SUMMARY
To identify FDA-approved agents targeting leukemic cells, we performed a chemical screen on two human leukemic cell lines and identified the antimicrobial tigecycline. A genome-wide screen in yeast identified mitochondrial translation inhibition as the mechanism of tigecycline-mediated lethality. Tigecycline selectively killed leukemia stem and progenitor cells compared to their normal counterparts and also showed anti-leukemic activity in mouse models of human leukemia. ShRNA-mediated knockdown of EF-Tu mitochondrial translation factor in leukemic cells reproduced the anti-leukemia activity of tigecycline. These effects were derivative of mitochondrial biogenesis which, together with an increased basal oxygen consumption, proved to be enhanced in AML versus normal hematopoietic cells and were also important for their difference in tigecycline sensitivity.
INTRODUCTION
Acute Myeloid Leukemia (AML) comprises a genetically and clinically heterogeneous group of aggressive hematological neoplasms characterized by clonal proliferation of malignant precursors with a reduced capacity to differentiate into mature cellular components (Löwenberg et al., 1999). While there have been recent advances in the treatment of some hematological malignancies, the therapy of AML has remained essentially unchanged for over 20 years. For patients diagnosed when older than 60, the prognosis is particularly poor, with a 2-year survival probability of less than 10 percent (Löwenberg et al., 1998). Thus, further research is warranted into developing therapeutic strategies for the treatment of this disease.
The anti-cancer effects of inhibiting cytoplasmic translation have been previously reported, but the impact of inhibiting mitochondrial translation is less well understood. Mitochondrial DNA (mt-DNA) is composed of a double-stranded circular genome 16.6 kb in length without introns (Lang et al., 1999). It encodes two rRNAs, 22 t-RNAs and 13 of the 90 proteins in the mitochondrial respiratory chain. The 13 mt-DNA encoded proteins are translated by mitochondrial ribosomes within the mitochondrial matrix. (Gaur et al., 2008; Hunter and Spremulli, 2004; Zhang and Spremulli, 1998). Mitochondrial ribosomes differ from eukaryotic cytosolic ribosomes in their structure and chemical properties (O’Brien, 2003). In addition they use unique protein translation machinery including distinct initiation and elongation factors.
To identify therapeutic strategies that target both leukemia stem cells (LSCs) and bulk AML cells, we compiled a library of on and off-patent drugs, and screened these for their ability to reduce the viability of leukemia cell lines that display the stem cell properties of differentiation self-renewal, and marrow engraftment. Based on the results of this screen, we investigated the critical dependence of early as well as late stage primary human AML cells on mitochondrial protein translation.
RESULTS
Chemical screen for compounds targeting leukemic cells identifies the antimicrobial tigecycline
Because of their known toxicology and pharmacology, off- and even on-patent drugs can be rapidly repurposed for new indications. To search among such compounds for those with potential anti-human AML activity, we compiled a library of 312 such drugs focused mainly on anti-microbials and metabolic regulators with well-characterized pharmacokinetics and toxicology, and wide therapeutic windows. We then screened this library to identify agents that reduced the viability of cells from two human AML cell lines, TEX and M9-ENL1, that display features of leukemia stem cells (see Figure S1). These two lines were chosen for our first screen because of their stem cell properties including hierarchal differentiation and self-renewal(Barabé et al., 2007; Warner et al., 2005). Figure 1A shows dose-response curves for 5 compounds that did not have any previously recognized anti-cancer activity but displayed some anti-leukemic activity against at least one of these 2 cell lines after a 72 hour period of exposure. Interestingly, salinomycin was recently shown to have specific activity against breast cancer stem cells (Gupta et al., 2009). The second most active drug was tigecycline, which we then chose to analyze further.
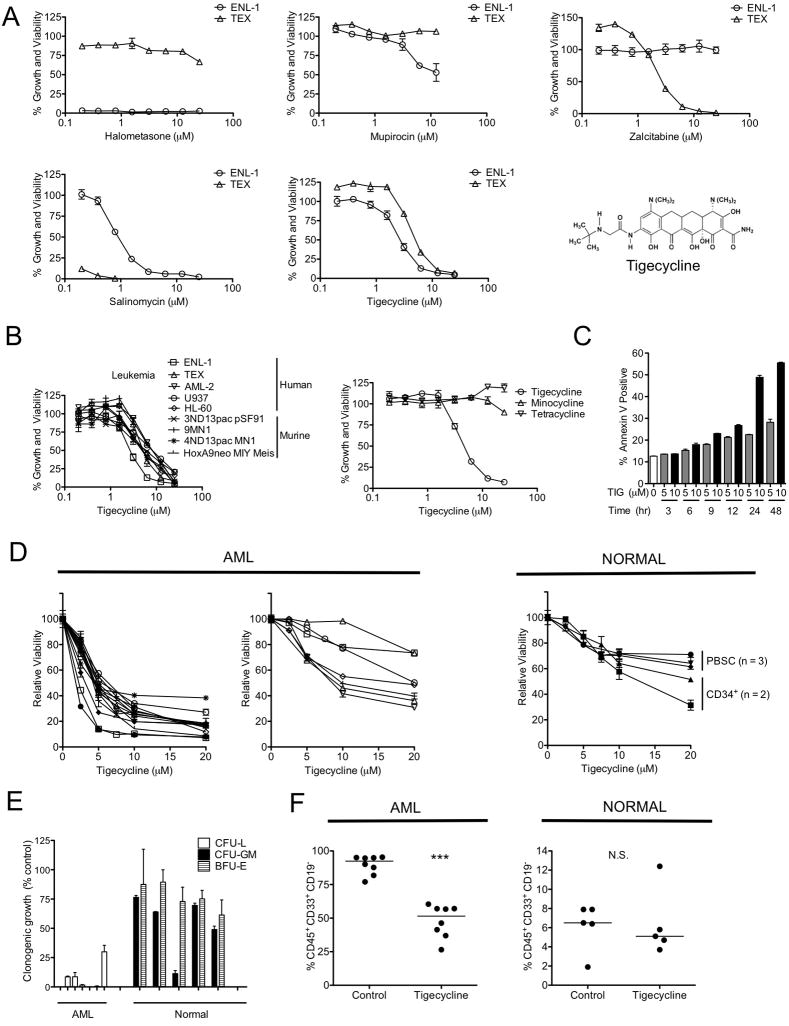
Figure 1. Chemical screen for compounds targeting leukemic cells identifies antimicrobial tigecycline.
(A) Drugs were added to TEX and ENL-1 cells (3 experiments each). Viability of cells after 72 hrs was determined by MTS staining and the results expressed as a percent of matching DMSO-treated controls (B) TEX, human and murine leukemia cells were incubated in triplicate experiments with drugs at concentrations shown for 72 hrs and viability was determined by MTS (human cells) or ViaCount (murine cells) staining and results expressed as a percent of results for untreated cells. (C) TEX cells were exposed to 5 or 10 μM of tigecycline (TIG) and Annexin V staining by flow cytometry was used to discriminate viable cells. Data represent the mean of Annexin V positive cells from a representative experiment (n=3). (D) Primary AML (1° AML) (n=20) and normal hematopoietic cells (n=5) were treated with increasing concentrations of tigecycline for 48 hrs. The proportion of viable cells was measured by Annexin-PI flow cytometry to calculate the yield of viable cells shown as a percent viable DMSO-treated cells in the same experiment. (E) Primary AML (n=7) and normal hematopoietic cells (n=5) were treated with 5 μM tigecycline and plated in clonogenic growth assays. Values shown are the percent of colonies obtained compared to DMSO-treated cells. (F) Cells from an AML patient and Lin− CD34+ enriched human cord blood cells were treated with 5 μM tigecycline or DMSO for 48 hrs in vitro and then injected into femurs of irradiated NOD/SCID mice preconditioned with anti-CD122. Six weeks later, the percent of human CD45+CD33+CD19− cells in femurs was measured by FACS. ***P < 0.0001, N.S. not significant P > 0.05 as determined by the unpaired student’s t test. Error bars represent mean +/− S.D. See also Table S1 and Figure S1.
To determine the effect of tigecycline on a broader spectrum of leukemia cell lines, a panel of human and murine leukemia cells was similarly treated with increasing concentrations of tigecycline. IC50s ranging from 3 to 8 μM were obtained for the various leukemia cell lines (Figure 1B). Tigecycline-induced cell death was confirmed by Annexin V/PI staining (Fig 1C). Of note, although tigecycline is a structural analogue of minocycline and tetracycline, TEX cells were not sensitive to either minocycline or tetracycline at concentrations up to 25 μM (Figure 1B).
