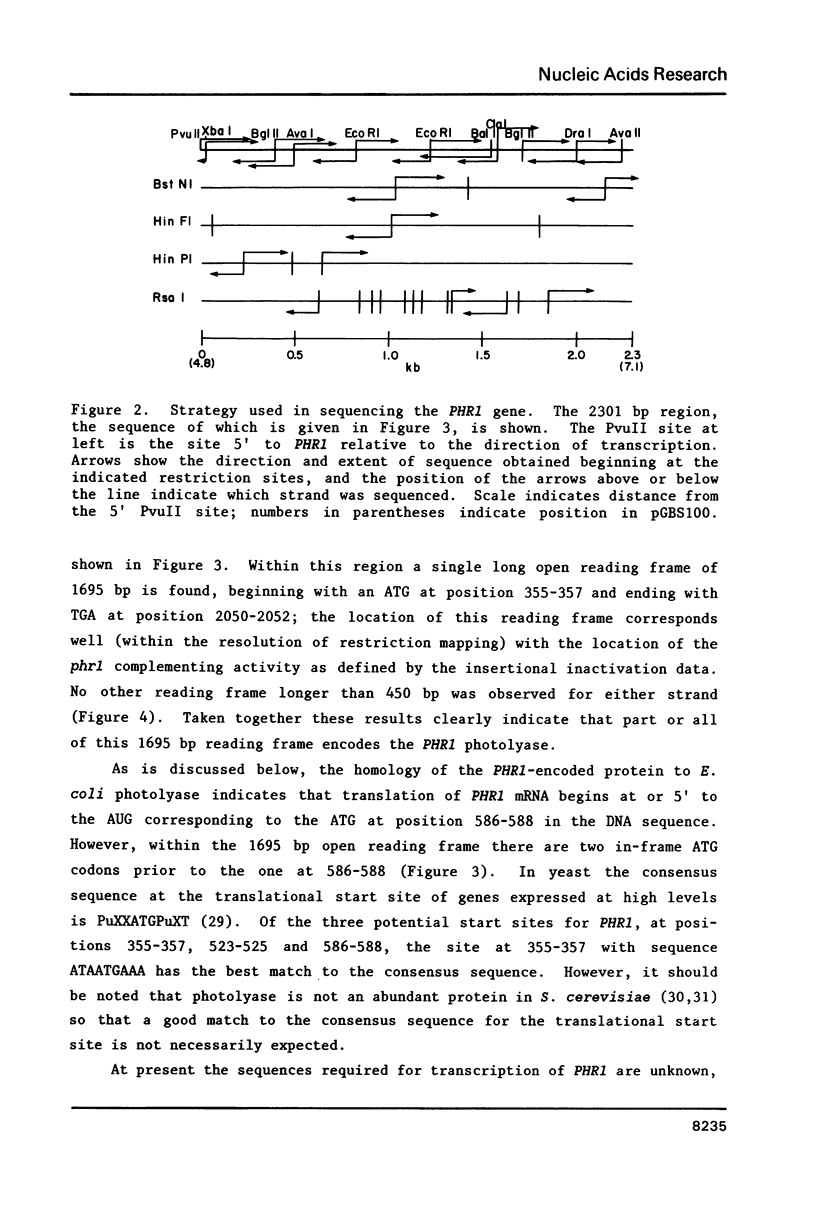
The nucleotide sequence of a 2301 base pair region of Saccharomyces cerevisiae DNA containing the PHR1 gene is reported. Within this region a single open reading frame of 1695 base pairs was found; using the insertional inactivation technique it was shown that part or all of this open reading frame specifies the PHR1-encoded photolyase. The amino acid sequence of the 565 amino acid long polypeptide predicted from the PHR1 nucleotide sequence was compared to the amino acid sequence of E. coli photolyase. Overall the sequence homology was 36.5%; however, two short regions near the amino terminus as well as the carboxy-terminal 150 amino acids display significantly greater sequence homology. The presence of these strongly conserved regions suggests that the yeast and E. coli photolyase possess common structural and functional domains involved in substrate and/or chromophore binding.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Boatwright D. T., Madden J. J., Denson J., Werbin H. Yeast DNA photolyase: molecular weight, subunit structure, and reconstruction of active enzyme from its subunits. Biochemistry. 1975 Dec 16;14(25):5418–5421. doi: 10.1021/bi00696a006. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F. Similar amino acid sequences: chance or common ancestry? Science. 1981 Oct 9;214(4517):149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- Eker A. P., Dekker R. H., Berends W. Photoreactivating enzyme from Streptomyces griseus-IV. On the nature of the chromophoric cofactor in Streptomyces griseus photoreactivating enzyme. Photochem Photobiol. 1981 Jan;33(1):65–72. doi: 10.1111/j.1751-1097.1981.tb04298.x. [DOI] [PubMed] [Google Scholar]
- Eker A. P., Fichtinger-Schepman A. M. Studies on a DNA photoreactivating enzyme from Streptomyces griseus. II. Purification of the enzyme. Biochim Biophys Acta. 1975 Jan 6;378(1):54–63. doi: 10.1016/0005-2787(75)90136-7. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Harm H., Rupert C. S. Analysis of photoenzymatic repair of UV lesions in DNA by single light flashes. I. In vitro studies with Haemophilus influenzae transforming DNA and yeast photoreactivating enzyme. Mutat Res. 1968 Nov-Dec;6(3):355–370. doi: 10.1016/0027-5107(68)90053-5. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Hughes C., Meynell G. G. Rapid screening for plasmid DNA. Mol Gen Genet. 1977 Mar 7;151(2):175–179. doi: 10.1007/BF00338692. [DOI] [PubMed] [Google Scholar]
- Iwatsuki N., Joe C. O., Werbin H. Evidence that deoxyribonucleic acid photolyase from baker's yeast is a flavoprotein. Biochemistry. 1980 Mar 18;19(6):1172–1176. doi: 10.1021/bi00547a021. [DOI] [PubMed] [Google Scholar]