Tigecycline kills primary AML bulk and progenitor cells more effectively than normal hematopoietic cells
We next compared the ability of tigecycline to kill cells from 20 primary AML samples (18 from newly diagnosed patients and 2 from patients with relapsed, treatment-refractory disease, see Table S1) and normal human hematopoietic cells within 48 hours of exposure in vitro. Bulk low-density normal human hematopoietic cells showed an LD50 of at least 10 μM, including the CD34+ cells isolated from 2 of these samples (Figure 1D). Cells from 7 of the 20 AML patients studied displayed a similar sensitivity to tigecycline (LD50 >10 μM) but, in the other 13 cases, a much greater sensitivity to tigecycline was observed (LD50 <5 μM). The CD34+38− subset of AML progenitor cells was also similarly sensitive to tigecycline (see Figure S1). Notably, no differences in cytogenetic risk or disease status were evident between the sensitive and insensitive groups and both of the samples from relapsed, treatment-refractory patients were sensitive to tigecycline.
To compare the effect of tigecycline on functionally defined subsets of primitive human AML and normal hematopoietic cell populations, additional experiments were performed. Incorporation of 5 μM tigecycline into the assay medium reduced the clonogenic growth of primary AML patient samples (n=7) by 93±4% (Figure 1E). In contrast, tigecycline had only a minimal effect on the clonogenic growth of normal hematopoietic cells assayed using the same protocol, including erythroid progenitors (BFU-E) (n=5). To assess the effects of tigecycline on AML and normal hematopoietic stem cells, we treated primary AML or normal Lin− CD34+-enriched normal human hematopoietic cord blood cells with 5 μM tigecycline or DMSO (as a control) for 48 hours in vitro and then compared the number of human cells produced after 6 weeks after transplantation into NOD/SCID mice (8 mice per group). This tigecycline treatment protocol reduced the repopulating ability of the primary AML cells tested (p<0.0001, student’s t test), but had no effect on the repopulating activity of normal hematopoietic cells (Figure 1F).
Thus for a majority of AML patients, including some with treatment refractory disease, tigecycline effectively targets all compartments of leukemic cells including the leukemia stem cells and does so at concentrations that appear pharmacologically achievable and that do not have a similar negative effect on normal hematopoietic cells.
Haplo-Insufficiency Profiling in S. Cerevisiae identifies mitochondrial translation as target of tigecycline in eukaryotic cells
Tigecycline is currently used clinically as a broad-spectrum antibiotic due to its high affinity and potent inhibition of the bacterial ribosome (Olson et al., 2006; Stein and Craig, 2006). To determine the mechanism of tigecycline’s activity in eukaryotic cells, we used Haplo-Insufficiency Profiling (HIP), a well-validated chemical genomics platform developed in the yeast S. Cerevisiae. The HIP assay allows an unbiased in vivo quantitative measure of the relative drug sensitivity of all ~6,000 yeast proteins in a single assay, and results in a list of candidate protein targets (Giaever et al., 1999; Hoon et al., 2008). Under standard fermentation conditions in rich media (YP), where the primary mode of metabolism is glycolysis, yeast growth was relatively insensitive to tigecycline. In contrast, yeast grown in respiratory conditions that depend on oxidative phosphorylation exhibited increased sensitivity and dose-dependent inhibition by tigecycline (Figure S2). Because growth inhibition is a necessary criterion for the HIP assay, all subsequent experiments were performed in respiratory media (YPGE).
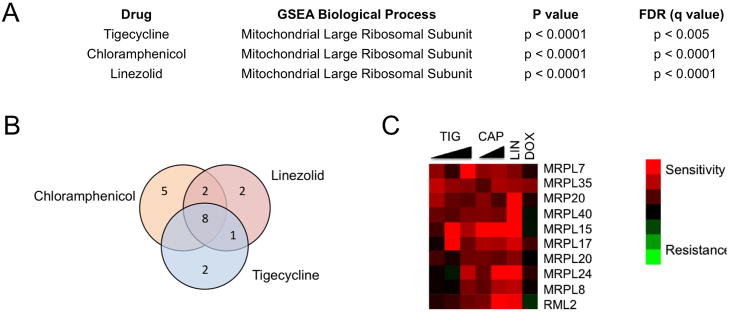
The rank-ordered gene list of drug sensitive strains generated from the HIP assays was analyzed using Gene Set Enrichment Analysis (GSEA) to identify Gene Ontology (GO) biological processes that were enriched in the tigecycline screens. The most significantly enriched GO process was the mitochondrial ribosome (p-value <0.0001, FDR q-value < 0.005) (Figure 2A). No other significant GO processes were enriched in the tigecycline screen. In the HIP assay, when the target is a large complex, no single gene in the complex stands out from the rest. For comparison, we also screened the known mammalian mitochondrial translation inhibitors chlorampheniciol (McKee et al., 2006) and linezolid (Nagiec et al., 2005), and the anthracycline family-member doxorubicin, which displays broad mechanisms of anti-cancer activity (Swift et al., 2006; Tewey et al., 1984; Wallin et al., 2010). As expected, chloramphenicol and linezolid yielded similar results to tigecylcine, while the doxorubucin GO enrichment analysis revealed a mechanism distinct from tigecycline (Figure 2B, C) (none of the doxorubicin GO terms passed our significance filter p-value <0.001, FDR q-value 0.1). Taken together, the yeast genomic screens suggest that tigecycline acts to inhibit growth and viability of eukaryotic cells through interference with mitochondrial protein translation. This finding is consistent with tigecycline’s known ability to inhibit bacterial protein synthesis by reversibly binding bacterial ribosomal RNA thereby preventing docking of the tRNA with its codon into the A site of the ribosome complex(Olson et al., 2006).
Figure 2. Haplo-Insufficiency Profiling in S. Cerevisiae identifies mitochondrial translation as target of tigecycline in eukaryotic cells.
(A) A pool of ~6000 S. Cerevisiae heterozygote mutant strains were cultured in the presence or absence of tigecycline, chloramphenicol (CAP), linezolid (LIN) or doxorubicin (DOX) in YPGE media and those showing altered growth responses relative to control cells identified. Gene Set Enrichment Analysis (GSEA) processes are shown. (B) Commonly enriched genes involved in mitochondrial translation identified from this GSEA analysis are shown in the Venn diagram (B) and the heat map (C). Red color denotes higher gene enrichment in the presence of drug relative to control cells. See also Figure S2.
Tigecycline inhibits mitochondrial translation in established and primary leukemia cells
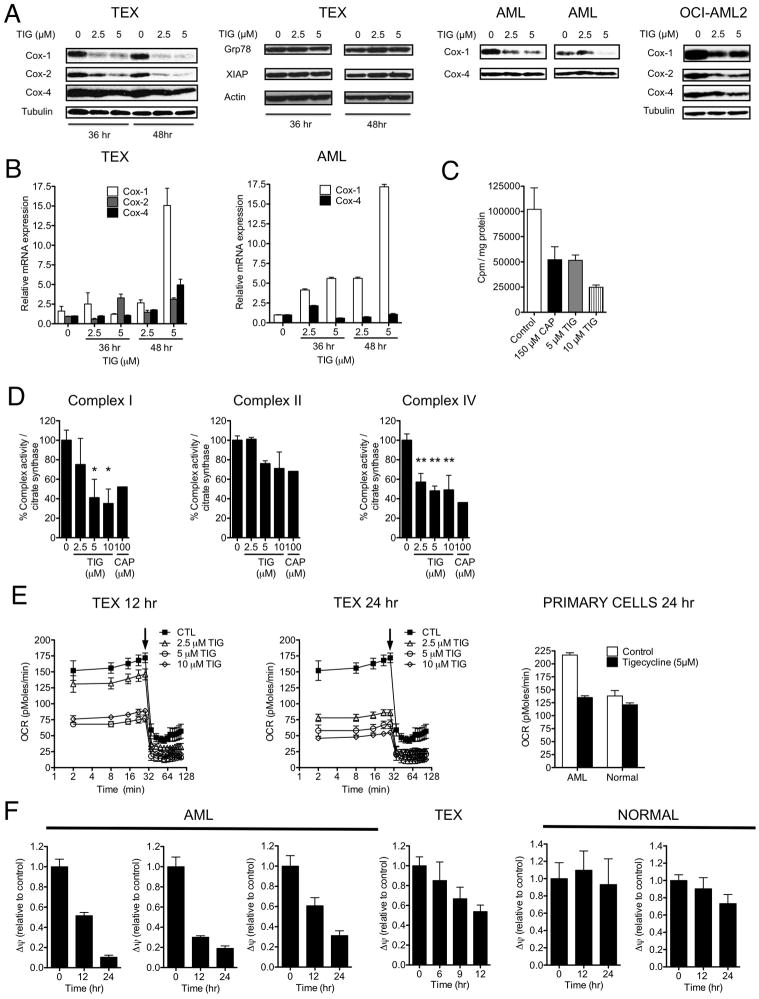
To determine whether the specific toxicity of tigecycline on leukemic cells is mediated by a similar mechanism, we next asked whether their exposure to tigecycline alters expression of proteins whose translation is known to be dependent on cytosolic and mitochondrial ribosomes. In a first set of experiments, TEX, OCI-AML2, primary AML cells were incubated for 48 hours in increasing concentrations of tigecycline and effects on Cytochrome C Oxidase-1, 2 and 4 (Cox-1, 2 and 4) levels measured at the end of that time. Cox-1 and Cox-2 are subunits of respiratory complex IV in the electron transport chain in mitochondria and are translated by mitochondrial ribosomes (Tam et al., 2008). Cox-4 is a component of the same respiratory complex, but is encoded by the nuclear genome and translated by nuclear ribosomes. Tigecycline treatment caused a preferential decrease of Cox-1 and Cox-2 as compared to Cox-4 (Figure 3A). Tigecycline also did not alter the expression of other proteins translated by cytosolic ribosomes including grp78 and the short half-life protein XIAP. The reductions in Cox-1 and Cox-2 protein levels were associated with increases in their mRNA expression with less change in Cox-4 mRNA levels in the same cells. (Figure 3B) This result is consistent with a previous report (Chrzanowska-Lightowlers et al., 1994) in which inhibition of mitochondrial translation was found to be accompanied by an increase in the expression of mitochondrially encoded mRNA. Finally, we demonstrated that tigecycline directly inhibits mitochondrial translation using cell-free assays with mitochondria isolated from leukemia cells (Figure 3C). Thus, taken together, our findings support a tigecycline-mediated inhibition of mitochondrial translation as its anti-leukemic cell mechanism of action.
Figure 3. Tigecycline inhibits mitochondrial protein translation.
(A) Effects of increasing concentrations of tigecycline on protein levels of Cox-1, Cox-2, Cox-4, Grp78, XIAP, Actin and Tubulin in TEX, OCI-AML2 and 2 AML patients’ cells treated for 48 hrs. (B) Effects of increasing concentrations of tigecycline on Cox-1, Cox-2, and Cox-4 mRNA expression in TEX and AML patient cells treated for 48 hrs. Transcript levels were determined by quantitative RT-PCR and normalized relative to 18S. Data is shown as mean fold change compared to untreated controls (n=3). (C) Mitochondrial isolates from OCI-AML2 cells were treated with buffer control, tigecycline or chloramphenicol for 5 min at 30°C, followed by the addition of [3H]-leucine. Incorporation of [3H]-leucine was measured after 60 min. (D) Effect of increasing concentrations of tigecycline and chloramphenicol on Complex I, II and IV enzyme activities relative to citrate synthase activity in TEX cells treated for 72 hrs. Values shown are average of 3 independent experiments. *P < 0.05, ** P < 0.005, as determined by Tukey’s test after One-way ANOVA analysis. (E) Oxygen consumption was measured in TEX, AML patient and normal hematopoietic cells treated for 12 or 24 hrs with tigecycline. The arrow denotes addition of 1.2 μM oligomycin. Values shown are average of 3 independent experiments. (F) Effect of increasing concentrations of tigecycline on mitochondrial membrane potential (Δψ) in TEX, AML patients’ and normal hematopoietic cells treated for 12 or 24 hrs and then stained with JC-1 dye for flow cytometry. Shown are the average Red/Green ratios derived for tigecycline-treated cells expressed as a percent of that in DMSO-treated control cells from the same experiments (n=3 per cell type). Error bars represent mean +/− S.D. See also Figure S3.
We next asked whether a similar exposure to tigecycline would affect the enzymatic activity of respiratory complexes I and IV, both of which contain proteins translated on mitochondrial ribosomes, and by comparison to the respiratory chain complex II, which does not contain mitochondrially-encoded subunits in its sub-structure (Ott and Herrmann, 2009). Tigecycline significantly decreased the enzyme activity of respiratory complexes I and IV, but had less effect on the enzymatic activity of the complex II (Figure 3D). These findings were mirrored by treatment with chloramphenicol, a known mitochondrial protein synthesis inhibitor. Consistent with its effects on the mitochondrial respiratory chain, tigecycline decreased oxygen consumption in TEX and primary AML but not normal hematopoietic cells at concentrations associated with, but at times preceding cell death (Figure 3E).
The mitochondrial respiratory chain generates an electrochemical proton gradient that establishes the mitochondrial membrane potential (Ramzan et al., 2010) used to drive ATP generation by complex V (ATP synthase). Therefore, we examined the effect of tigecycline treatment on the mitochondrial membrane potential as determined by staining with the carbocyanine dye JC-1 (Smiley et al., 1991). TEX cells and 3 different primary AML samples showed a decreased mitochondrial membrane potential after tigecycline treatment (5 μM), at times preceding the onset of cell death (Figure 3F). In contrast, loss of mitochondrial membrane potential was not seen in normal hematopoietic cells from after a similar in vitro incubation with tigecycline. The preferential effect of tigecycline on collapsing the membrane potential of leukemia cells may help explain the preferential cytotoxicity of tigecycline for AML cells over normal cells.
A byproduct of the mitochondrial electron transport chain is the generation of reactive oxygen species (ROS). Respiratory chain inhibitors such as rotenone and Na azide have been previously shown to induce rapid increases in ROS generation leading to cell death (Li et al., 2003; Park et al., 2007; Turrens and Boveris, 1980). Therefore, we explored the role of tigecycline on ROS generation in leukemia cells. Tigecycline did not increase ROS generation in TEX cells at time-points up to 24 hours (Figure S3). In contrast, various mitochondrial complex enzyme inhibitors produced rapid increases in ROS levels (Figure S3). Therefore, we postulate that the kinetics of tigecycline-induced inhibition of mitochondrial translation and respiratory complex activity produce functional effects distinct from agents that inhibit or uncouple the respiratory chain.
Inhibition of mitochondrial translation is functionally important for the selective toxicity of tigecycline on leukemia cells
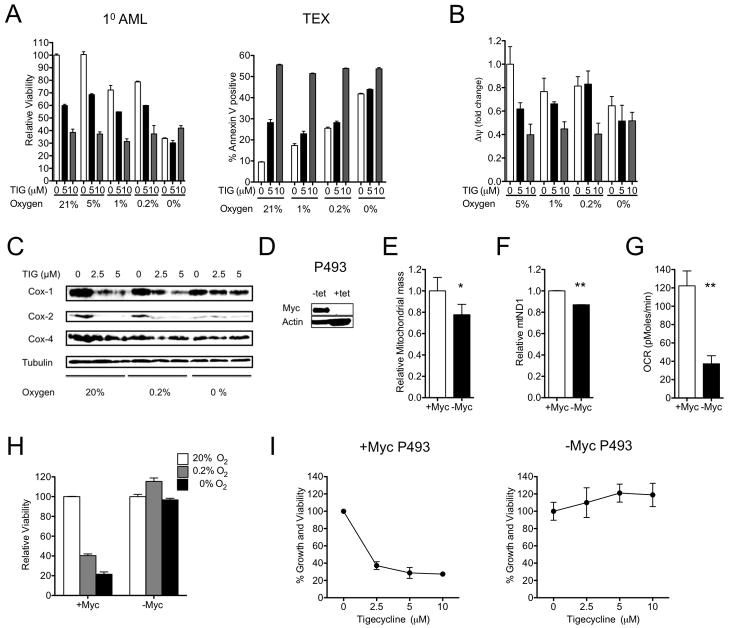
To determine whether tigecycline’s effects on the viability of leukemia cells are dependent on inhibition of mitochondrial function, we evaluated its anti-leukemic activity under different conditions of hypoxia (20% – 0.2% O2) and anoxia (0% O2) (Figure 4A). Incubation of both TEX and primary AML cells under anoxic conditions alone was toxic and in parallel reduced the mitochondrial membrane potential in these cells (Figure 4A, B). However, co-exposure to tigecycline under anoxic conditions caused no further cell loss and changes in mitochondrially-translated COX subunits I, II determined by immunoblotting 48 hours after tigecycline treatment, were no longer evident (Figure 4C). Nevertheless, these experiments demonstrated that tigecycline remained active against leukemia cells and primary patient AML samples under oxygen concentrations of 1–5% that are present in the bone marrow of patients with AML (Fiegl et al., 2009). Taken together, these results demonstrate that the ability of tigecycline to kill leukemia cells is dependent on oxygen availability and an intact mitochondrial respiratory chain.
Figure 4. Inhibition of mitochondrial translation is functionally important for tigecycline-induced death of leukemia cells.
(A) TEX and primary AML cells were treated under different oxygen concentrations for 48 hrs. Viability (Annexin-PI) and mitochondrial membrane potential (B) (Δψ, Red/Green ratio of JC-1) were assessed by flow cytometry. Results are shown relative to DMSO treated control. (C) Total proteins were extracted from TEX cells and analyzed by immunoblotting for Cox1, Cox2, Cox4, and Tubulin. (D) P493 lymphoma cells carrying a tetracycline-repressible human Myc construct were cultured in the presence and absence of 0.1 μg/ml (0.22 μM) of tetracycline for 96 hrs. Total proteins were extracted and analyzed by immunoblotting for Myc and actin. (E) Mitochondrial mass was measured by incubating cells with mitotracker Green FM dye, and subsequent flow cytometry. Median fluorescence intensity is shown relative to wild-type p493 cells. (F) DNA was extracted from cells and qPCR was used to measure levels of mitochondrial ND1 relative to human globulin (HGB). ND1/HGB ratio is shown relative to wild-type p493 cells. (G) Oxygen consumption was measured and is shown after 15 min incubation in cell chambers. *P < 0.05, **P < 0.005 as determined by unpaired student’s t test. (H) P493 cells with or without repressed Myc were plated in different oxygen concentrations (20%, 0.2%, and 0%) for 72 hrs. The proportion of viable cells was measured by Annexin-PI flow cytometry and calculated as the percent of viable cells compared to 20% oxygen control condition. (I) P493 cells with or without repressed MYC were washed and then treated with increasing concentrations of tigecycline for 48 hrs. After treatment, the number of viable cells was determined by trypan blue staining. Data represent the mean number of viable cells from 1 of 3 independent experiments. Error bars represent mean +/− S.D.