- Jorns M. S., Sancar G. B., Sancar A. Identification of a neutral flavin radical and characterization of a second chromophore in Escherichia coli DNA photolyase. Biochemistry. 1984 Jun 5;23(12):2673–2679. doi: 10.1021/bi00307a021. [DOI] [PubMed] [Google Scholar]
- Jorns M. S., Sancar G. B., Sancar A. Identification of oligothymidylates as new simple substrates for Escherichia coli DNA photolyase and their use in a rapid spectrophotometric enzyme assay. Biochemistry. 1985 Apr 9;24(8):1856–1861. doi: 10.1021/bi00329a008. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Madden J. J., Werbin H. Use of membrane binding technique to study the kinetics of yeast deoxyribonucleic acid photolyase reactions. Formation of enzyme-substrate complexes in the dark and their photolysis. Biochemistry. 1974 May 7;13(10):2149–2154. doi: 10.1021/bi00707a024. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minato S., Werbin H. Spectral properties of the chromophoric material associated with the deoxyribonucleic acid photoreactivating enzyme isolated from baker's yeast. Biochemistry. 1971 Nov 23;10(24):4503–4508. doi: 10.1021/bi00800a025. [DOI] [PubMed] [Google Scholar]
- Müller F., Brüstlein M., Hemmerich P., Massey V., Walker W. H. Light-absorption studies on neutral flavin radicals. Eur J Biochem. 1972 Feb;25(3):573–580. doi: 10.1111/j.1432-1033.1972.tb01730.x. [DOI] [PubMed] [Google Scholar]
- Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol Gen Genet. 1981;184(3):471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- RUPERT C. S. Photoenzymatic repair of ultraviolet damage in DNA. II. Formation of an enzyme-substrate complex. J Gen Physiol. 1962 Mar;45:725–741. doi: 10.1085/jgp.45.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Weber S., Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci U S A. 1985 Jan;82(1):168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. Escherichia coli DNA photolyase is a flavoprotein. J Mol Biol. 1984 Jan 15;172(2):223–227. doi: 10.1016/s0022-2836(84)80040-6. [DOI] [PubMed] [Google Scholar]
- Sancar A., Smith F. W., Sancar G. B. Purification of Escherichia coli DNA photolyase. J Biol Chem. 1984 May 10;259(9):6028–6032. [PubMed] [Google Scholar]
- Sancar G. B. Expression of a Saccharomyces cerevisiae photolyase gene in Escherichia coli. J Bacteriol. 1985 Feb;161(2):769–771. doi: 10.1128/jb.161.2.769-771.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. B., Sancar A., Little J. W., Rupp W. D. The uvrB gene of Escherichia coli has both lexA-repressed and lexA-independent promoters. Cell. 1982 Mar;28(3):523–530. doi: 10.1016/0092-8674(82)90207-0. [DOI] [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Lorence M. C., Rupert C. S., Sancar A. Sequences of the Escherichia coli photolyase gene and protein. J Biol Chem. 1984 May 10;259(9):6033–6038. [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Sancar A. Binding of Escherichia coli DNA photolyase to UV-irradiated DNA. Biochemistry. 1985 Apr 9;24(8):1849–1855. doi: 10.1021/bi00329a007. [DOI] [PubMed] [Google Scholar]