We next assessed tigecycline sensitivity of leukemic cells with genetically altered mitochondrial biogenesis. Previous studies have shown that Myc plays an important role in promoting mitochondrial biogenesis in a Burkitt’s lymphoma model (Li et al., 2005). Consistent with previous reports, inducible repression of Myc in p493 Burkitt’s cells resulted in decreased mitochondrial mass (p<0.05, t test) (Figure 4D, E), mitochondrial DNA copy number (p<0.005, t test) (Figure 4F) and oxygen consumption rate (p<0.005, t test) (Figure 4G). Furthermore, Myc-repressed p493 cells with decreased mitochondrial mass were sensitive to hypoxia conditions, highlighting their dependence on oxidative metabolism (Figure 4H). We used these cells to evaluate the effects of reduced mitochondrial biogenesis on the cytotoxicity of tigecycline. Tigecycline treatment reduced the growth and viability of control p493 cells with functional Myc. In contrast, p493 cells with decreased mitochondrial mass following Myc repression were resistant to tigecycline (Figure 4I). These results further support the notion that tigecycline’s anti-leukemic mechanism of action is dependent on inhibition of mitochondrial function. However, we recognize that Myc repression can affect processes beyond mitochondrial biogenesis including cell cycle and non-mitochondrial translation that may also impact sensitivity to tigecycline.
Genetic and chemical inhibition of mitochondrial translation displays anti-leukemia properties
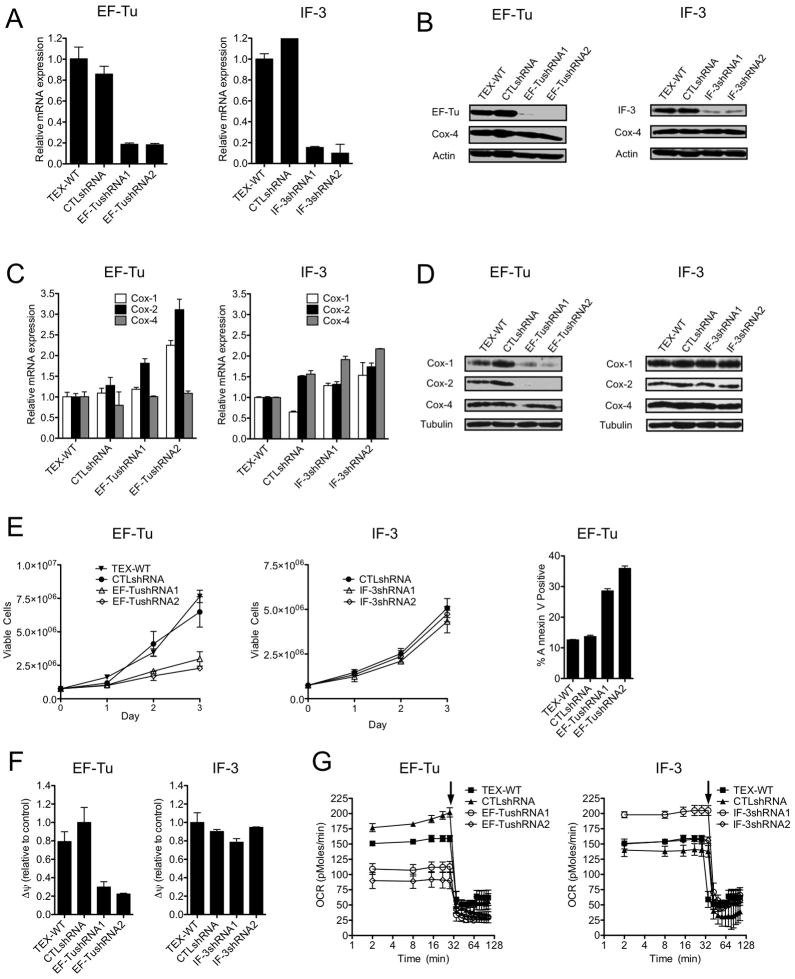
To further explore the anti-leukemic activity of mitochondrial translation inhibition, we asked whether genetic strategies would produce similar anti-leukemic effects as seen with tigecycline. Protein translation in mitochondria is regulated by a series of initiation and elongation factors specific to this organelle (Spremulli et al., 2004). Mitochondrial Initiation Factor 3 (IF-3) plays an active role in the initiation of mitochondrial translation (Christian and Spremulli, 2009). Mitochondrial elongation factor Tu (EF-Tu) is responsible for bringing aminoacyl-tRNAs in complex with GTP to the decoding site on the mitochondrial ribosome (Spremulli et al., 2004). We evaluated the effects of lentiviral vector-mediated shRNA knock-down of IF-3 or EF-Tu in TEX cells. Target knockdown was confirmed by QRT-PCR and immunoblotting using 2 independent shRNA for each gene (Figure 5A, B). Compared to control shRNA, knockdown of EF-Tu increased mRNA expression and decreased protein expression of Cox-1 and Cox-2 (Figure 5C, D), but did not change Cox-4 protein or mRNA levels. Similar to tigecycline, EF-Tu knockdown reduced the growth and viability of TEX cells (Figure 5E), and was associated with decreased mitochondrial membrane potential and oxygen consumption (Figures 5F, G), with no change in ROS production (Figure S4). In contrast to the effects of EF-Tu knockdown, IF-3 knockdown did not alter levels of Cox-1 and Cox-2 protein and mRNA (Figure 5C, D), did not reduce mitochondrial membrane potential or oxygen consumption (Figure 5F, G), and did not alter the cell growth and viability of TEX cells (Figure 5E). These results support inhibition of mitochondrial translation as a therapeutic strategy against human leukemic cells. These results also demonstrate that some but not all components of the mitochondrial protein translation machinery are necessary to maintain mitochondrial translation.
Figure 5. Alternative genetic and chemical strategies to inhibit mitochondrial translation have anti-leukemia effects.
(A) TEX cells were infected with EF-Tu or IF-3 targeting shRNAs or control sequences in lentiviral vectors. Six days post-transduction, EF-Tu and IF-3 mRNA expression relative to 18S (A) and protein expression determinations (B) were made by qRT-PCR and immunoblotting, respectively. (C) Effects on mRNA expression of Cox-1, Cox-2, Cox-4 and Tubulin were determined by q-RT-PCR using 18S RNA as an internal standard (1 of 3 representative experiments shown). (D) Effects on expression of Cox-1, Cox-2, Cox-4 and Tubulin protein were determined by immunoblotting (a representative experiment is shown). (E) Viable cells were measured by trypan blue staining and cell death by Annexin-V staining. Data from 1 of 3 independent experiments are shown. Additional cells treated in the same way were used to measure effects on other parameters. (F) Effects on mitochondrial membrane potential (Δψ) were determined by staining cells with the JC-1 dye and then determining Red/Green ratios by flow cytometric analysis. (G) Effects on oxygen consumption were determined as described in the supplemental experimental procedures. Arrow denotes addition of 1.2 μM oligomycin. Results for 1 of 2 experiments with similar outcomes are shown. Error bars represent mean +/− S.D. See also Figure S4.
To further assess the possibility that other strategies of inhibiting mitochondrial translation might also have anti-leukemic potential, we treated TEX leukemia cells with increasing concentrations of chloramphenicol and linezolid, compounds known to inhibit mitochondrial translation in mammalian cells at high concentrations in vitro (McKee et al., 2006; Nagiec et al., 2005). A 4-day incubation with either agent decreased the viability and proliferation of TEX cells, but at higher drug concentrations than required for tigecycline-induced killing (Figure S4). Both drugs also reduced expression of mitochondrially-translated Cox-1 and Cox-2 subunits but did not affect levels of Cox-4. Furthermore, chloramphenicol (50 μM) reduced the clonogenic growth of cells from 2 primary AML patient samples to a greater degree than that of normal hematopoietic progenitors. These results suggest that tigecycline is a more potent inhibitor of mammalian mitochondrial ribosomes compared to chloramphenicol or linezolid, consistent with its more potent inhibition of bacterial protein synthesis (Contreras and Vázquez, 1977; Olson et al., 2006; Shinabarger et al., 1997), and provide an explanation for tigecycline’s selective anti-leukemic effects at pharmacologically achievable concentrations. Overall, our findings in these chemical and genetic experiments validate inhibition of mitochondrial translation as a plausible therapeutic strategy for AML.
AML cells have increased mitochondrial biogenesis and basal oxygen consumption compared to normal hematopoietic cells
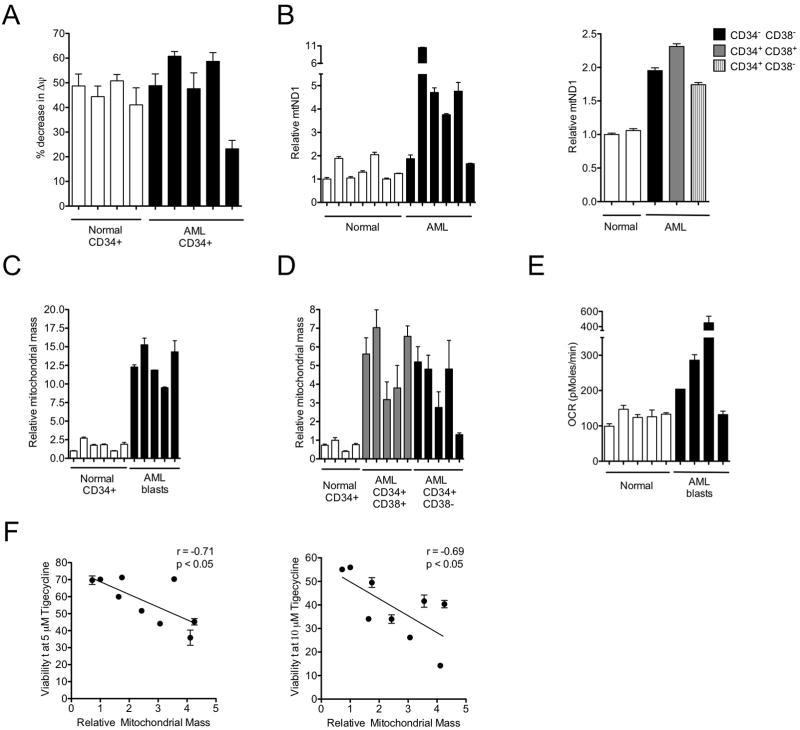
To investigate the basis of leukemic cell hypersensitivity to mitochondrial translation inhibition, we assessed baseline mitochondrial characteristics of primary normal hematopoietic and AML cells. There was no difference in resting mitochondrial membrane potential between leukemic and normal hematopoietic progenitor cells that could account for their differential sensitivity to tigecycline (Figure 6A). We then evaluated mitochondrial DNA copy number, which has previously been used as an estimate of mitochondrial mass (Xing et al., 2008) and the energy demand of a cell (Capps et al., 2003). Cells from 7 AML patients, including both CD34+/CD38+ and CD34+/CD38− subsets had higher mitochondrial DNA copy number compared to normal hematopoietic cells (Figure 6B). Determination of mitochondrial mass using Mitotracker green FM, which stains mitochondria regardless of resting mitochondrial membrane potential (Pendergrass et al., 2004), again showed higher values for the AML cells (n=5) than for normal CD34+ hematopoietic cells (n=6) (Figure 6C), including both the CD34+/CD38+ and CD34+/CD38− subsets of leukemic cells (Figure 6D). Consistent with these findings, rates of oxygen consumption were higher in primary AML cells (n=4) compared to normal hematopoietic cells (n=5) (Figure 6E).
Figure 6. Mitochondrial characteristics of AML and normal hematopoietic cells.
(A) Baseline mitochondrial membrane potential values for AML and normal CD34+ cells before and after uncoupling the potential with CCCP, as determined by staining the cells with DilC1 (5). (B) Left panel shows mitochondrial DNA copy number determined in 7 AML patients and 6 normal hematopoietic samples. Right panel shows mitochondrial DNA copy number determined in a primary AML sample after CD34+ and CD38+ fluorescence-activated cell sorting and 2 normal hematopoietic samples. DNA was extracted from cells and qPCR was performed for mitochondrial ND1 relative to human globulin (HGB). The ND1/HGB ratio is shown relative to cells from one normal sample. (C) Mitochondrial mass values for AML blasts (C), CD34+/CD38+ cells and CD34+/CD38− cells (D) and normal CD34+ cells were determined by flow cytometric analysis of cells stained with Mitotracker Green FM. Median fluorescence intensity (MFI) values are shown by comparison to the MFI measured for one of the normal samples. (E) Comparison of resting oxygen consumption rates of primary AML cells (n=4) and normal hematopoietic cells (n=5). (F) Correlation analysis of mitochondrial mass (Mitotracker Green FM staining) and in vitro toxicity to tigecycline (Annexin-V/PI staining) of primary AML cells (n=11) based on results obtained at doses of 5 and 10 μM. *P < 0.05, as determined by Pearson correlation coefficient. Error bars represent mean +/− S.D.
To investigate whether baseline mitochondrial mass differences in AML patient samples are related to their in vitro hypersensitivity to tigecycline, baseline mitochondrial mass measurements were performed on the leukemic cells from 9 AML patients and compared to their individual sensitivities to 5 and 10 μM tigecycline (Figure 6F). Mitochondrial mass was significantly negatively correlated with in vitro sensitivity to tigecycline after 48 hours (5 μM dose, r = −0.71, p <0.05, 10 μM dose, r = −0.69, p <0.05 pearson correlation). Thus, samples with the greatest mitochondrial mass were most sensitive to tigecycline treatment in vitro. Taken together, these results suggest that AML progenitors and stem cells are more metabolically active and dependent on mitochondrial function than are normal hematopoietic cells, and provide a mechanism to explain the observed differential activity of mitochondrial translation inhibition in leukemic and normal hematopoietic cells at all levels of differentiation (Figure 1D).
Tigecycline shows anti-AML activity in xenograft models of human leukemia
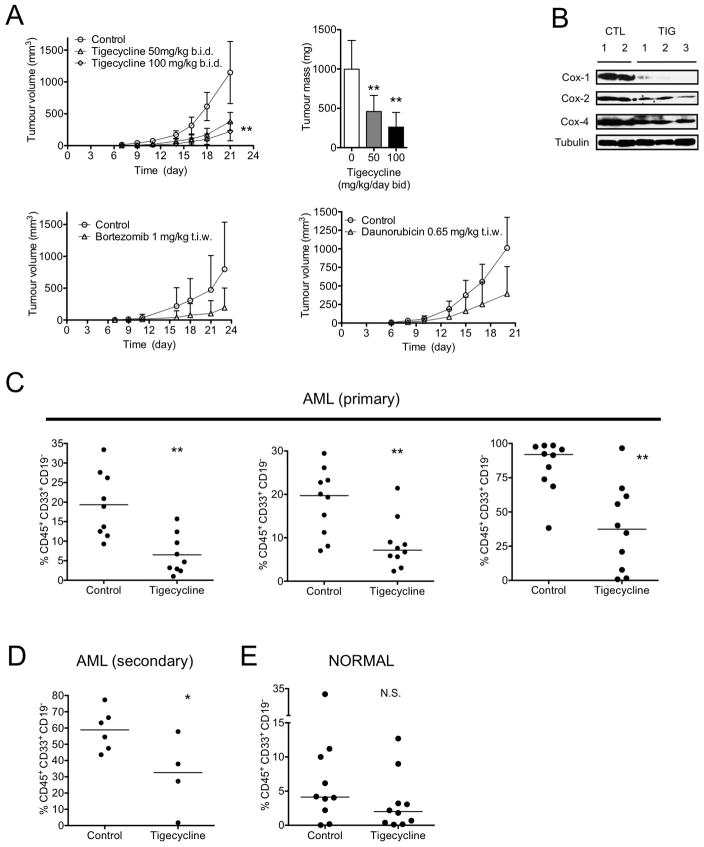
To assess the in vivo anti-leukemia efficacy of mitochondrial translation inhibition using xenograft models, we first evaluated the pharmacokinetics of tigecycline in mice (Figure S5). Based on these studies, we chose a treatment schedule of twice daily intraperitoneal (i.p.) injections. In a first experimental design, OCI-AML2 cells were transplanted subcutaneously into severe combined immune deficiency (SCID) mice and treatment was started 7 days later when tumors were already palpable (8 mice per group). Compared to the vehicle control, tigecycline significantly delayed tumor growth and showed equivalent or greater potency than daunorubicin or bortezomib at their maximally tolerated doses (Figure 7A) (p<0.005, Tukey’s t test after one-way ANOVA). Treatment with tigecycline did not alter the appearance or behavior of the mice. Moreover, at the conclusion of the experiment 3 weeks post transplant, there were no gross changes to the organs at necropsy. Tumors excised from mice treated for 5 days with tigecycline (50 mg/kg b.i.d.) showed a greater reduction of expression of mitochondrially-translated Cox-1 and Cox-2 relative to nuclear-translated Cox-4, compared to vehicle-treated controls (Figure 7B). Thus, tigecycline inhibits growth and mitochondrial function of this AML cell line in vivo.