- Sancar G. B., Smith F. W., Sancar A. Identification and amplification of the E. coli phr gene product. Nucleic Acids Res. 1983 Oct 11;11(19):6667–6678. doi: 10.1093/nar/11.19.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Johnston J., Chang C., Mortimer R. K. Cloning and mapping of Saccharomyces cerevisiae photoreactivation gene PHR1. Mol Cell Biol. 1984 Sep;4(9):1864–1870. doi: 10.1128/mcb.4.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow J. K., Bollum F. J. The minimum size of the substrate for yeast photoreactivating enzyme. Biochim Biophys Acta. 1968 Apr 22;157(2):233–237. doi: 10.1016/0005-2787(68)90077-4. [DOI] [PubMed] [Google Scholar]
- Setlow R. B., Carrier W. L., Bollum F. J. Pyrimidine dimers in UV-irradiated poly dI:dC. Proc Natl Acad Sci U S A. 1965 May;53(5):1111–1118. doi: 10.1073/pnas.53.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Drenth J., Schulz G. E. Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J Mol Biol. 1983 Jul 5;167(3):725–739. doi: 10.1016/s0022-2836(83)80106-5. [DOI] [PubMed] [Google Scholar]
- Wilcox D. R., Prakash L. Incision and postincision steps of pyrimidine dimer removal in excision-defective mutants of Saccharomyces cerevisiae. J Bacteriol. 1981 Nov;148(2):618–623. doi: 10.1128/jb.148.2.618-623.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E., Friedberg E. C. Molecular cloning and nucleotide sequence analysis of the Saccharomyces cerevisiae RAD1 gene. Mol Cell Biol. 1984 Oct;4(10):2161–2169. doi: 10.1128/mcb.4.10.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui A., Langeveld S. A. Homology between the photoreactivation genes of Saccharomyces cerevisiae and Escherichia coli. Gene. 1985;36(3):349–355. doi: 10.1016/0378-1119(85)90190-8. [DOI] [PubMed] [Google Scholar]
- Yasui A., Laskowski W. Determination of the number of photoreactivating enzyme molecules per haploid Saccharomyces cells. Int J Radiat Biol Relat Stud Phys Chem Med. 1975 Dec;28(6):511–518. doi: 10.1080/09553007514551371. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zwetsloot J. C., Vermeulen W., Hoeijmakers J. H., Yasui A., Eker A. P., Bootsma D. Microinjected photoreactivating enzymes from Anacystis and Saccharomyces monomerize dimers in chromatin of human cells. Mutat Res. 1985 Jul;146(1):71–77. doi: 10.1016/0167-8817(85)90057-4. [DOI] [PubMed] [Google Scholar]