Figure 7. Tigecycline has in vivo activity in models of human leukemia in mice.
(A) Human leukemia (OCI-AML2) cells were injected subcutaneously into the flank of SCID mice. Seven days later, when tumors were palpable, mice were treated with tigecycline (50 mg/kg or 100 mg/kg twice daily by i.p. injection), bortezomib (1 mg/kg t.i.w.), daunorubicin (0.65 mg/kg t.i.w.) or vehicle control (n = 10 per group). Three weeks after injection of cells, mice were sacrificed, tumors excised and the volume and mass of the tumors were measured. The tumor mass and the mean volume are shown. **P < 0.005, as determined by Tukey’s test after one-way ANOVA analysis. (B) Tumors from two control mice, and three tigecycline-treated mice were excised after 5 days of treatment and total proteins were extracted and analyzed by immunoblotting for Cox-1, Cox-2, Cox-4, and tubulin. (C) Primary cells from 3 AML patients and Lin− CD34+ enriched human cord blood cells (E) (Normal) were injected intra-femorally into irradiated female NOD/SCID mice. Three weeks after injection, the mice were treated with tigecycline (100 mg/kg by i.p. injection daily) or vehicle control (n = 10 per group) for three weeks. Following treatment, human leukemia cell engraftment in the femur was measured by flow cytometric analysis of human CD45+CD19−CD33+ cells. **P < 0.005 as determined by student’s t test. (D) Cells from mice transplanted with one AML patient experiment were used to assess secondary engraftment in a second generation of NOD/SCID mice. Equal numbers of viable leukemia cells from the bone marrow of control and tigecycline-treated mice were pooled and aliquots injected into irradiated NOD/SCID mice, which were not treated with tigecycline. Six weeks later, human leukemia cell engraftment in the femur was measured by flow cytometric analysis for human CD45+CD19−CD33+ cells. Line represents median of engrafted human cells. *P < 0.05, N.S. not significant P > 0.05 as determined by student’s t test. Error bars represent mean +/− S.D. See also Figure S5.
Xenotransplantation in NOD/SCID mice is a robust model system to assay stem cells and test the efficacy of anti-leukemia therapies in vivo(Bonnet and Dick, 1997; Jin et al., 2006; Lapidot et al., 1994). We transplanted pre-conditioned NOD/SCID mice intra-femorally with primary AML cells from 3 patients and Lin− CD34+ enriched human cord blood cells, and then evaluated the effects of a 3-week course of tigecycline started 3 weeks post-transplant (10 mice per group). Tigecycline-treated mice had significantly lower levels of leukemic engraftment compared to control-treated mice without evidence of toxicity (Figure 7C) (p<0.005, student’s t test). Treatment with tigecycline did not alter the appearance or behavior of the mice nor produce gross changes to the organs at necropsy. There were also no alterations in serum levels of liver or muscle/cardiac enzmes (see Figure S5).
Importantly, leukemic cells harvested from the bone marrow of tigecycline-treated primary mice generated smaller leukemic grafts in untreated secondary mice, compared to cells harvested from control-treated primary mice (Figure 7D) (p<0.05, student’s t test) indicating that tigecycline was active against AML stem cells. In contrast, tigecycline treatment in vivo did not reduce engraftment of normal myeloid cells, indicating preferential activity against LSCs over normal hematopoietic cells (Figure 7E).
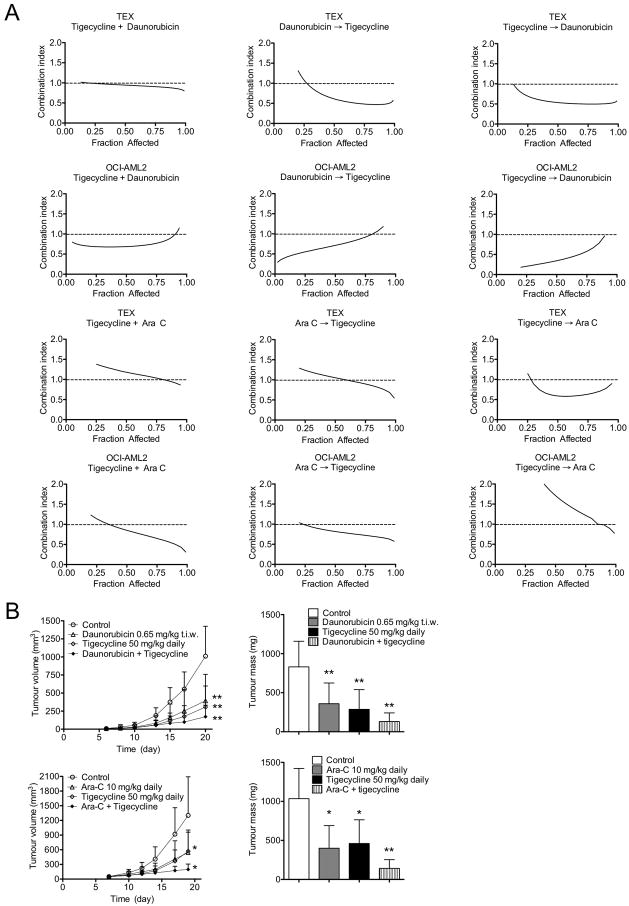
We also evaluated the efficacy of tigecycline in combination with daunorubicin or cytarabine, 2 standard chemotherapeutic agents used for the treatment of AML. TEX and OCI-AML2 leukemia cells were treated in vitro with increasing concentrations of tigecycline alone or in combination with daunorubicin or cytarabine, and growth and viability were assessed (Figure 8A). Data were analyzed using the Calcusyn median effect model, where the combination index (CI) indicates synergism (CI<0.9), additivity (CI=0.9–1.1) or antagonism (CI>1.1). Tigecycline and daunorubicin added together showed an additive or synergistic effect (CI=0.75 – 1.0). However, when tigecycline was added either before or after daunorubicin, the combination was clearly synergistic (CI values at ED50 < 0.8). Treatment with tigecycline in combination with cytarabine was additive or synergistic (CI=0.75 – 1.3) regardless of drug sequence. We then tested the efficacy of the tigecycline/daunorubicin and tigecycline/cytarabine combinations in the OCI-AML2 xenograft model (8 mice per group). Mice treated with the 2 drug combinations showed reduced tumor growth by comparison to those receiving single agents (Figure 8B) (p<0.005, Tukey’s t test after one-way ANOVA). Combination treatments did not alter the gross histology of heart, muscle, brain, spleen and kidney (Figure S6). There were also no alterations in serum levels of liver or muscle/cardiac enzmes. These results suggest that combination therapy with tigecycline may enhance the anti-leukemic efficacy of standard chemotherapeutic agents in patients.
Figure 8. Tigecycline has in vivo anti-leukemia activity in combination with AML agents daunorubicin and cytarabine.
(A) The effect of a 72 hour exposure of TEX and OCI-AML2 cells to different concentrations of tigecycline in combination with daunorubicin or cytarabine on the viability of the cells was measured by MTS assay after 72 hours of incubation. Data were analyzed with Calcusyn software to generate a Combination index versus Fractional effect (cell death) plot showing the effect of the combination of tigecycline with daunorubicin or cytarabine. CI < 1 indicates synergism. (B) Human leukemia (OCI-AML2) cells were injected subcutaneously into the flank of SCID mice. Six days after injection, when tumors were palpable, mice were treated with tigecycline (50 mg/kg daily by i.p. injection) and/or daunorubicin (0.65 mg/kg t.i.w. by i.p. injection) and/or cytarabine (10 mg/kg daily by i.p. injection) or vehicle control (7 mice per treatment group). Another 2 weeks later, mice were sacrificed, tumors excised and the volume and mass of the tumors were measured and mean values determined. The tumor mass and the mean volume are shown. *P < 0.05, ** P < 0.005, as determined by Tukey’s test after One-way ANOVA analysis. Error bars represent mean +/− S.D. See also Figure S6.
DISCUSSION
One approach to develop therapies for AML is to target both the LSCs and bulk AML cells. Here we report that the antimicrobial tigecycline has toxicity for human AML cells at all stages of development in both in vitro and in vivo preclinical models, while sparing normal hematopoietic cells. In addition, we provide detailed evidence that this drug acts by inhibiting mitochondrial translation because of a dependence of leukemic cells on this aspect of cell behavior.
Tigecycline is an anti-microbial agent of the glycylcycline class and is active against a range of gram-positive and gram-negative bacteria, particularly drug-resistant pathogens (Stein and Craig, 2006). From a screen of the effects of tigecycline on yeast mutants that cover most of the yeast genome, we correctly identified the inhibition of mitochondrial-based translation as the mechanism used by tigecycline to inhibit eukaryotic cells. This finding, in turn, led to our discovery that leukemic cells owe their heightened sensitivity to tigecycline due to an increased dependence on mitochondrial function. These results underscore the incredible power of yeast screens to reveal critical pathways that underlie effects seen in drug screens.
To interrogate the role of mitochondrial functions in leukemic cells and their potential for specific anti-leukemic targeting strategies, we used a combination of genetic, chemical, biochemical and biologic approaches. Knockdown of initiation (IF-3) and elongation (EF-Tu) factors in leukemia cells provided genetic confirmation of the prediction that specific inhibition of mitochondrial translation in leukemic cells would mimic the effects of tigecycline, although IF-3 knockdown did not. Currently, it is not fully understood why IF-3 knockdown does not inhibit mitochondrial translation. IF-3 stimulates mitochondrial translation by altering the ribosomal subunit dissociation equilibrium, and doesn’t directly stimulate the binding of tRNA molecules (Koc and Spremulli, 2002).We speculate that in the absence of IF-3, functional translation still occurs due to compensation by other members of the translation machinery. Thus, some factors involved in mitochondrial translation appear more critical than others for the integrity of this process. Accordingly, future investigations exploring the possible role of mitochondrial translation in other cancers will also likely need to assess the functional importance of various initiation and elongation factors.
The impact of inhibiting mitochondrial protein synthesis and the oxidative phosphorylation pathway in leukemia has not been fully assessed. Interestingly, the 13 mtDNA-encoded subunits of the electron transport chain are important for functional regulation of oxidative phosphorylation (Fukuda et al., 2007). The Warburg hypothesis proposes that malignant cells rely on glycolysis and are significantly less dependent on oxidative phosphorylation for survival (Warburg, 1956). Yet, more recent studies indicate that some tumors are highly dependent on oxidative phosphorylation for survival (Funes et al., 2007; Moreno-Sánchez et al., 2007; Rodriguez-Enriquez et al., 2006). Our data suggest that LSCs are unique in their mitochondrial characteristics, sensitivity to inhibition of mitochondrial protein synthesis, and their reliance on oxidative phosphorylation. Recently it was demonstrated that leukemia cells have increased rates of fatty acid oxidation (Samudio et al., 2010) and inhibition of fatty acid oxidation targeted both leukemia stem cells and their “mature” blast progeny. These findings complement those we now report. Electrons generated from the oxidation of fatty acids ultimately flow through the mitochondrial respiratory chain. As such, reducing the components of the mitochondrial respiratory chain via mitochondrial translation inhibition would limit the ability of leukemia cells to derive energy from fatty acid oxidation thus offering an explanation of how inhibition of either of these processes might specifically constrain the survival or growth of leukemic cells.
The differences in the mitochondrial characteristics of primary AML cells and their normal counterparts are also noteworthy. AML bulk, stem cells and their progeny had a greater mitochondrial mass and higher rates of oxygen consumption compared to normal hematopoietic progenitor cells as shown by multiple endpoints. Notably, AML CD34+/CD38+ and bulk subsets had higher mitochondrial mass than CD34+/CD38− cells, reflecting the metabolic differences in rapidly diving AML blasts. However, all AML subsets had notably higher mitochondrial mass than normal hematopoietic cells. Moreover, AML cells with the highest mitochondrial mass were the most sensitive to tigecycline suggesting a biological correlation between these two parameters. The fact that normal hematopoietic cells have a low mitochondrial mass is consistent with this finding and may explain the general preferential sensitivity of AML cells to inhibition of mitochondrial protein synthesis. While this correlation does not imply a cause and effect relationship, mitochondrial mass may serve to identify potential subgroups of AML patients most likely to respond to a therapeutic strategy that targets their functions. The robust preclinical anti-leukemia activity documented with tigecycline using a variety of in vitro and in vivo models and its known toxicology and pharmacology in humans and animals, support rapidly advancing this drug into clinical trial for leukemia to evaluate proof-of-mechanism and proof-of-concept. In conclusion, a combination of small-molecule screens and yeast mutant screens, coupled with follow-up studies of primary AML cell responses in vitro and in vivo has enabled mitochondrial translation inhibition to be identified as a therapeutic target for human AML with the antimicrobial tigecycline as a lead candidate. Investigation of the mechanistic basis of the selective sensitivity of AML cells to agents that inhibit this process has revealed an elevated state of mitochondrial biogenesis in AML cells upon which AML cells at all stages of differentiation apparently depend. Thus, in spite of the genetic and biological diversity of spontaneously arising human AML clones, some common biochemical pathways accessible to selective targeting appear to still exist and await therapeutic exploitation.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are available in the supplemental experimental procedures.
Primary AML and normal hematopoietic cells
Primary human AML samples were isolated from peripheral blood samples from consenting patients with AML, who had at least 80% malignant cells among the low –density cells isolated by Ficoll density centrifugation. Primary low-density normal hematopoietic cells were similarly obtained from healthy consenting volunteers donating peripheral blood stem cells for allogeneic stem cell transplantation after G-CSF mobilization. Primary cells were cultured at 37°C in IMDM, supplemented with 20% fetal bovine serum (FBS), and appropriate antibiotics. The collection and use of human tissue for this study were approved by the University Health Network institutional review board and Review Ethics Board of the University of British Columbia.
Yeast genomic screen
To identify the primary mechanism of drug action, HIP in yeast was used to profile the fitness of ~6000 heterozygous deletion strains (Giaever et al., 2004; Smith et al., 2010) in the presence of our compounds. Because wild-type yeast growth was more sensitive in respiratory media, the yeast heterozygous deletion pools were grown in YP media supplemented with 2% glycerol and 1% ethanol. The fitness assay on the deletion strains was performed as described (Pierce et al., 2006) with the following modifications: 1) for barcode amplification, 0.2 μg of genomic DNA was used in a 50 μl PCR reaction containing 1uM mix of up- or down-tag primers and 82% (v/v) of High Fidelity Platinum PCR Supermix (Invitrogen, Carlsbad, California); 2) 34 amplification cycles were used for the PCR using an extension temperature of 68° C for 2 minutes except for a final 10 minutes in the last cycle 3) after 10–16 hours of hybridization the arrays were washed in a GeneChip Fluidic Station 450 (Affymetrix, Santa Clara, CA) using the GeneFlex_Sv3_450 protocol with one additional wash cycle before the staining. The Affymetrix GeneChip Command Console Software was used to extract the intensity values from the arrays and the fitness defects were calculated for each deletion strains as log2 ratios (mean signal intensity of control/mean signal intensity of drug).
Yeast growth rate measurements
The growth rate of wild-type yeast(Giaever et al., 2002) was determined by measuring the ratio of the area under the curve (AUC) after 20 generations of growth plus drug to AUC in vehicle alone (2% DMSO).
Assessment of tigecycline’s anti-leukemia activity in mouse models of human leukemia
All animal studies were carried out according to the regulations of the Canadian Council on Animal Care and with the approval of the Ontario Cancer Institute Animal Ethics Review board. Tumor xenograft (OCI-AML2) experiments were performed using SCID mice. Primary AML and normal cord blood engraftment in the mouse bone marrow was performed using irradiated NOD-SCID mice. Details on mouse models and subsequent analysis can be found in Supplemental Experimental Procedures.
Statistical Analysis
All data are expressed as mean and standard deviation (SD) to indicate data variability. Statistical analyses were performed by unpaired student’s t test, one-way ANOVA and post-hoc Tukey’s test, as indicated. Differences were considered statistically significant at p <0.05.
Supplementary Material
HIGHLIGHTS.
Inhibition of mitochondrial translation has anti-leukemia activity
AML cells have higher mitochondrial biogenesis than normal hematopoietic cells
Mitochondrial translation inhibition is a novel therapeutic strategy for AML
Tigecycline is a potential therapeutic agent for AML therapy
SIGNIFICANCE.
An important role of cytosolic translation has been well documented in the context of cancer, but the functional importance of mitochondrial translation in leukemia has not been well studied. Here, we used chemical and genetic approaches to show that mitochondrial translation inhibition selectively kills leukemic vs normal cells, including those defined functionally as progenitors and stem cells. This may be attributable to the higher rate of mitochondrial biogenesis found in leukemic cells. Given these results and the known pharmacology and toxicology of tigecycline in humans, targeting mitochondrial translation inhibition as a therapeutic strategy in human leukemia is attractive.
Acknowledgments
This work was supported by the Canadian Stem Cell Network, the Canadian Institutes for Health Research, the Leukemia and Lymphoma Society, the National Institutes of Health (NCI 1R01CA157456), the Terry Fox Foundation, MaRS Innovation, the Ontario Institute of Cancer Research with funding provided by the Ontario Ministry of Research and Innovation, the Princess Margaret Hospital Foundation, and the Ministry of Long Term Health and Planning in the Province of Ontario. MS holds a Canada Graduate Scholarship from the Canadian Institutes of Health Research. ADS is a Leukemia and Lymphoma Society Scholar in Clinical Research.
Footnotes
Accession Number
Yeast genomic screen microarray data are deposited at Array Express Archive (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-814.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barabé F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007:600–604. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Capps GJ, Samuels DC, Chinnery PF. A model of the nuclear control of mitochondrial DNA replication. J Theor Biol. 2003:565–583. doi: 10.1006/jtbi.2003.3207. [DOI] [PubMed] [Google Scholar]
- Christian BE, Spremulli LL. Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria. Biochemistry. 2009:3269–3278. doi: 10.1021/bi8023493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Lightowlers ZM, Preiss T, Lightowlers RN. Inhibition of mitochondrial protein synthesis promotes increased stability of nuclear-encoded respiratory gene transcripts. J Biol Chem. 1994:27322–27328. [PubMed] [Google Scholar]
- Contreras A, Vázquez D. Cooperative and antagonistic interactions of peptidyl-tRNA and antibiotics with bacterial ribosomes. Eur J Biochem. 1977:539–547. doi: 10.1111/j.1432-1033.1977.tb11422.x. [DOI] [PubMed] [Google Scholar]
- Crandon JL, Kim A, Nicolau DP. Comparison of tigecycline penetration into the epithelial lining fluid of infected and uninfected murine lungs. J Antimicrob Chemother. 2009;64:837–839. doi: 10.1093/jac/dkp301. [DOI] [PubMed] [Google Scholar]
- Fiegl M, Samudio I, Clise-Dwyer K, Burks JK, Mnjoyan Z, Andreeff M. CXCR4 expression and biologic activity in acute myeloid leukemia are dependent on oxygen partial pressure. Blood. 2009;113:1504–1512. doi: 10.1182/blood-2008-06-161539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim J-w, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- Funes JM, Quintero M, Henderson S, Martinez D, Qureshi U, Westwood C, Clements MO, Bourboulia D, Pedley RB, Moncada S, Boshoff C. Transformation of human mesenchymal stem cells increases their dependency on oxidative phosphorylation for energy production. Proc Natl Acad Sci U S A. 2007;104:6223–6228. doi: 10.1073/pnas.0700690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur R, Grasso D, Datta PP, Krishna PDV, Das G, Spencer A, Agrawal RK, Spremulli L, Varshney U. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol Cell. 2008:180–190. doi: 10.1016/j.molcel.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci U S A. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet. 1999:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, Fung E, Proctor M, Shokat KM, Zhang C, Davis RW, et al. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat Chem Biol. 2008:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- Hunter SE, Spremulli LL. Mutagenesis of glutamine 290 in Escherichia coli and mitochondrial elongation factor Tu affects interactions with mitochondrial aminoacyl-tRNAs and GTPase activity. Biochemistry. 2004:6917–6927. doi: 10.1021/bi036068j. [DOI] [PubMed] [Google Scholar]
- Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- Koc EC, Spremulli LL. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J Biol Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim J-w, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J Biol Chem. 2003:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- Löwenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999:1051–1062. doi: 10.1056/NEJM199909303411407. [DOI] [PubMed] [Google Scholar]
- Löwenberg B, Suciu S, Archimbaud E, Haak H, Stryckmans P, de Cataldo R, Dekker AW, Berneman ZN, Thyss A, van der Lelie J, et al. Mitoxantrone versus daunorubicin in induction-consolidation chemotherapy--the value of low-dose cytarabine for maintenance of remission, and an assessment of prognostic factors in acute myeloid leukemia in the elderly: final report. European Organization for the Research and Treatment of Cancer and the Dutch-Belgian Hemato-Oncology Cooperative Hovon Group. J Clin Oncol. 1998:872–881. doi: 10.1200/JCO.1998.16.3.872. [DOI] [PubMed] [Google Scholar]
- McKee EE, Ferguson M, Bentley AT, Marks TA. Inhibition of mammalian mitochondrial protein synthesis by oxazolidinones. Antimicrob Agents Chemother. 2006:2042–2049. doi: 10.1128/AAC.01411-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Sánchez R, Rodríguez-Enríquez S, Marín-Hernández A, Saavedra E. Energy metabolism in tumor cells. FEBS J. 2007:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- Muralidharan G, Micalizzi M, Speth J, Raible D, Troy S. Pharmacokinetics of tigecycline after single and multiple doses in healthy subjects. Antimicrob Agents Chemother. 2005:220–229. doi: 10.1128/AAC.49.1.220-229.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec EE, Wu L, Swaney SM, Chosay JG, Ross DE, Brieland JK, Leach KL. Oxazolidinones inhibit cellular proliferation via inhibition of mitochondrial protein synthesis. Antimicrob Agents Chemother. 2005:3896–3902. doi: 10.1128/AAC.49.9.3896-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien TW. Properties of human mitochondrial ribosomes. IUBMB Life. 2003:505–513. doi: 10.1080/15216540310001626610. [DOI] [PubMed] [Google Scholar]
- Olson MW, Ruzin A, Feyfant E, Rush TS, O’Connell J, Bradford PA. Functional, biophysical, and structural bases for antibacterial activity of tigecycline. Antimicrob Agents Chemother. 2006:2156–2166. doi: 10.1128/AAC.01499-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Herrmann JM. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbamcr.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Park WH, Han YW, Kim SH, Kim SZ. An ROS generator, antimycin A, inhibits the growth of HeLa cells via apoptosis. J Cell Biochem. 2007:98–109. doi: 10.1002/jcb.21280. [DOI] [PubMed] [Google Scholar]
- Pendergrass W, Wolf N, Poot M. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A. 2004:162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- Pierce SE, Fung EL, Jaramillo DF, Chu AM, Davis RW, Nislow C, Giaever G. A unique and universal molecular barcode array. Nat Methods. 2006;3:601–603. doi: 10.1038/nmeth905. [DOI] [PubMed] [Google Scholar]
- Ramzan R, Staniek K, Kadenbach B, Vogt S. Mitochondrial respiration and membrane potential are regulated by the allosteric ATP-inhibition of cytochrome c oxidase. Biochim Biophys Acta. 2010:1672–1680. doi: 10.1016/j.bbabio.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Vital-Gonzalez PA, Flores-Rodriguez FL, Marin-Hernandez A, Ruiz-Azuara L, Moreno-Sanchez R. Control of cellular proliferation by modulation of oxidative phosphorylation in human and rodent fast-growing tumor cells. Toxicol Appl Pharmacol. 2006;215:208–217. doi: 10.1016/j.taap.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, Andreeff M. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinabarger DL, Marotti KR, Murray RW, Lin AH, Melchior EP, Swaney SM, Dunyak DS, Demyan WF, Buysse JM. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob Agents Chemother. 1997:2132–2136. doi: 10.1128/aac.41.10.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley ST, Reers M, Mottola-Hartshorn C, Lin M, Chen A, Smith TW, Steele GD, Chen LB. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci USA. 1991:3671–3675. doi: 10.1073/pnas.88.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Ammar R, Nislow C, Giaever G. A survey of yeast genomic assays for drug and target discovery. Pharmacol Ther. 2010:156–164. doi: 10.1016/j.pharmthera.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spremulli LL, Coursey A, Navratil T, Hunter SE. Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog Nucleic Acid Res Mol Biol. 2004:211–261. doi: 10.1016/S0079-6603(04)77006-3. [DOI] [PubMed] [Google Scholar]
- Stein GE, Craig WA. Tigecycline: a critical analysis. Clin Infect Dis. 2006:518–524. doi: 10.1086/505494. [DOI] [PubMed] [Google Scholar]
- Swift LP, Rephaeli A, Nudelman A, Phillips DR, Cutts SM. Doxorubicin-DNA adducts induce a non-topoisomerase II-mediated form of cell death. Cancer Res. 2006:4863–4871. doi: 10.1158/0008-5472.CAN-05-3410. [DOI] [PubMed] [Google Scholar]
- Tam EWY, Feigenbaum A, Addis JBL, Blaser S, Mackay N, Al-Dosary M, Taylor RW, Ackerley C, Cameron JM, Robinson BH. A novel mitochondrial DNA mutation in COX1 leads to strokes, seizures, and lactic acidosis. Neuropediatrics. 2008:328–334. doi: 10.1055/s-0029-1202287. [DOI] [PubMed] [Google Scholar]
- Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- Turrens JF, Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem J. 1980:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin JJ, Guan J, Prior WW, Edgar KA, Kassees R, Sampath D, Belvin M, Friedman LS. Nuclear phospho-Akt increase predicts synergy of PI3K inhibition and doxorubicin in breast and ovarian cancer. Sci Transl Med. 2010:48ra66. doi: 10.1126/scitranslmed.3000630. [DOI] [PubMed] [Google Scholar]
- WARBURG O. On the origin of cancer cells. Science. 1956:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warner JK, Wang JCY, Takenaka K, Doulatov S, McKenzie JL, Harrington L, Dick JE. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia. 2005:1794–1805. doi: 10.1038/sj.leu.2403917. [DOI] [PubMed] [Google Scholar]
- Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, et al. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Spremulli LL. Roles of residues in mammalian mitochondrial elongation factor Ts in the interaction with mitochondrial and bacterial elongation factor Tu. J Biol Chem. 1998:28142–28148. doi: 10.1074/jbc.273.43.28142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.